Impact of Ring Finger Protein 20 and Its Downstream Regulation on Renal Tubular Injury in a Unilateral Nephrectomy Mouse Model Fed a High-Fat Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Biochemical and Histopathological Studies
2.3. RNA Sequencing and Analysis
2.4. Cell Culture Treatment
2.5. Lipid Accumulation
2.6. Western Blot
2.7. Statistical Analysis
3. Results
3.1. Changes in Lipids and Kidney Function Caused by a High-Fat Diet and Uninephrectomy
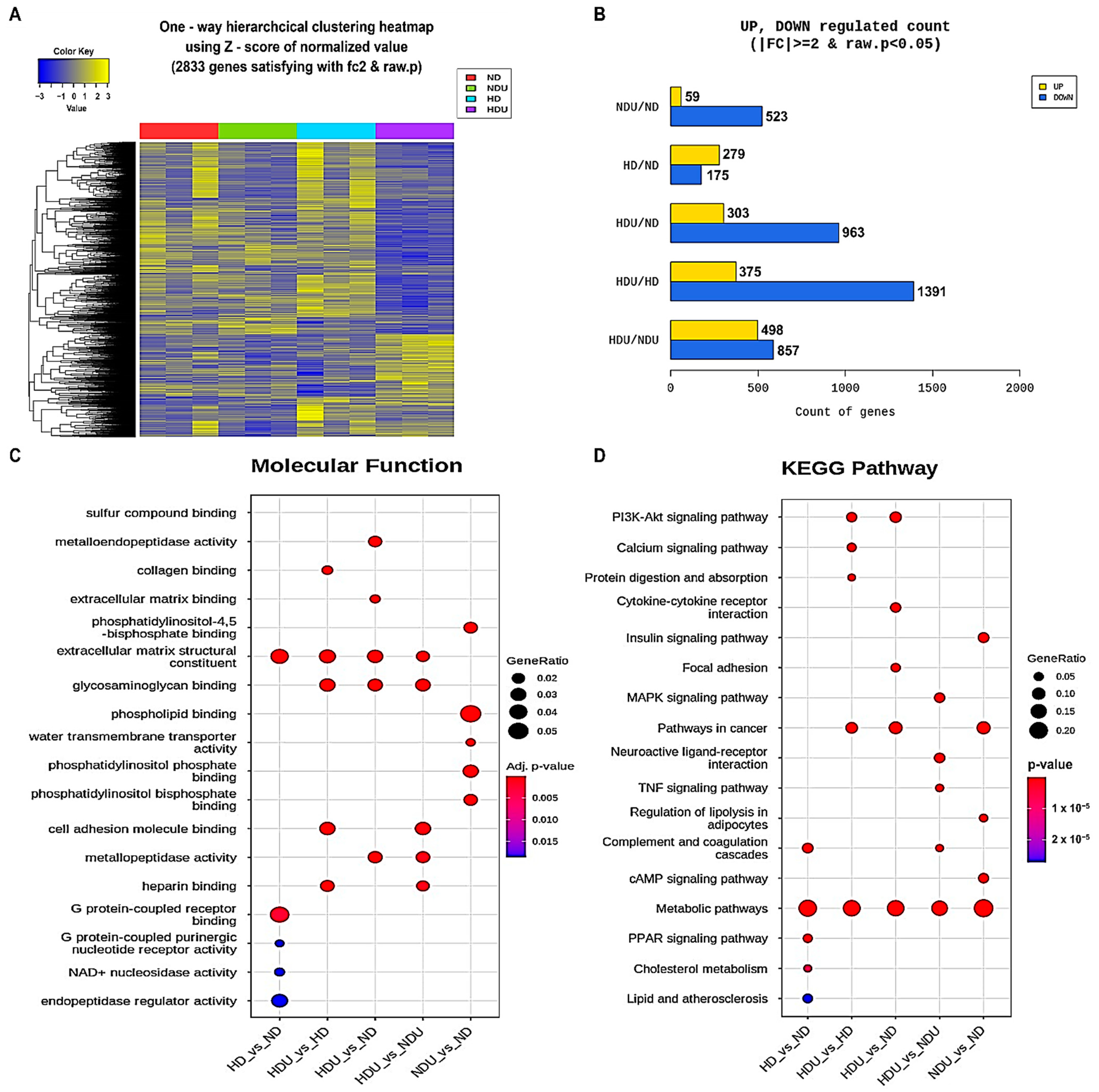
3.2. Genome-Wide Transcriptome Analysis Associated with the Acceleration of Kidney Injury Progression by High-Fat Diet and Uninephrectomy
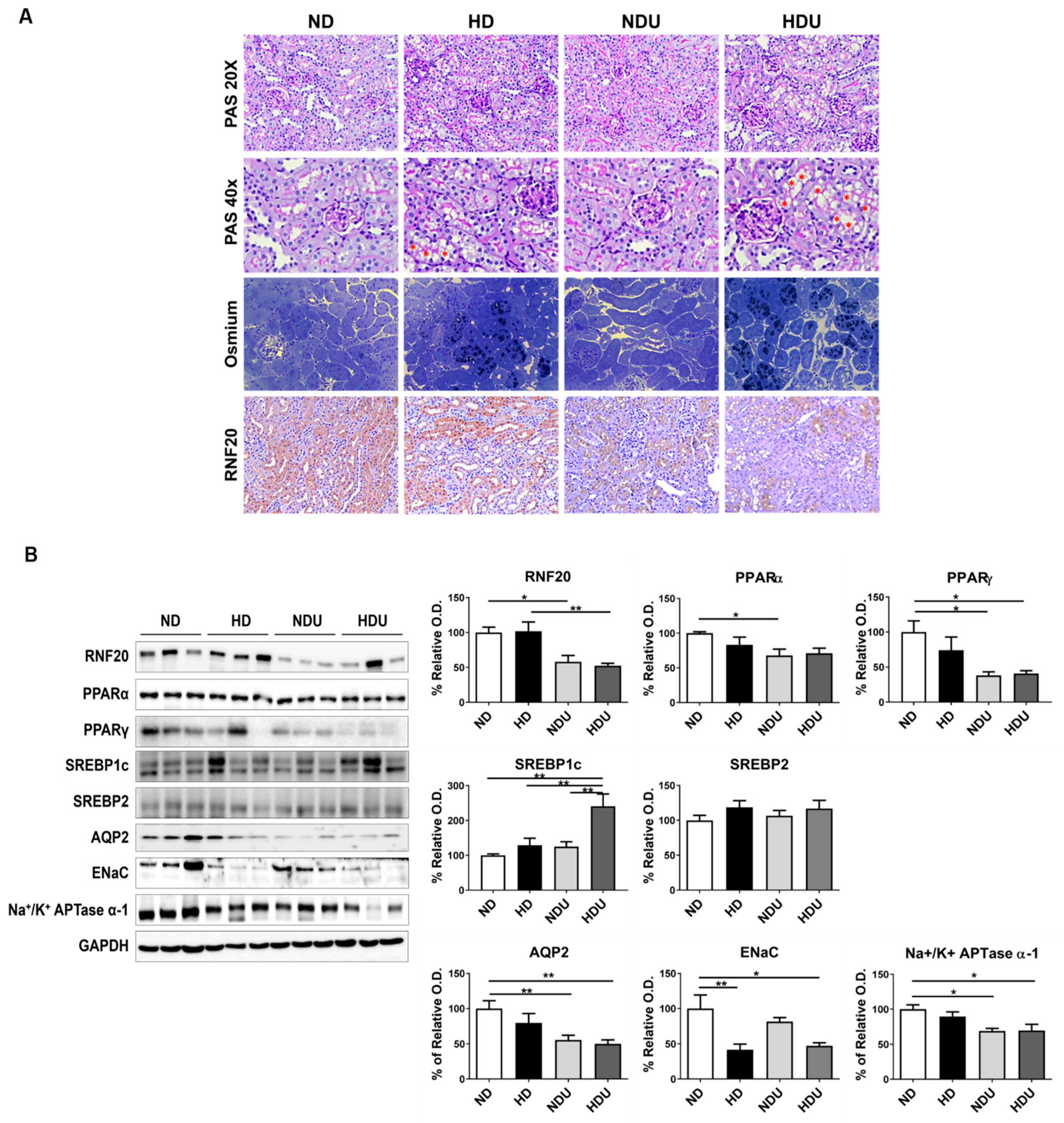
3.3. Changes in Protein Expression by Kidney Cell Types Due to High-Fat Diet and Unilateral Kidney Injury
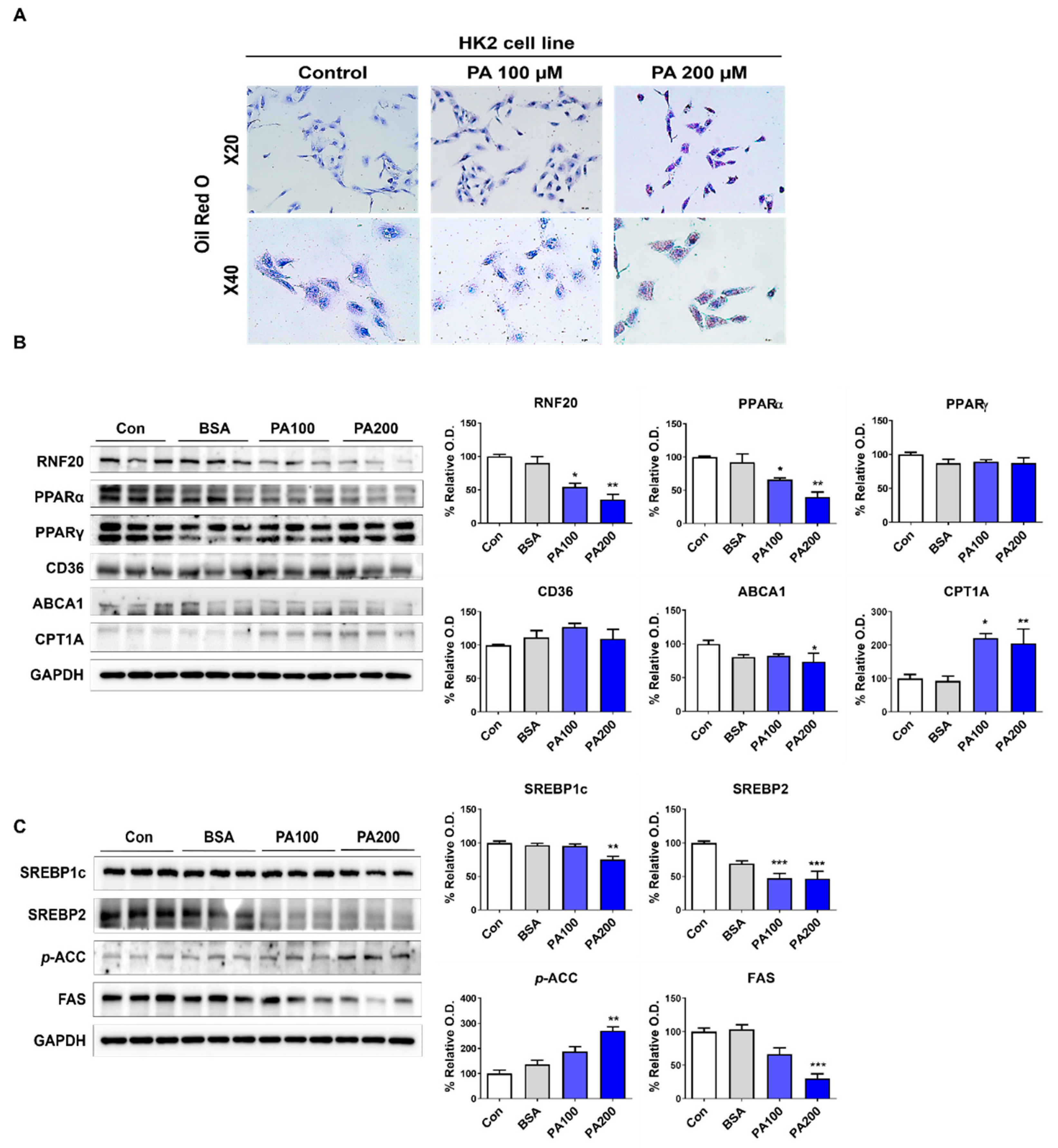
3.4. Lipid Accumulation Due to RNF20-Related Lipid Metabolism Changes in PT Cells
3.5. Lipid-Induced Changes in Nuclear Receptor Factors and Water Channel-Related Proteins in CD Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Escasany, E.; Izquierdo-Lahuerta, A.; Medina-Gómez, G. Chapter 7—Kidney Damage in Obese Subjects: Oxidative Stress and Inflammation. In Obesity; del Moral, A.M., Aguilera García, C.M., Eds.; Academic Press: New York, USA, 2018; pp. 135–162. [Google Scholar] [CrossRef]
- Gai, Z.; Wang, T.; Visentin, M.; Kullak-Ublick, G.A.; Fu, X.; Wang, Z. Lipid Accumulation and Chronic Kidney Disease. Nutrients 2019, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Stasi, A.; Cosola, C.; Caggiano, G.; Cimmarusti, M.T.; Palieri, R.; Acquaviva, P.M.; Rana, G.; Gesualdo, L. Obesity-Related Chronic Kidney Disease: Principal Mechanisms and New Approaches in Nutritional Management. Front. Nutr. 2022, 9, 925619. [Google Scholar] [CrossRef] [PubMed]
