Dietary Impacts on Changes in Diversity and Abundance of the Murine Microbiome during Progression and Treatment of Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Diet
2.3. Experimental Design
2.4. Sample Collection
2.5. DNA Isolation and Processing
2.6. Morphological Analysis
2.7. Microbiome Data Analysis
3. Results
3.1. Morphometric Measures
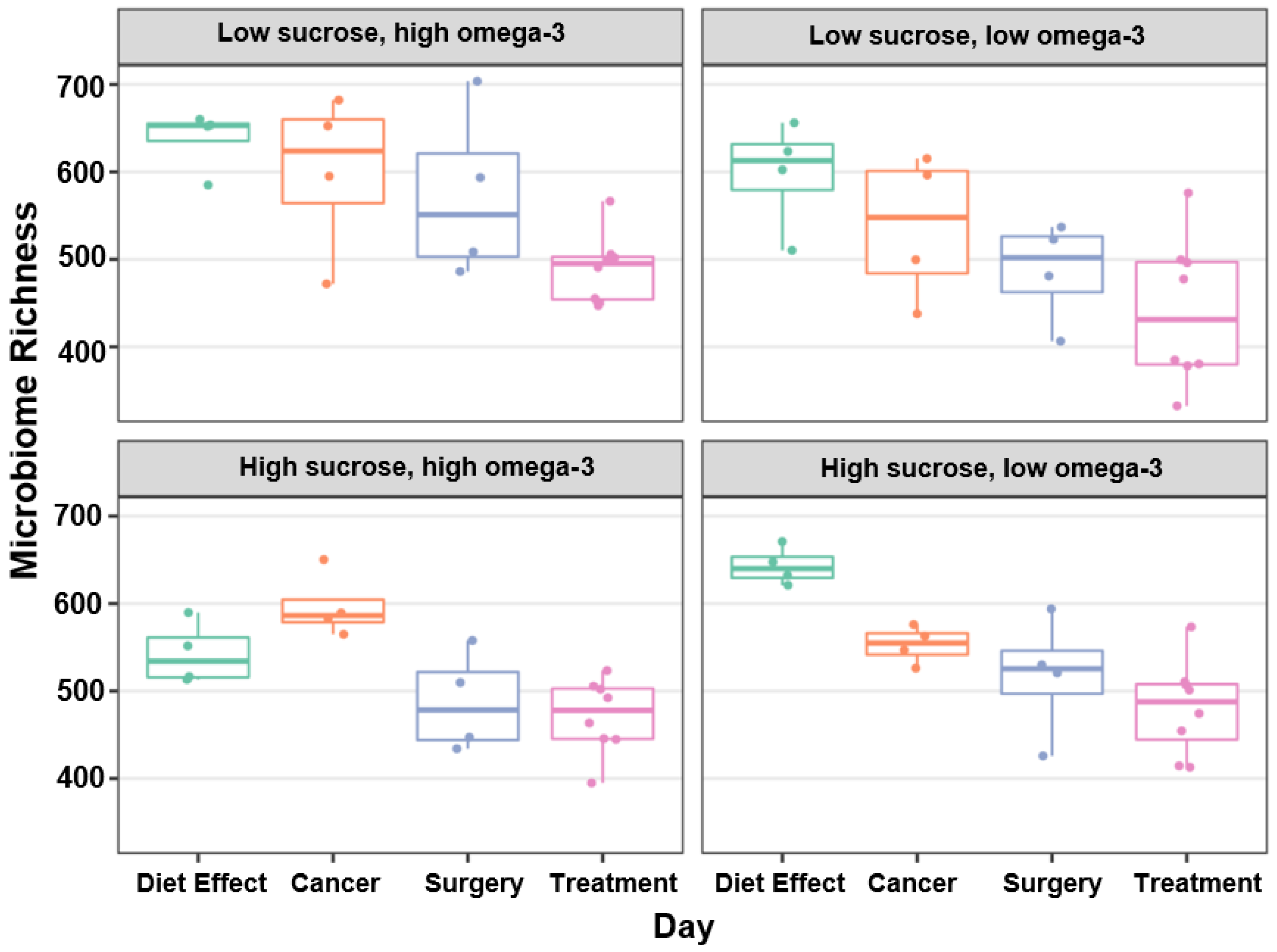
3.2. Microbial Diversity, Richness, and Abundance
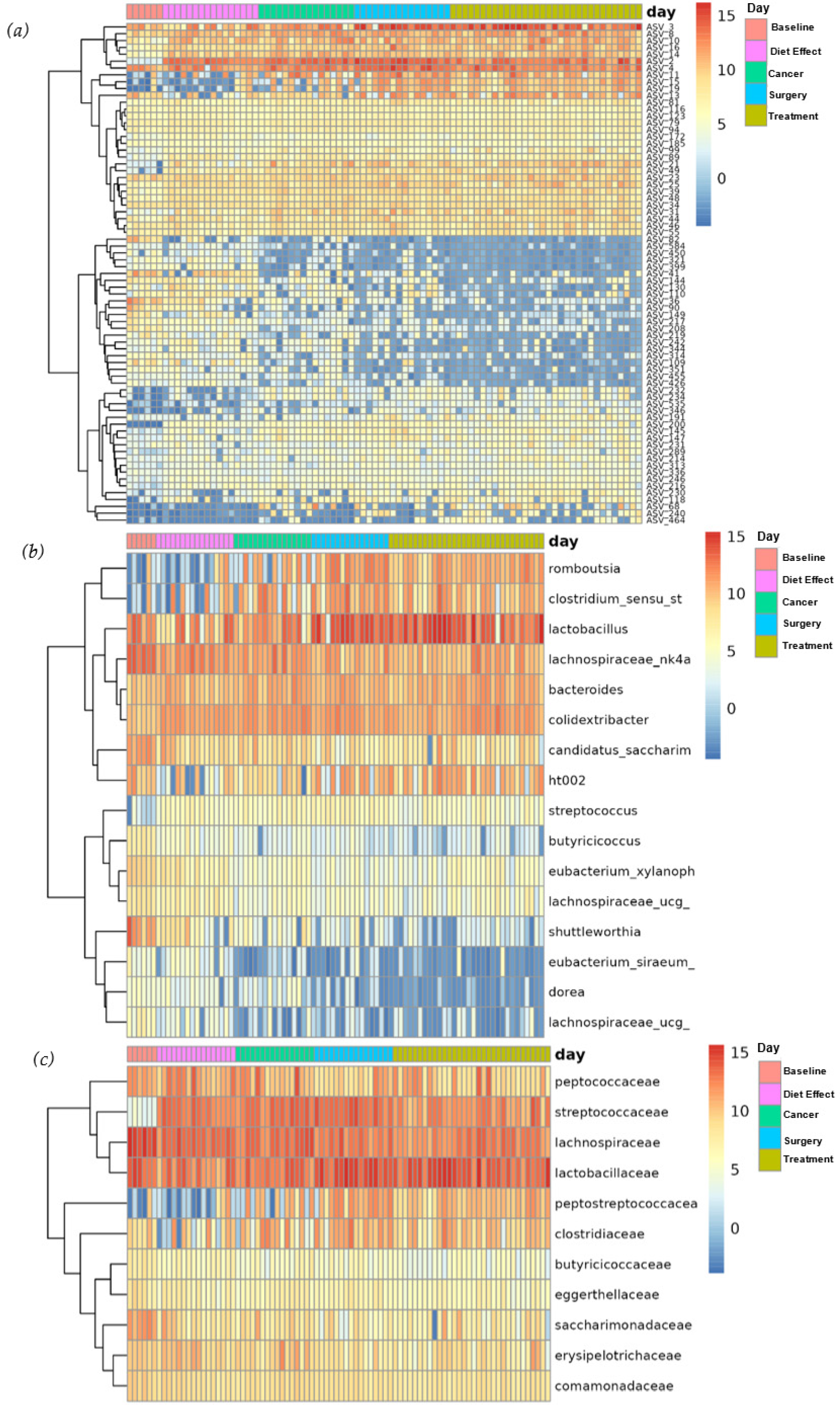
3.3. Overall Microbiome Composition
3.4. Differential Abundance
