Mothers’ Breast Milk Composition and Their Respective Infant’s Gut Microbiota Differ between Five Distinct Rural and Urban Regions in Vietnam
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Cohort and Sampling
2.2. DNA Isolation
2.3. PCR Amplification and 16S rRNA Gene Illumina Sequencing
2.4. 16S rRNA Gene Sequence Analysis and Statistics
2.5. Dietary Intake Analysis
2.6. Blood Parameters
2.7. Breast Milk Component Analysis
3. Results
3.1. Demographics of the Cohort
3.2. Quality Control and Overall Microbiota Composition
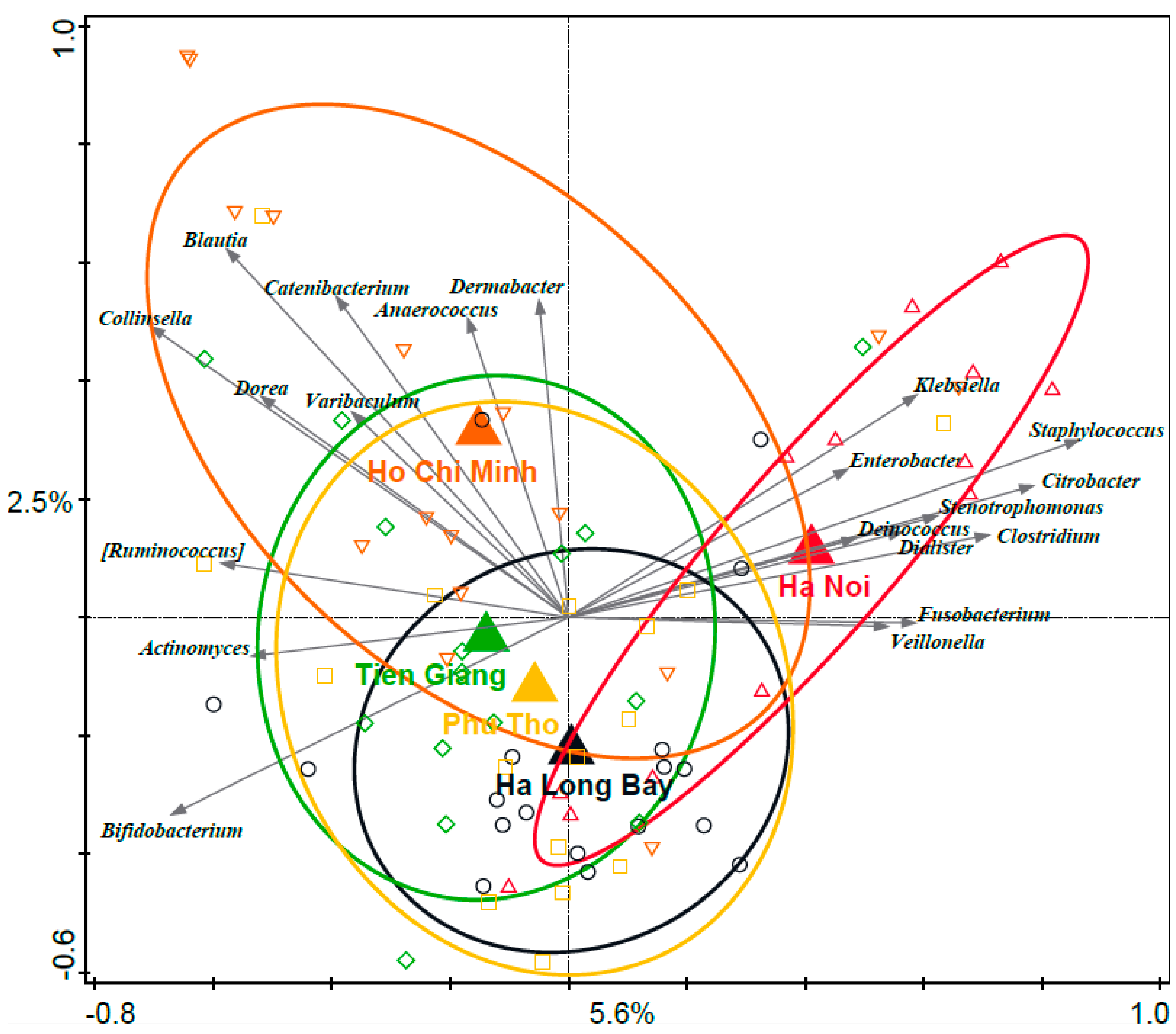
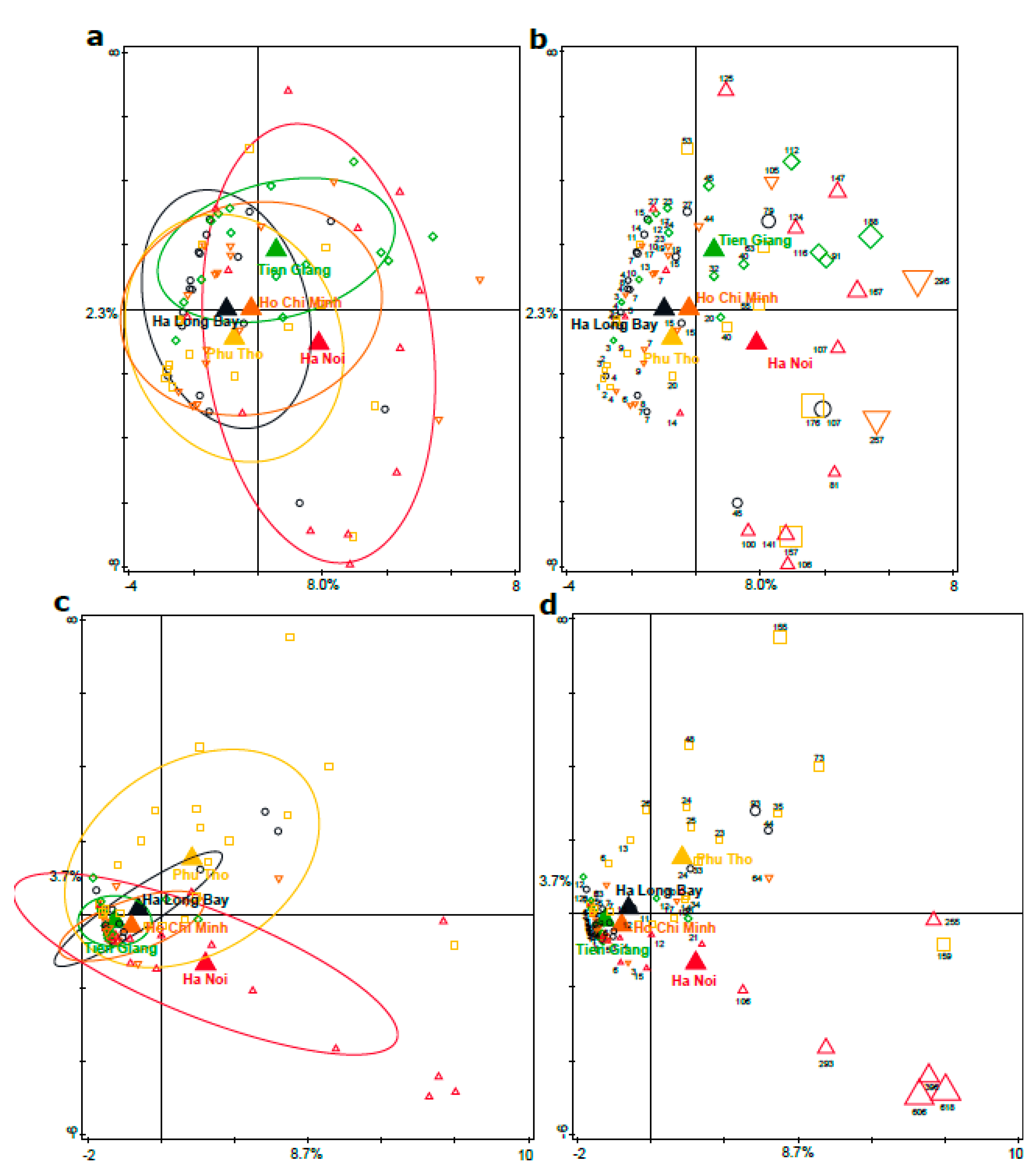
3.3. Associations between Distinct Geographical Areas and Infant Fecal Microbiota Composition
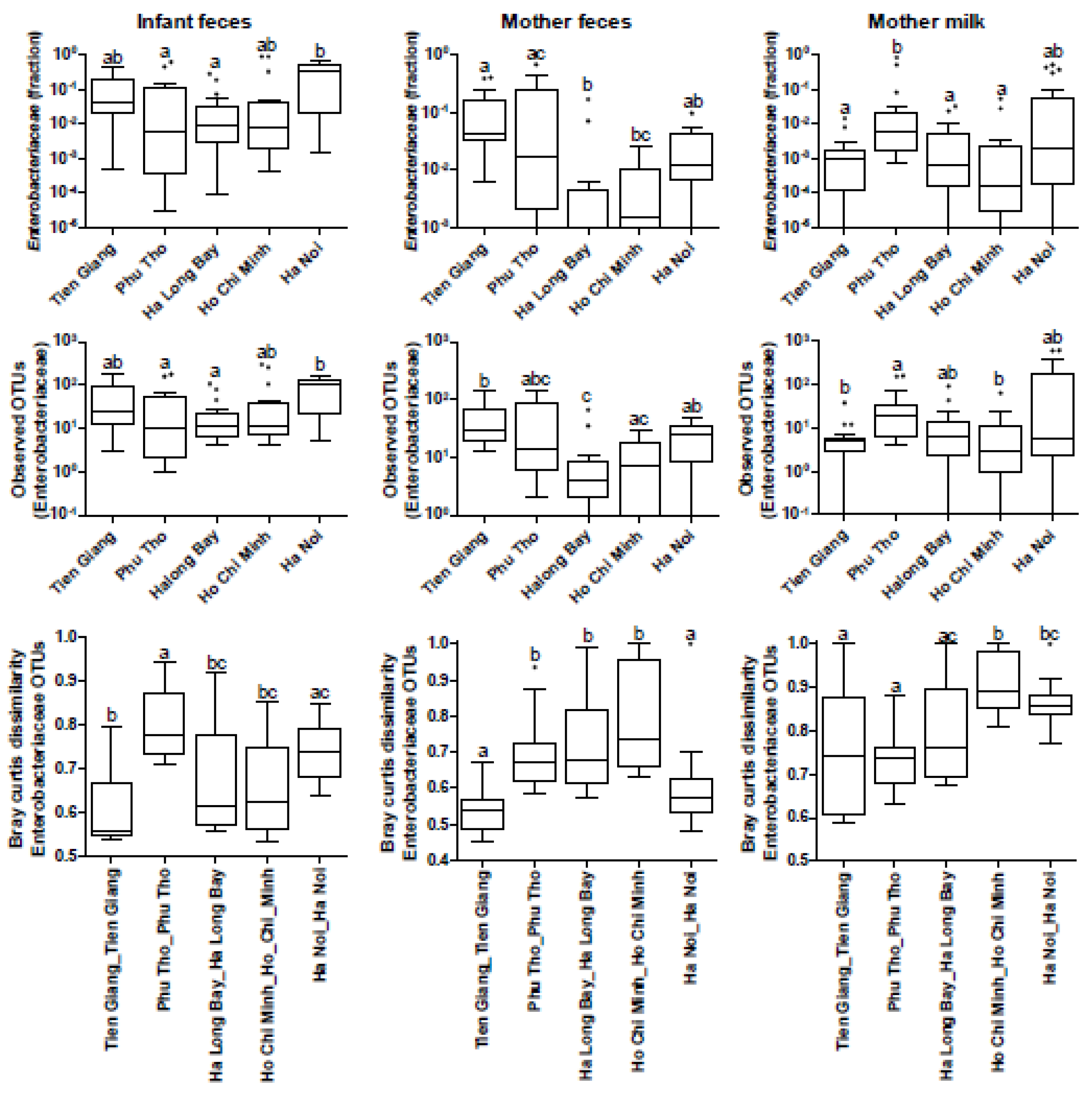
3.4. Phylogenetic Analyses of the Enterobacteriaceae as Present in the Infant Microbiota and Maternal Breast Milk Samples
3.5. Associations between Geography and Breast Milk Nutrient Composition
3.6. Assessment of Maternal Transmission of Fecal and/or Breast Milk Microbiota to the Infant Fecal Microbiota
3.7. Associations between Breast Milk Nutrients and Infant Microbiota Composition
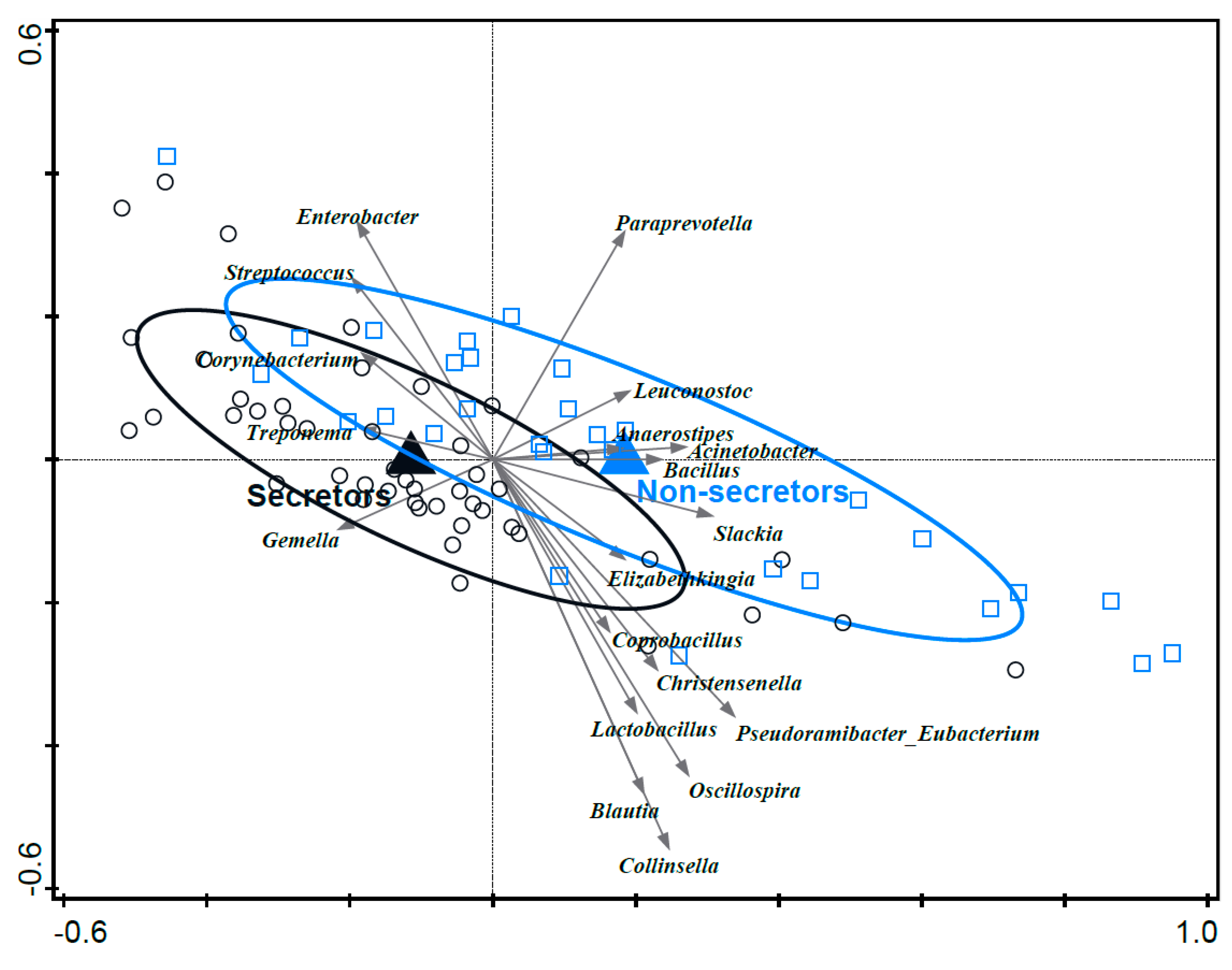
3.8. Associations between Mother’s Secretor Status and Infant Fecal Microbiota Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef]
- Moore, R.E.; Townsend, S.D. Temporal development of the infant gut microbiome. Open Biol. 2019, 9, 190128. [Google Scholar] [CrossRef]
- Bunesova, V.; Lacroix, C.; Schwab, C. Mucin Cross-Feeding of Infant Bifidobacteria and Eubacterium hallii. Microb. Ecol. 2018, 75, 228–238. [Google Scholar] [CrossRef]
- Carr, L.E.; Virmani, M.D.; Rosa, F.; Munblit, D.; Matazel, K.S.; Elolimy, A.A.; Yeruva, L. Role of Human Milk Bioactives on Infants’ Gut and Immune Health. Front. Immunol. 