- Herrington, W.G.; Smith, M.; Bankhead, C.; Matsushita, K.; Stevens, S.; Holt, T.; Hobbs, F.D.R.; Coresh, J.; Woodward, M. Body-mass index and risk of advanced chronic kidney disease: Prospective analyses from a primary care cohort of 1.4 million adults in England. PLoS ONE 2017, 12, e0173515. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xing, C.; Li, G.; Dai, X.; Gao, X.; Zhuang, Y.; Cao, H.; Hu, G.; Guo, X.; Yang, F. The key role of proteostasis at mitochondria-associated endoplasmic reticulum membrane in vanadium-induced nephrotoxicity using a proteomic strategy. Sci. Total Environ. 2023, 869, 161741. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.E. Proximal Tubulopathy: Prime Mover and Key Therapeutic Target in Diabetic Kidney Disease. Diabetes 2017, 66, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.; Palm, F.; Layton, A.T. A model of mitochondrial O2 consumption and ATP generation in rat proximal tubule cells. Am. J. Physiol. Ren. Physiol. 2020, 318, F248–F259. [Google Scholar] [CrossRef] [PubMed]
- Forst, A.L.; Reichold, M.; Kleta, R.; Warth, R. Distinct Mitochondrial Pathologies Caused by Mutations of the Proximal Tubular Enzymes EHHADH and GATM. Front. Physiol. 2021, 12, 715485. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Higgins, P.J.; Zhang, W. Development and Diseases of the Collecting Duct System. Results Probl. Cell Differ. 2017, 60, 165–203. [Google Scholar] [CrossRef]
- Shiloh, Y.; Shema, E.; Moyal, L.; Oren, M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 2011, 585, 2795–2802. [Google Scholar] [CrossRef]
- Zhang, M.; Su, L.; Wang, W.; Li, C.; Liang, Q.; Ji, F.; Jiao, J. Endothelial cells regulated by RNF20 orchestrate the proliferation and differentiation of neural precursor cells during embryonic development. Cell Rep. 2022, 40, 111350. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, J.; Cao, C.; Liang, X.; Yang, S.; Liu, L.; Tao, C.; Zhao, J.; Wang, Y. RNF20 affects porcine adipocyte differentiation via regulation of mitotic clonal expansion. Cell Prolif. 2021, 54, e13131. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, G.Y.; Jang, H.; Choe, S.S.; Koo, S.