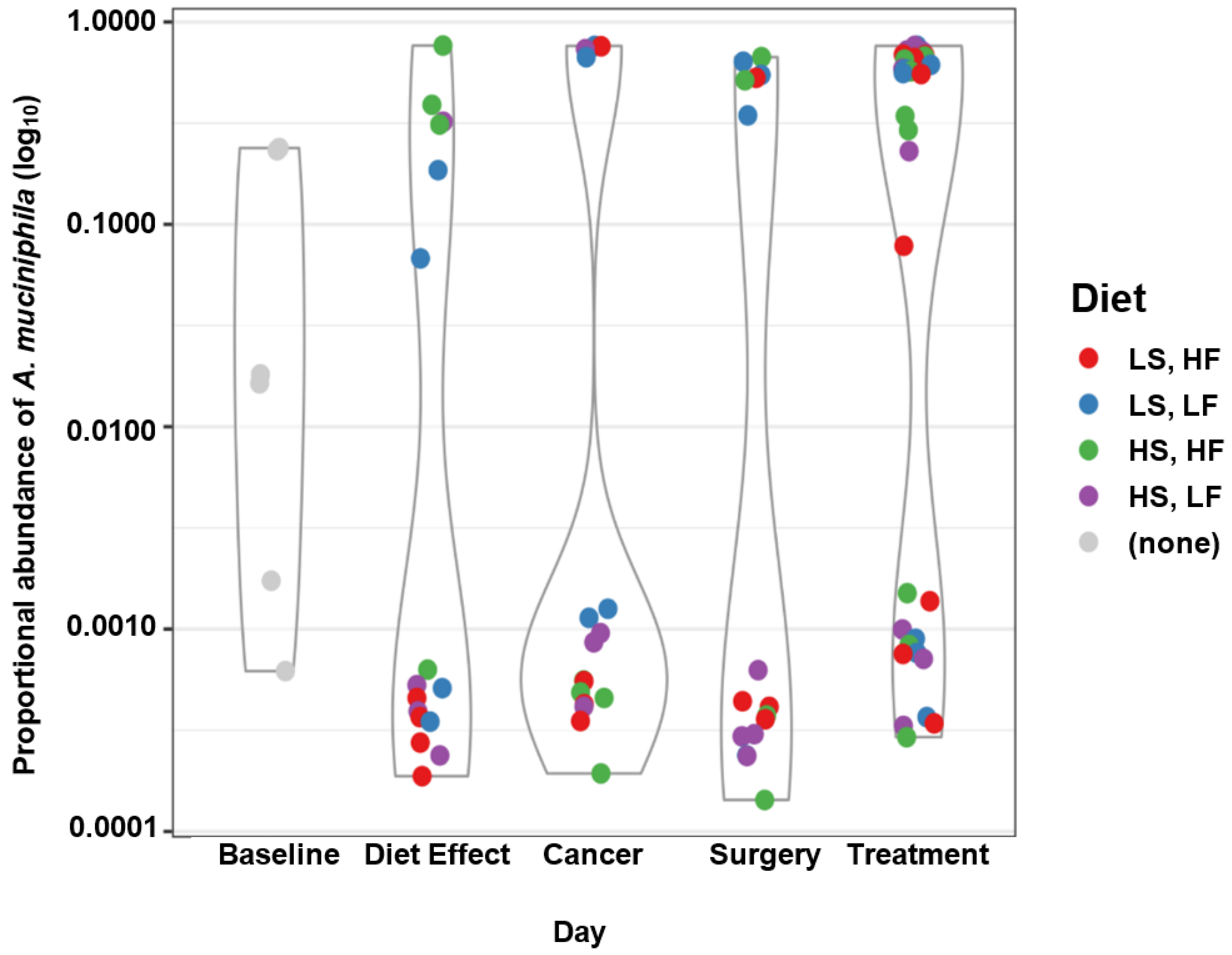
3.5. Taxa of Interest
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA. Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute SEER Cancer Stat Facts: Female Breast Cancer. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 9 June 2022).