2021, 12, 604080. [Google Scholar] [CrossRef]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles across Specific Geographic Locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Macias, E.; Selma-Royo, M.; Garcia-Mantrana, I.; Calatayud, M.; Gonzalez, S.; Martinez-Costa, C.; Collado, M.C. Maternal Diet Shapes the Breast Milk Microbiota Composition and Diversity: Impact of Mode of Delivery and Antibiotic Exposure. J. Nutr. 2021, 151, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gallego, C.; Garcia-Mantrana, I.; Salminen, S.; Collado, M.C. The human milk microbiome and factors influencing its composition and activity. Semin. Fetal Neonatal Med. 2016, 21, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Samuel, T.M.; Zhou, Q.; Giuffrida, F.; Munblit, D.; Verhasselt, V.; Thakkar, S.K. Nutritional and Non-nutritional Composition of Human Milk Is Modulated by Maternal, Infant, and Methodological Factors. Front. Nutr. 2020, 7, 576133. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A review of the factors that influence composition. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef]
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; Moore, S.E.; et al. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. [Google Scholar] [CrossRef]
- Ruan, C.; Liu, X.; Man, H.; Ma, X.; Lu, G.; Duan, G.; DeFrancesco, C.A.; Connor, W.E. Milk composition in women from five different regions of China: The great diversity of milk fatty acids. J. Nutr. 1995, 125, 2993–2998. [Google Scholar] [CrossRef]
- Boudry, G.; Charton, E.; Le Huerou-Luron, I.; Ferret-Bernard, S.; Le Gall, S.; Even, S.; Blat, S. The Relationship between Breast Milk Components and the Infant Gut Microbiota. Front. Nutr. 2021, 8, 629740. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, X.; Zhou, B.; Jiang, A.C.; Chai, L. An updated review of worldwide levels of docosahexaenoic and arachidonic acid in human breast milk by region. Public Health Nutr. 2016, 19, 2675–2687. [Google Scholar] [CrossRef]