-H.; Kim, J.B. Ring finger protein20 regulates hepatic lipid metabolism through protein kinase A-dependent sterol regulatory element binding protein1c degradation. Hepatology 2014, 60, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.G.; Lee, J.H.; Ji, Y.; Sohn, J.H.; Lee, D.; Kim, D.W.; Yoon, S.G.; Shin, K.C.; Park, J.; Seong, J.K.; et al. RNF20 Functions as a Transcriptional Coactivator for PPARγ by Promoting NCoR1 Degradation in Adipocytes. Diabetes 2020, 69, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Wang, B.; Guan, Y.-F. Nuclear Receptor Regulation of Aquaporin-2 in the Kidney. Int. J. Mol. Sci. 2016, 17, 1105. [Google Scholar] [CrossRef] [PubMed]
- Corrales, P.; Izquierdo-Lahuerta, A.; Medina-Gómez, G. Maintenance of Kidney Metabolic Homeostasis by PPAR Gamma. Int. J. Mol. Sci. 2018, 19, 2063. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Feng, W.; Zhao, X.; Zhang, P.; Li, S.; Li, Z.; Lin, Y.; Liang, B.; Li, C.; Wang, W.; et al. Statins ameliorate cholesterol-induced inflammation and improve AQP2 expression by inhibiting NLRP3 activation in the kidney. Theranostics 2020, 10, 10415–10433. [Google Scholar] [CrossRef] [PubMed]
- Mount, P.; Davies, M.; Choy, S.-W.; Cook, N.; Power, D. Obesity-Related Chronic Kidney Disease—The Role of Lipid Metabolism. Metabolites 2015, 5, 720–732. [Google Scholar] [CrossRef]
- Bulbul, M.C.; Dagel, T.; Afsar, B.; Ulusu, N.N.; Kuwabara, M.; Covic, A.; Kanbay, M. Disorders of Lipid Metabolism in Chronic Kidney Disease. Blood Purif. 2018, 46, 144–152. [Google Scholar] [CrossRef]
- Sethi, G.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P. Role of RNF20 in cancer development and progression—A comprehensive review. Biosci. Rep. 2018, 38, BSR20171287. [Google Scholar] [CrossRef]
- Liang, X.; Tao, C.; Pan, J.; Zhang, L.; Liu, L.; Zhao, Y.; Fan, Y.; Cao, C.; Liu, J.; Zhang, J.; et al. Rnf20 deficiency in adipocyte impairs adipose tissue development and thermogenesis. Protein Cell 2021, 12, 475–492. [Google Scholar] [CrossRef]