- Lund, B.M.; O’Brien, S.J. The Occurrence and Prevention of Foodborne Disease in Vulnerable People. Foodborne Pathog. Dis. 2011, 8, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Kendall, P.A.; Hillers, V.N.; Medeiros, L.C. Qualitative Studies of the Food Safety Knowledge and Perceptions of Transplant Patients. J. Food Prot. 2010, 73, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, L.C.; Chen, G.; Kendall, P.; Hillers, V.N. Food Safety Issues for Cancer and Organ Transplant Patients. Nutr. Clin. Care 2004, 7, 141–148. [Google Scholar] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States-Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Bow, E.J. Infection Risk and Cancer Chemotherapy: The Impact of the Chemotherapeutic Regimen in Patients with Lymphoma and Solid Tissue Malignancies. J. Antimicrob. Chemother. 1998, 41, 1–5. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The Microbiome in Cancer Immunotherapy: Diagnostic Tools and Therapeutic Strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef]
- Liu, J.; Liu, C.; Yue, J. Radiotherapy and the Gut Microbiome: Facts and Fiction. Radiat. Oncol. 2021, 16, 9. [Google Scholar] [CrossRef]
- Panebianco, C.; Andriulli, A.; Pazienza, V. Pharmacomicrobiomics: Exploiting the Drug-Microbiota Interactions in Anticancer Therapies. Microbiome 2018, 6, 92. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Nyberg, K. Gut Microbiome Correlates with Response to Immunotherapy in Metastatic Melanoma. ASCO-SITC Clin. Immuno-Oncol. Symp. 2017, 1–2. [Google Scholar]
- Sethi, V.; Vitiello, G.A.; Saxena, D.; Miller, G.; Dudeja, V. The Role of the Microbiome in Immunologic Development and Its Implication For Pancreatic Cancer Immunotherapy. Gastroenterology 2019, 156, 2097–2115.e2. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Bailey, M.T. The Contributing Role of the Intestinal Microbiota in Stressor-Induced Increases in Susceptibility to Enteric Infection and Systemic Immunomodulation. Horm. Behav. 2012, 62, 286–294. [Google Scholar] [CrossRef]
- Jamar, G.; Ribeiro, D.A.; Pisani, L.P. High-Fat or High-Sugar Diets as Trigger Inflammation in the Microbiota-Gut-Brain Axis. Crit. Rev. Food Sci. Nutr. 2021, 61, 836–854. [Google Scholar] [CrossRef]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Bäckhed, F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef]
- Szczyrek, M.; Bitkowska, P.; Chunowski, P.; Czuchryta, P.; Krawczyk, P.; Milanowski, J. Diet, Microbiome, and Cancer Immunotherapy—A Comprehensive Review. Nutrients 2021, 13, 2217. [Google Scholar] [CrossRef]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.; Salvador, C.; Skibola, C.; Tollefsbol, T.O. Influences of Diet and the Gut Microbiome on Epigenetic Modulation in Cancer and Other Diseases. Clin. Epigenetics 2015, 7, 112. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2015, 14, 20–32. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; Van Den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; De Vos, W.M.; Kleerebezem, M. The Human Small Intestinal Microbiota Is Driven by Rapid Uptake and Conversion of Simple Carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.; Backhed, F.; Fulton, L.; Gordon, J. Marked Alterations in the Distal Gut Microbiome Linked to Diet-Induced Obesity. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Razin, S. Molecular properties of mollicutes: A synopsis. In Molecular and Diagnostic Procedures in Mycoplasmology; Razin, S., Tully, J.G., Eds.; Academic Press: Cambridge, MA, USA, 1995; pp. 1–25. ISBN 9780125838054. [Google Scholar]