- Roy, S.; Dhar, P.; Ghosh, S. Comparative evaluation of essential fatty acid composition of mothers’ milk of some urban and suburban regions of West Bengal, India. Int. J. Food Sci. Nutr. 2012, 63, 895–901. [Google Scholar] [CrossRef]
- Taylor, R.; Keane, D.; Borrego, P.; Arcaro, K. Effect of Maternal Diet on Maternal Milk and Breastfed Infant Gut Microbiomes: A Scoping Review. Nutrients 2023, 15, 1420. [Google Scholar] [CrossRef] [PubMed]
- Ayeni, F.A.; Biagi, E.; Rampelli, S.; Fiori, J.; Soverini, M.; Audu, H.J.; Cristino, S.; Caporali, L.; Schnorr, S.L.; Carelli, V.; et al. Infant and Adult Gut Microbiome and Metabolome in Rural Bassa and Urban Settlers from Nigeria. Cell Rep. 2018, 23, 3056–3067. [Google Scholar] [CrossRef] [PubMed]
- Brushett, S.; Sinha, T.; Reijneveld, S.A.; de Kroon, M.L.A.; Zhernakova, A. The Effects of Urbanization on the Infant Gut Microbiota and Health Outcomes. Front. Pediatr. 2020, 8, 408. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Gomez, A.; Petrzelkova, K.J.; Burns, M.B.; Yeoman, C.J.; Amato, K.R.; Vlckova, K.; Modry, D.; Todd, A.; Jost Robinson, C.A.; Remis, M.J.; et al. Gut Microbiome of Coexisting BaAka Pygmies and Bantu Reflects Gradients of Traditional Subsistence Patterns. Cell Rep. 2016, 14, 2142–2153. [Google Scholar] [CrossRef]
- Obregon-Tito, A.J.; Tito, R.Y.; Metcalf, J.; Sankaranarayanan, K.; Clemente, J.C.; Ursell, L.K.; Zech Xu, Z.; Van Treuren, W.; Knight, R.; Gaffney, P.M.; et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 2015, 6, 6505. [Google Scholar] [CrossRef]
- Winglee, K.; Howard, A.G.; Sha, W.; Gharaibeh, R.Z.; Liu, J.; Jin, D.; Fodor, A.A.; Gordon-Larsen, P. Recent urbanization in China is correlated with a Westernized microbiome encoding increased virulence and antibiotic resistance genes. Microbiome 2017, 5, 121. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Vaidya, Y.H.; Patel, S.H.; Patel, R.J.; Pandit, R.J.; Joshi, C.G.; Kunjadia, A.P. Human milk microbiome in urban and rural populations of India. Meta Gene 2017, 13, 13–22. [Google Scholar] [CrossRef]