- Lee, J.H.; Jeon, Y.G.; Lee, K.H.; Lee, H.W.; Park, J.; Jang, H.; Kang, M.; Lee, H.S.; Cho, H.J.; Nam, D.H.; et al. RNF20 Suppresses Tumorigenesis by Inhibiting the SREBP1c-PTTG1 Axis in Kidney Cancer. Mol. Cell Biol. 2017, 37, e00265-17. [Google Scholar] [CrossRef] [PubMed]
- Geiger, M.A.; Guillaumon, A.T.; Paneni, F.; Matter, C.M.; Stein, S. Role of the Nuclear Receptor Corepressor 1 (NCOR1) in Atherosclerosis and Associated Immunometabolic Diseases. Front. Immunol. 2020, 11, 569358. [Google Scholar] [CrossRef] [PubMed]
- Luan, Z.L.; Zhang, C.; Ming, W.H.; Huang, Y.Z.; Guan, Y.F.; Zhang, X.Y. Nuclear receptors in renal health and disease. EBioMedicine 2022, 76, 103855. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gu, Z. The Role of Peroxisome Proliferator-Activated Receptors in Kidney Diseases. Front. Pharmacol. 2022, 13, 832732. [Google Scholar] [CrossRef]
- Sharma, V.; Patial, V. Peroxisome proliferator-activated receptor gamma and its natural agonists in the treatment of kidney diseases. Front. Pharmacol. 2022, 13, 991059. [Google Scholar] [CrossRef]
- Kwon, T.H.; Frøkiær, J.; Nielsen, S. Regulation of aquaporin-2 in the kidney: A molecular mechanism of body-water homeostasis. Kidney Res. Clin. Pract. 2013, 32, 96–102. [Google Scholar] [CrossRef]


| Primary Antibody | Host | Dilution | Cat. No. | Source |
|---|---|---|---|---|
| PPARγ (C26H12) | Rabbit | 1:1000 | 2435s | Cell Signaling Technology, Danvers, MA, USA |
| LXRα | Rabbit | 1:1000 | ab176323 | Abcam, Cambridge, UK |
| CD36 | Rabbit | 1:1000 | ab133625 | Abcam, Cambridge, UK |
| ABCA1 | Mouse | 1:1000 | ab18180 | Abcam, Cambridge, UK |
| PPARα | Mouse | 1:500 | SC-398394 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
| CPT1A | Mouse | 1:1000 | ab128568 | Abcam, Cambridge, UK |
| Fibronectin | Rabbit | 1:1000 | ab2413 | Abcam, Cambridge, UK |
| RNF20 | Rabbit | 1:1000 | Ab32629 | Abcam, Cambridge, UK |
| AQP2 | Mouse | 1:1000 | SC-515770 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
| ENaC | Rabbit | 1:1000 | PA1-920A | Invitrogen, Grand Island, NY, USA |