- Xuan, C.; Shamonki, J.M.; Chung, A.; DiNome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial Dysbiosis Is Associated with Human Breast Cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- USDA. Dietary Guidelines for Americans 2020–2025; United States Department of Agriculture: Washington, DC, USA, 2020. [Google Scholar]
- Ormiston, K.; Gaudier-Diaz, M.M.; TinKai, T.; Fitzgerald, J.; Cole, R.M.; Andridge, R.; Lustberg, M.; DeVries, A.C.; Orchard, T. Effects of Plant-Based versus Marine-Based Omega-3 Fatty Acids and Sucrose on Brain and Liver Fatty Acids in a Mouse Model of Chemotherapy. Nutr. Neurosci. 2021, 25, 2650–2658. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Q.; Dang, R.L.; Tang, M.M.; Cai, H.L.; Li, H.D.; Liao, D.H.; He, X.; Cao, L.J.; Xue, Y.; Jiang, P. Long Chain Omega-3 Polyunsaturated Fatty Acid Supplementation Alleviates Doxorubicin-Induced Depressive-like Behaviors and Neurotoxicity in Rats: Involvement of Oxidative Stress and Neuroinflammation. Nutrients 2016, 8, 243. [Google Scholar] [CrossRef]
- Ma, T.; Liaset, B.; Hao, Q.; Petersen, R.K.; Fjære, E.; Ngo, H.T.; Lillefosse, H.H.; Ringholm, S.; Sonne, S.B.; Treebak, J.T.; et al. Sucrose Counteracts the Anti-Inflammatory Effect of Fish Oil in Adipose Tissue and Increases Obesity Development in Mice. PLoS ONE 2011, 6, e21647. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2019. CA. Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Rodriguez-Palacios, A.; Harding, A.; Menghini, P.; Himmelman, C.; Retuerto, M.; Nickerson, K.P.; Lam, M.; Croniger, C.M.; McLean, M.H.; Durum, S.K.; et al. The Artificial Sweetener Splenda Promotes Gut Proteobacteria, Dysbiosis, and Myeloperoxidase Reactivity in Crohn’s Disease-Like Ileitis. Inflamm. Bowel Dis. 2018, 24, 1005–1020. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J.; Retuerto, M.; Sikaroodi, M.; Brown, R.E.; Jurevic, R.; Salata, R.A.; Lederman, M.M.; Gillevet, P.M.; Ghannoum, M.A. Oral Mycobiome Analysis of HIV-Infected Patients: Identification of Pichia as an Antagonist of Opportunistic Fungi. PLoS Pathog. 2014, 10, e1003996. [Google Scholar] [CrossRef]
- Mair, P.; Wilcox, R. Robust Statistical Methods in R Using the WRS2 Package. Behav. Res. Methods 2020, 52, 464–488. [Google Scholar] [CrossRef]
- Ohio Supercomputer Center Ohio Supercomputer Center 1987.
- Andrews, S. Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 2 March 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize Analysis Results Formultiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing. R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 2 March 2022).
- R Core Team, R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing, Vienna, Austria. 2017. Available online: https://www.R-project.org/ (accessed on 3 March 2022).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic Analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. CRAN—Package Vegan. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 2 March 2022).