- Vangay, P.; Ward, T.; Gerber, J.S.; Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015, 17, 553–564. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Stoutjesdijk, E.; Ten Kate, G.A.; Schaafsma, A.; Dijck-Brouwer, J.; Muskiet, F.A.J.; Dijkhuizen, L. Regional variations in human milk oligosaccharides in Vietnam suggest FucTx activity besides FucT2 and FucT3. Sci. Rep. 2018, 8, 16790. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Zeller, G.; Sunagawa, S.; Pelletier, E.; Alberti, A.; Levenez, F.; Tramontano, M.; Driessen, M.; Hercog, R.; Jung, F.E.; et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017, 35, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- ter Braak, C.; Šmilauer, P. Canoco Reference Manual and User’s Guide: Software for Ordination (Version 5.0); Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Gibson, R.S. Principles of Nutritional Assessment, 2nd ed.; Oxford University Press: New York, NY, USA; Oxford, UK, 2005; 908p. [Google Scholar]
- Vietnamese Food Composition Table; Hanoi Medical Publishing House: Hanoi, Vietnam, 2007.
- Hampel, D.; Shahab-Ferdows, S.; Domek, J.M.; Siddiqua, T.; Raqib, R.; Allen, L.H. Competitive chemiluminescent enzyme immunoassay for vitamin B12 analysis in human milk. Food Chem. 2014, 153, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Stoutjesdijk, E.; Schaafsma, A.; Nhien, N.V.; Khor, G.L.; Kema, I.P.; Hollis, B.W.; Dijck-Brouwer, D.A.J.; Muskiet, F.A.J. Milk vitamin D in relation to the ‘adequate intake’ for 0–6-month-old infants: A study in lactating women with different cultural backgrounds, living at different latitudes. Br. J. Nutr. 2017, 118, 804–812. [Google Scholar] [CrossRef]