| SREBP1 | Mouse | 1:500 | SC-365513 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
| SREBP2 | Mouse | 1:500 | SC-13552 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
| Acetyl-CoA Carboxylase (C83B10) | Rabbit | 1:1000 | 3676S | Cell Signaling Technology, Danvers, MA, USA |
| Phospho-Acetyl-CoA Carboxylase (Ser79) | Rabbit | 1:1000 | 3661s | Cell Signaling Technology, Danvers, MA, USA |
| Fatty Acid Synthase (C20G5) | Rabbit | 1:1000 | 3180s | Cell Signaling Technology, Danvers, MA, USA |
| Na+/K+ATPase α1 | Mouse | 1:1000 | 05-369 | Sigma-Aldrich, St. Louis, MO, USA |
| GAPDH | Rabbit | 1:5000 | 2118 | Cell Signaling Technology, Danvers, MA, USA |
| Weeks | ND | NDU | HD | HDU |
|---|---|---|---|---|
| Before 2 weeks | 20.0 ± 0.2 | 20.2 ± 0.2 | 19.0 ± 0.4 | 20.3 ± 0.4 |
| Uninephrectomy o.p. weeks | 24.4 ± 0.8 | 23.4 ± 0.4 | 23.7 ± 1.1 | 25.5 ± 0.6 |
| After 2 weeks | 27.7 ± 0.9 | 23.7 ± 0.5 | 25.7 ± 0.9 | 26.0 ± 0.7 |
| After 4 weeks | 29.1 ± 0.8 | 25.4 ± 0.5 | 29.0 ± 1.2 | 28.5 ± 0.7 |
| After 6 weeks | 31.5 ± 0.8 | 28.1 ± 0.5 | 34.7 ± 2.0 | 32.4 ± 0.7 |
| After 8 weeks | 33.4 ± 1.0 | 29.3 ± 0.6 | 35.8 ± 2.3 | 34.4 ± 0.8 |
| After 12 weeks | 34.6 ± 1.1 | 29.6 ± 1.0 | 35.8 ± 2.5 | 34.8 ± 1.3 |
| Average | ND | NDU | HD | HDU |
|---|---|---|---|---|
| Blood urea nitrogen (mg/dL) | 23.7 ± 2.1 | 34.3 ± 0.6 ** | 20.0 ± 1.8 | 43.3 ± 3.4 #,$$$ |
| Creatinine (mg/dL) | 0.17 ± 0.01 | 0.24 ± 0.01 * | 0.17 ± 0.01 | 0.23 ± 0.03 $ |
| Cholesterol (mg/dL) | 113.0 ± 2.5 | 101.2 ± 2.8 | 158.3 ± 2.4 *** | 164.6 ± 6.7 ### |
| LDL cholesterol (mg/dL) | 23.2 ± 3.4 | 12.2 ± 0.6 | 46.3 ± 4.5 *** | 37.7 ± 2.5 ### |
| HDL cholesterol (mg/dL) | 75.4 ± 3.1 | 82.5 ± 2.3 | 90.0 ± 7.3 | 121.2 ± 9.3 ###,$$$ |
| TG (mg/dL) | 140.2 ± 14.5 | 106.2 ± 7.5 | 123.7 ± 22.7 | 140.7 ± 53.7 |
| Glucose (mg/dL) | 202.4 ± 17.1 | 248.1 ± 10.1 | 199.3 ± 30.7 | 235.2 ± 30.4 |
| Albumin (g/dL) | 2.7 ± 0.2 | 3.5 ± 0.0 ** | 2.8 ± 0.2 | 3.2 ± 0.2 |
| Protein (g/dL) | 4.3 ± 0.1 | 4.8 ± 0.0 * | 4.6 ± 0.1 | 4.6 ± 0.1 |