- Willis, A.; Bunge, J. Estimating Diversity via Frequency Ratios. Biometrics 2015, 71, 1042–1049. [Google Scholar] [CrossRef]
- Willis, A.; Bunge, J.; Whitman, T. Improved Detection of Changes in Species Richness in High Diversity Microbial Communities. J. R. Stat. Soc. Ser. C Appl. Stat. 2017, 66, 963–977. [Google Scholar] [CrossRef]
- Willis, A.D.; Martin, B.D. Estimating Diversity in Networked Ecological Communities. Biostatistics 2022, 23, 207–222. [Google Scholar] [CrossRef]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the Analysis of High-Throughput Sequencing Datasets: Characterizing RNA-Seq, 16S RRNA Gene Sequencing and Selective Growth Experiments by Compositional Data Analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Araujo, D.V.; Watson, G.A.; Oliva, M.; Heirali, A.; Coburn, B.; Spreafico, A.; Siu, L.L. Bugs as Drugs: The Role of Microbiome in Cancer Focusing on Immunotherapeutics. Cancer Treat. Rev. 2021, 92, 102125. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.R.; Moustafa, M.A.A.; Saad, B.M.; Hamed, R.; Moustafa, A.R.A. Impact of Diet on Human Gut Microbiome and Disease Risk. New Microbes New Infect. 2021, 41, 100845. [Google Scholar] [CrossRef]
- Jensen, T.; Abdelmalek, M.F.; Sullivan, S.; Nadeau, K.J.; Green, M.; Roncal, C.; Nakagawa, T.; Kuwabara, M.; Sato, Y.; Kang, D.H.; et al. Fructose and Sugar: A Major Mediator of Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2018, 68, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Borton, M.A.; Sabag-Daigle, A.; Wu, J.; Solden, L.M.; O’Banion, B.S.; Daly, R.A.; Wolfe, R.A.; Gonzalez, J.F.; Wysocki, V.H.; Ahmer, B.M.M.; et al. Chemical and Pathogen-Induced Inflammation Disrupt the Murine Intestinal Microbiome. Microbiome 2017, 5, 47. [Google Scholar] [CrossRef]
- Goubet, A.G.; Daillère, R.; Routy, B.; Derosa, L.; Roberti, P.M.; Zitvogel, L. The Impact of the Intestinal Microbiota in Therapeutic Responses against Cancer. Comptes Rendus Biol. 2018, 341, 284–289. [Google Scholar] [CrossRef]
- Hooper, L.V.; MacPherson, A.J. Immune Adaptations That Maintain Homeostasis with the Intestinal Microbiota. Nat. Rev. Immunol. 2010, 10, 159–169. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The Microbiome and Innate Immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Łaniewski, P.; Ilhan, Z.E.; Herbst-Kralovetz, M.M. The Microbiome and Gynaecological Cancer Development, Prevention and Therapy. Nat. Rev. Urol. 2020, 17, 232–250. [Google Scholar] [CrossRef]
- Blaustein, R.A.; Seed, P.C.; Hartmann, E.M. Biotransformation of Doxorubicin Promotes Resilience in Simplified Intestinal Microbial Communities. mSphere 2021, 6, e00068-21. [Google Scholar] [CrossRef]
- Huang, J.; Wei, S.; Jiang, C.; Xiao, Z.; Liu, J.; Peng, W.; Zhang, B.; Li, W. Involvement of Abnormal Gut Microbiota Composition and Function in Doxorubicin-Induced Cardiotoxicity. Front. Cell. Infect. Microbiol. 2022, 12, 155. [Google Scholar] [CrossRef]
- Muls, A.; Andreyev, J.; Lalondrelle, S.; Taylor, A.; Norton, C.; Hart, A. The Impact of Cancer Treatment on the Gut and Vaginal Microbiome in Women with a Gynecological Malignancy. Int. J. Gynecol. Cancer 2017, 27, 1550–1559. [Google Scholar] [CrossRef]
- Flórez, A.B.; Sierra, M.; Ruas-Madiedo, P.; Mayo, B. Susceptibility of Lactic Acid Bacteria, Bifidobacteria and Other Bacteria of Intestinal Origin to Chemotherapeutic Agents. Int. J. Antimicrob. Agents 2016, 48, 547–550. [Google Scholar] [CrossRef]