- Volmer, M.; Meiborg, G.; Muskiet, F.A. Simultaneous capillary gas chromatographic profiling of medium-and long-chain fatty acid methyl esters with split injection. Correction for injection-related discrimination by the ‘bracketing’ method. J. Chromatogr. 1988, 434, 385–394. [Google Scholar] [CrossRef]
- van der Steege, G.; Muskiet, F.A.; Martini, I.A.; Hutter, N.H.; Boersma, E.R. Simultaneous quantification of total medium- and long-chain fatty acids in human milk by capillary gas chromatography with split injection. J. Chromatogr. 1987, 415, 1–11. [Google Scholar] [CrossRef]
- Kortman, G.A.; Raffatellu, M.; Swinkels, D.W.; Tjalsma, H. Nutritional iron turned inside out: Intestinal stress from a gut microbial perspective. FEMS Microbiol. Rev. 2014, 38, 1202–1234. [Google Scholar] [CrossRef]
- Choi, Y.K.; Kim, J.M.; Lee, J.E.; Cho, M.S.; Kang, B.S.; Choi, H.; Kim, Y. Association of Maternal Diet with Zinc, Copper, and Iron Concentrations in Transitional Human Milk Produced by Korean Mothers. Clin. Nutr. Res. 2016, 5, 15–25. [Google Scholar] [CrossRef]
- Le Luu, T. Remarks on the current quality of groundwater in Vietnam. Environ. Sci. Pollut. Res. Int. 2019, 26, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Minh, N.N.Q.; Toi, P.V.; Qui, L.M.; Tinh, L.B.B.; Ngoc, N.T.; Kim, L.T.N.; Uyen, N.H.; Hang, V.T.T.; Chinh B’Krong, N.T.T.; Tham, N.T.; et al. Antibiotic use and prescription and its effects on Enterobacteriaceae in the gut in children with mild respiratory infections in Ho Chi Minh City, Vietnam. A prospective observational outpatient study. PLoS ONE 2020, 15, e0241760. [Google Scholar] [CrossRef] [PubMed]
- Danko, D.; Bezdan, D.; Afshin, E.E.; Ahsanuddin, S.; Bhattacharya, C.; Butler, D.J.; Chng, K.R.; Donnellan, D.; Hecht, J.; Jackson, K.; et al. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell 2021, 184, 3376–3393. [Google Scholar] [CrossRef] [PubMed]
- Bolte, L.A.; Vich Vila, A.; Imhann, F.; Collij, V.; Gacesa, R.; Peters, V.; Wijmenga, C.; Kurilshikov, A.; Campmans-Kuijpers, M.J.E.; Fu, J.; et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021, 70, 1287–1298. [Google Scholar] [CrossRef]