| ALT (U/L) | 46.2 ± 9.7 | 20.2 ± 0.9 * | 35.3 ± 5.0 | 36.3 ± 8.5 |
| AST (U/L) | 137.4 ± 40.1 | 64.0 ± 6.3 | 142.0 ± 18.5 | 132.8 ± 26.7 |
| Log-Fold Change | |||||||
|---|---|---|---|---|---|---|---|
| Gene Name Abbreviation | Gene Full Name | Pathway | NDU vs. ND | HD vs. ND | HDU vs. ND | HDU vs. HD | HDU vs. NDU |
| Rnf20 | Ring finger protein 20 | Protein modification and ubiquitination | 1.08 | −1.02 | −1.03 | −1.00 | −1.11 |
| Ube2ql1 | Ubiquitin-conjugating enzyme E2Q family-like 1 | Ubiquitin-mediated proteolysis | −2.57 * | −1.16 | −3.07 ** | −2.65 ## | −1.2 |
| NcoR1 | Nuclear receptor corepressor 1 | Nuclear receptor transcription pathway | 1.35 ** | −1.02 | −1.03 | −1.02 | −1.40 $$ |
| NcoR2 | Nuclear receptor corepressor 2 | Nuclear receptor transcription pathway | −1.13 | −1.09 | −1.06 | 1.03 | 1.06 |
| Pparα | Peroxisome proliferator-activated receptor alpha | PPAR signaling | 1.38 * | 2.06 ** | 1.83 ** | −1.13 | 1.32 |
| Pparζ | Peroxisome proliferator-activator receptor delta | PPAR signaling | −1.06 | −2.23 ** | −1.54 * | 1.45 | −1.45 |
| Pparγ | Peroxisome proliferator-activated receptor gamma | PPAR signaling | −9.45 ** | 1.28 | −2.76 | −3.53 | 3.42 |
| Angptl4 | Angiopoietin-like 4 | PPAR signaling | 1.93 * | 2.17 ** | 3.62 ** | 1.67 # | 1.88 $ |
| Apoa1 | Apolipoprotein A-I | PPAR signaling | −1.33 | −3.18 ** | 2.20 * | 7.01 ## | 2.94 $$ |
| Apoc3 | Apolipoprotein C-III | PPAR signaling | −1.13 | −3.53 ** | 1.28 | 4.51 ## | 1.45 |
| Hmgcs2 | 3-hydroxy-3-methylglutaryl- coenzyme A synthase 2 | PPAR signaling | 2.95 ** | 4.19 ** | 2.92 ** | −1.44 | −1.01 |
| Pck1 | Phosphoenolpyruvate carboxykinase 1, cytosolic | PPAR signaling | 1.22 | 2.53 ** | 3.45 ** | 1.36 | 2.84 $$ |
| Plin4 | perilipin 4 | PPAR signaling | −13.5 ** | 2.83 | −2.47 | −6.98 # | 5.47 |
| Pltp | Phospholipid transfer protein | PPAR signaling | −1.67 | 3.06 * | −2.3 | −7.04 ## | −1.38 |
| Cpt1b | Carnitine palmitoyltransferase 1b | PPAR signaling Fatty acid degradation | −2.62 * | 1.82 | −1.35 | −2.47 | 1.94 |