- Tubiana-Hulin, M. How to Maximize the Efficacy of Taxanes in Breast Cancer. Cancer Chemother. Biol. Response Modif. 2005, 31, S3–S9. [Google Scholar] [CrossRef]
- Lee, K.A.; Luong, M.K.; Shaw, H.; Nathan, P.; Bataille, V.; Spector, T.D. The Gut Microbiome: What the Oncologist Ought to Know. Br. J. Cancer 2021, 125, 1197–1209. [Google Scholar] [CrossRef]
- Chen, J.; Douglass, J.; Prasath, V.; Neace, M.; Atrchian, S.; Manjili, M.H.; Shokouhi, S.; Habibi, M. The Microbiome and Breast Cancer: A Review. Breast Cancer Res. Treat. 2019, 178, 493–496. [Google Scholar] [CrossRef]
- Nguyen, T.L.A.; Vieira-Silva, S.; Liston, A.; Raes, J. How Informative Is the Mouse for Human Gut Microbiota Research? Dis. Model. Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.; Metcalf, J.; Wegener Parfrey, L.; Knight, R. Defining the Human Microbiome. N. Engl. J. Med. 2012, 70, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Lederer, A.K.; Chikhladze, S.; Kohnert, E.; Huber, R.; Müller, A. Current Insights: The Impact of Gut Microbiota on Postoperative Complications in Visceral Surgery—A Narrative Review. Diagnostics 2021, 11, 2099. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Sun, C.; Zheng, J.; Wen, C.; Ji, C.; Zhang, D.; Chen, Y.; Hou, Z.; Yang, N. Efficacy of Fecal Sampling as a Gut Proxy in the Study of Chicken Gut Microbiota. Front. Microbiol. 2019, 10, 2126. [Google Scholar] [CrossRef]



| Variable | Day | Diet × Day |
|---|---|---|
| Diet Effect 1 | −132.4 (15.7) *** | |
| Cancer | −164.3 (16.1) *** | −33.4 (15.3) * |
| Surgery | −219.1 (16.2) *** | −61.5 (15.5) *** |
| Treatment | −263.3 (11.6) *** | −139.8 (11.1) *** |
| LS, LF 2 | −36.3 (14.1) ** | |
| HS, HF | −90.5 (13.3) *** | |
| HS, LF | 11.6 (13.9) | |
| LS, LF × Cancer | −44.9 (34.3) | |
| HS, HF × Cancer | 86.9 (28.4) ** | |
| HS, LF × Cancer | −58.9 (29.1) * | |
| LS, LF × Surgery | −59.2 (32.0) + | |
| HS, HF × Surgery | 12.8 (32.0) | |
| HS, LF × Surgery | −80.6 (31.4) ** | |
| LS, LF × Treatment | −27.8 (22.5) | |
| HS, HF × Treatment | 68.4 (21.1) *** | |
| HS, LF × Treatment | −16.9 (22.3) | |
| AIC 3 | 968.5 | 906.7 |
| Log. Lik. | −478.268 | −436.327 |
| formula | estimate ~ day | estimate ~ diet × day |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paden, H.; Kurbatfinski, N.; Poelstra, J.W.; Ormiston, K.; Orchard, T.; Ilic, S. Dietary Impacts on Changes in Diversity and Abundance of the Murine Microbiome during Progression and Treatment of Cancer. Nutrients 2023, 15, 724. https://doi.org/10.3390/nu15030724
Paden H, Kurbatfinski N, Poelstra JW, Ormiston K, Orchard T, Ilic S. Dietary Impacts on Changes in Diversity and Abundance of the Murine Microbiome during Progression and Treatment of Cancer. Nutrients. 2023; 15(3):724. https://doi.org/10.3390/nu15030724
Chicago/Turabian StylePaden, Holly, Nikola Kurbatfinski, Jelmer W. Poelstra, Kate Ormiston, Tonya Orchard, and Sanja Ilic. 2023. "Dietary Impacts on Changes in Diversity and Abundance of the Murine Microbiome during Progression and Treatment of Cancer" Nutrients 15, no. 3: 724. https://doi.org/10.3390/nu15030724
APA StylePaden, H., Kurbatfinski, N., Poelstra, J. W., Ormiston, K., Orchard, T., & Ilic, S. (2023). Dietary Impacts on Changes in Diversity and Abundance of the Murine Microbiome during Progression and Treatment of Cancer. Nutrients, 15(3), 724. https://doi.org/10.3390/nu15030724