- Vijay, A.; Astbury, S.; Le Roy, C.; Spector, T.D.; Valdes, A.M. The prebiotic effects of omega-3 fatty acid supplementation: A six-week randomised intervention trial. Gut Microbes 2021, 13, 1863133. [Google Scholar] [CrossRef]
- Watson, H.; Mitra, S.; Croden, F.C.; Taylor, M.; Wood, H.M.; Perry, S.L.; Spencer, J.A.; Quirke, P.; Toogood, G.J.; Lawton, C.L.; et al. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut 2018, 67, 1974–1983. [Google Scholar] [CrossRef]
- Brenna, J.T.; Varamini, B.; Jensen, R.G.; Diersen-Schade, D.A.; Boettcher, J.A.; Arterburn, L.M. Docosahexaenoic and arachidonic acid concentrations in human breast milk worldwide. Am. J. Clin. Nutr. 2007, 85, 1457–1464. [Google Scholar] [CrossRef]
- Raiten, D.J.; Talbot, J.M.; Waters, J.H. Assessment of nutrient requirements for infant formulas. J. Nutr. 1998, 128 (Suppl. 11), 2059S–2293S. [Google Scholar]
- Lewis, Z.T.; Totten, S.M.; Smilowitz, J.T.; Popovic, M.; Parker, E.; Lemay, D.G.; Van Tassell, M.L.; Miller, M.J.; Jin, Y.S.; German, J.B.; et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015, 3, 13. [Google Scholar] [CrossRef]
- Smith-Brown, P.; Morrison, M.; Krause, L.; Davies, P.S. Mothers Secretor Status Affects Development of Childrens Microbiota Composition and Function: A Pilot Study. PLoS ONE 2016, 11, e0161211. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Tao, J.; Zhou, J.; Fan, Q.; Liu, M.; Hu, Y.; Xu, Y.; Zhang, L.; Yuan, J.; Li, W.; et al. Fucosylated Human Milk Oligosaccharides and N-Glycans in the Milk of Chinese Mothers Regulate the Gut Microbiome of Their Breast-Fed Infants during Different Lactation Stages. mSystems 2018, 3, e00206-18. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Stewart, C.J.; Laucirica, D.R.; Ajami, N.J.; Robertson, B.; Autran, C.A.; Shinge, D.; Rani, S.; Anandan, S.; Hu, L.; et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 2018, 9, 5010. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Salonen, A.; Hickman, B.; Kunz, C.; Sprenger, N.; Kukkonen, K.; Savilahti, E.; Kuitunen, M.; de Vos, W.M. Fucosylated oligosaccharides in mother’s milk alleviate the effects of caesarean birth on infant gut microbiota. Sci. Rep. 2018, 8, 13757. [Google Scholar] [CrossRef]
- Seferovic, M.D.; Mohammad, M.; Pace, R.M.; Engevik, M.; Versalovic, J.; Bode, L.; Haymond, M.; Aagaard, K.M. Maternal diet alters human milk oligosaccharide composition with implications for the milk metagenome. Sci. Rep. 2020, 10, 22092. [Google Scholar] [CrossRef]
- Sundquist, A.; Bigdeli, S.; Jalili, R.; Druzin, M.L.; Waller, S.; Pullen, K.M.; El-Sayed, Y.Y.; Taslimi, M.M.; Batzoglou, S.; Ronaghi, M. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC Microbiol. 2007, 7, 108. [Google Scholar] [CrossRef]


| Region | Typical Diet | Energy (kcal) | Protein (g) | Animal Protein (g) | Fat (g) | Plant-Based Fat (g) |
|---|---|---|---|---|---|---|
| Tien Giang | Freshwater fish | 2022 a | 87 b | 52 b | 52 b | 10 ab |
| Phu Tho | Rice | 1677 c | 65 a | 27 a | 36 a | 12 a |
| Ha Long Bay | Saltwater fish | 1711 ac | 72 ab | 32 a | 36 a | 12 a |
| Ha Noi | Westernized | 2053 b | 93 b | 60 b | 50 b | 4 b |
| Ho Chi Minh City | Westernized | 2214 b | 99 b | 59 b | 74 c | 23 ab |
| Breast Milk Nutrient | Explains (%) | p-Value | p-Value (FDR) |
|---|---|---|---|
| Zinc | 3.3 | 0.012 | 0.161 |
| Iron | 2.7 | 0.014 | 0.161 |
| Iodine | 2.2 | 0.068 | 0.355 |
| Potassium | 2.2 | 0.072 | 0.355 |
| Magnesium | 2.0 | 0.108 | 0.355 |
| Vitamin B12 | 1.9 | 0.100 | 0.355 |
| Selenium | 1.9 | 0.104 | 0.355 |
| Palmitoleic acid | 1.7 | 0.204 | 0.587 |
| Copper | 1.6 | 0.242 | 0.618 |
| Docosahexaenoic acid (DHA) | 1.5 | 0.294 | 0.619 |
| Oleic acid | 1.4 | 0.296 | 0.619 |
| Linoleic acid | 1.3 | 0.396 | 0.715 |