| Cpt1c | Carnitine palmitoyltransferase 1c | PPAR signaling Fatty acid metabolism | −1.13 | 1.06 | −2.04 ** | −2.17 ## | −1.80 $$ |
| Scd1 | Stearoyl-Coenzyme A desaturase 1 | PPAR signaling Fatty acid metabolism | −7.04 ** | −1.93 | −5.43 * | −2.81 | 1.3 |
| Scd2 | Stearoyl-Coenzyme A desaturase 2 | PPAR signaling Fatty acid metabolism | −1.91 | −1.24 | −2.70 ** | −2.18 # | −1.42 |
| Acacα | Acetyl-coenzyme A carboxylase alpha | Fatty acid metabolism | −9.70 ** | −2.13 | −5.82 * | −2.74 | 1.67 |
| Fasn | Fatty acid synthase | Fatty acid metabolism | −14.92 ** | −1.86 | −6.57 * | −3.54 | 2.27 |
| Hacd4 | 3-hydroxyacyl-CoA dehydratase 4 | Fatty acid metabolism | −4.48 * | 1.1 | −4.67 ** | −5.16 ## | −1.04 |
| Acaa2 | Acetyl-coenzyme A acyltransferase 2 | Fatty acid degradation | −1.04 | 1.42 | −1.54 * | −2.19 ## | −1.48 |
| Cyp2u1 | Cytochrome P450, family 2, subfamily u, polypeptide 1 | Fatty acid degradation | −1.08 | 1.57 * | −1.58 * | −2.48 ## | −1.46 |
| Atp1α2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | PT bicarbonate reclamation | −15.66 ** | 1.62 | −3.61 | −5.83 # | 4.34 |
| Atp1α3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | PT bicarbonate reclamation | −11.15 ** | −1.42 | −7.22 * | −5.09 | 1.55 |
| Atp1β2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | PT bicarbonate reclamation | −1.43 * | 1.02 | −2.51 ** | −2.55 ## | −1.75 $$ |
| Cebpβ | CCAAT/enhancer-binding protein (C/EBP), beta | TNF signaling | −1.46 | −1.2 | 2.22 ** | 2.67 ## | 3.23 $$ |
| Adcy3 | Adenylate cyclase 3 | Vasopressin-regulated water reabsorption | −3.35 * | 1.48 | −3.08 * | −4.57 ## | 1.09 |
| Aqp2 | Aquaporin 2 | Vasopressin-regulated water reabsorption | −2.37 ** | −1.2 | 1.1 | 1.32 | 2.61 $$ |
| Aqp4 | Aquaporin 4 | Vasopressin-regulated water reabsorption | −3.45 * | 1.14 | −1.33 | −1.51 | 2.6 |
| Arhgdiγ | Rho-GDP dissociation inhibitor gamma | Vasopressin-regulated water reabsorption | −4.20 ** | 1.5 | −1.08 | −1.62 | 3.88 $$ |
| Avpr2 | Arginine vasopressin receptor 2 | Vasopressin-regulated water reabsorption | −1.52 | −1.08 | −2.55 ** | −2.37 ## | −1.68 $ |
| Crebbp | CREB-binding protein | TGF-beta signaling, cell cycle, JAK-STAT signaling | −1.04 | −1.12 | −1.08 | 1.04 | −1.04 |
| Abca1 | ATP-binding cassette, subfamily A, member 1 | ABC transporters, Fat digestion and absorption, cholesterol metabolism | 1.03 | 1.38 | −1.39 | −1.92 ## | −1.43 |
| Acox1 | Acyl-coenzyme A oxidase 1 | Fatty acid degradation unsaturated fatty acid biosynthesis | 1.35 * | 1.11 | 1.46 ** | 1.32 # | 1.08 |
| Ar | Androgen receptor | Androgen receptor signaling | 1.36 * | 1.06 | 1.59 ** | 1.49 # | 1.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-J.; Oh, S.-H.; Lim, J.-H.; Cho, J.-H.; Jung, H.-Y.; Kim, C.-D.; Park, S.-H.; Kwon, T.-H.; Kim, Y.-L. Impact of Ring Finger Protein 20 and Its Downstream Regulation on Renal Tubular Injury in a Unilateral Nephrectomy Mouse Model Fed a High-Fat Diet. Nutrients 2023, 15, 4959. https://doi.org/10.3390/nu15234959
Kim Y-J, Oh S-H, Lim J-H, Cho J-H, Jung H-Y, Kim C-D, Park S-H, Kwon T-H, Kim Y-L. Impact of Ring Finger Protein 20 and Its Downstream Regulation on Renal Tubular Injury in a Unilateral Nephrectomy Mouse Model Fed a High-Fat Diet. Nutrients. 2023; 15(23):4959. https://doi.org/10.3390/nu15234959
Chicago/Turabian StyleKim, You-Jin, Se-Hyun Oh, Jeong-Hoon Lim, Jang-Hee Cho, Hee-Yeon Jung, Chan-Duck Kim, Sun-Hee Park, Tae-Hwan Kwon, and Yong-Lim Kim. 2023. "Impact of Ring Finger Protein 20 and Its Downstream Regulation on Renal Tubular Injury in a Unilateral Nephrectomy Mouse Model Fed a High-Fat Diet" Nutrients 15, no. 23: 4959. https://doi.org/10.3390/nu15234959
APA StyleKim, Y.-J., Oh, S.-H., Lim, J.-H., Cho, J.-H., Jung, H.-Y., Kim, C.-D., Park, S.-H., Kwon, T.-H., & Kim, Y.-L. (2023). Impact of Ring Finger Protein 20 and Its Downstream Regulation on Renal Tubular Injury in a Unilateral Nephrectomy Mouse Model Fed a High-Fat Diet. Nutrients, 15(23), 4959. https://doi.org/10.3390/nu15234959