| 2’FL | 1.3 | 0.404 | 0.715 |
| Vaccenic acid | 1.2 | 0.484 | 0.795 |
| Vitamin D (ARA) | 1.1 | 0.528 | 0.810 |
| Alpha-linoleic acid | 1.0 | 0.696 | 0.852 |
| Lauric acid | 1.0 | 0.752 | 0.852 |
| Palmitic acid | 0.9 | 0.722 | 0.852 |
| Calcium | 0.9 | 0.770 | 0.852 |
| Sodium | 0.9 | 0.764 | 0.852 |
| Eicosapentaenoic acid (EPA) | 0.9 | 0.778 | 0.852 |
| Myristic acid | 0.9 | 0.820 | 0.857 |
| Arachidonic acid | 0.6 | 0.978 | 0.978 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kortman, G.A.M.; Timmerman, H.M.; Schaafsma, A.; Stoutjesdijk, E.; Muskiet, F.A.J.; Nhien, N.V.; van Hoffen, E.; Boekhorst, J.; Nauta, A. Mothers’ Breast Milk Composition and Their Respective Infant’s Gut Microbiota Differ between Five Distinct Rural and Urban Regions in Vietnam. Nutrients 2023, 15, 4802. https://doi.org/10.3390/nu15224802
Kortman GAM, Timmerman HM, Schaafsma A, Stoutjesdijk E, Muskiet FAJ, Nhien NV, van Hoffen E, Boekhorst J, Nauta A. Mothers’ Breast Milk Composition and Their Respective Infant’s Gut Microbiota Differ between Five Distinct Rural and Urban Regions in Vietnam. Nutrients. 2023; 15(22):4802. https://doi.org/10.3390/nu15224802
Chicago/Turabian StyleKortman, Guus A. M., Harro M. Timmerman, Anne Schaafsma, Eline Stoutjesdijk, Frits A. J. Muskiet, Nguyen V. Nhien, Els van Hoffen, Jos Boekhorst, and Arjen Nauta. 2023. "Mothers’ Breast Milk Composition and Their Respective Infant’s Gut Microbiota Differ between Five Distinct Rural and Urban Regions in Vietnam" Nutrients 15, no. 22: 4802. https://doi.org/10.3390/nu15224802
APA StyleKortman, G. A. M., Timmerman, H. M., Schaafsma, A., Stoutjesdijk, E., Muskiet, F. A. J., Nhien, N. V., van Hoffen, E., Boekhorst, J., & Nauta, A. (2023). Mothers’ Breast Milk Composition and Their Respective Infant’s Gut Microbiota Differ between Five Distinct Rural and Urban Regions in Vietnam. Nutrients, 15(22), 4802. https://doi.org/10.3390/nu15224802





