Predicting Personalized Diets Based on Microbial Characteristics between Patients with Superficial Gastritis and Atrophic Gastritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Publicly Available Metagenomic and 16S rRNA Datasets
2.2. Experimental Settings for 16S rRNA Datasets
2.3. Subject Recruitment and Sample Collection
2.4. DNA Extraction of Gastric Antrum Tissue and MiSeq Sequencing
2.5. ASV Construction and Taxonomic Assignment
2.6. Adopted Machine Learning Methods
2.7. Prediction of Metagenomic Functions
2.8. Exogenous Metabolites and Whole Foods Prediction
2.9. Statistical Tests
3. Results
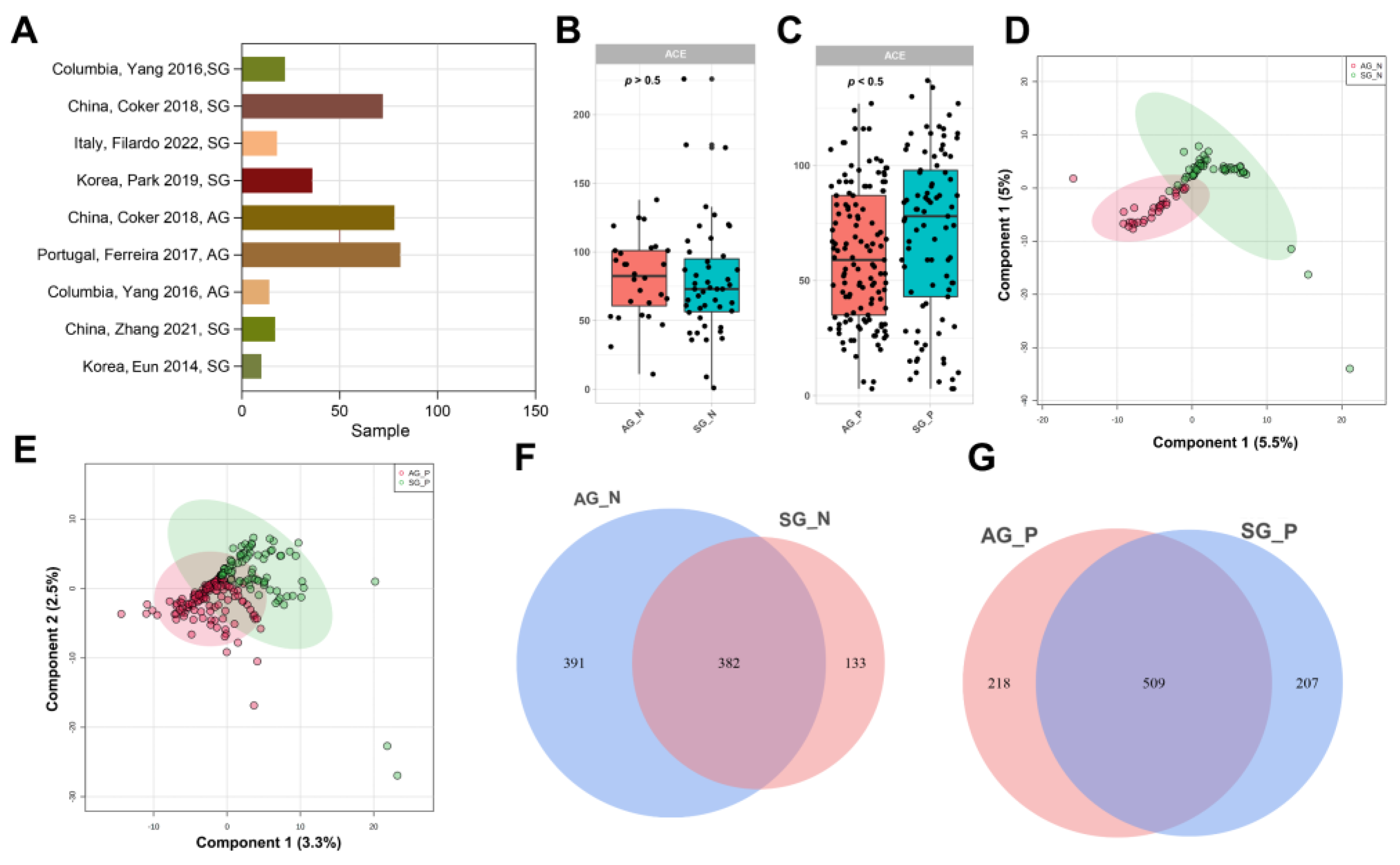
3.1. Published 16S rRNA Data of Gastritis Included in This Study
3.2. Gastric Microbial Diversity of CKD between SG and AG
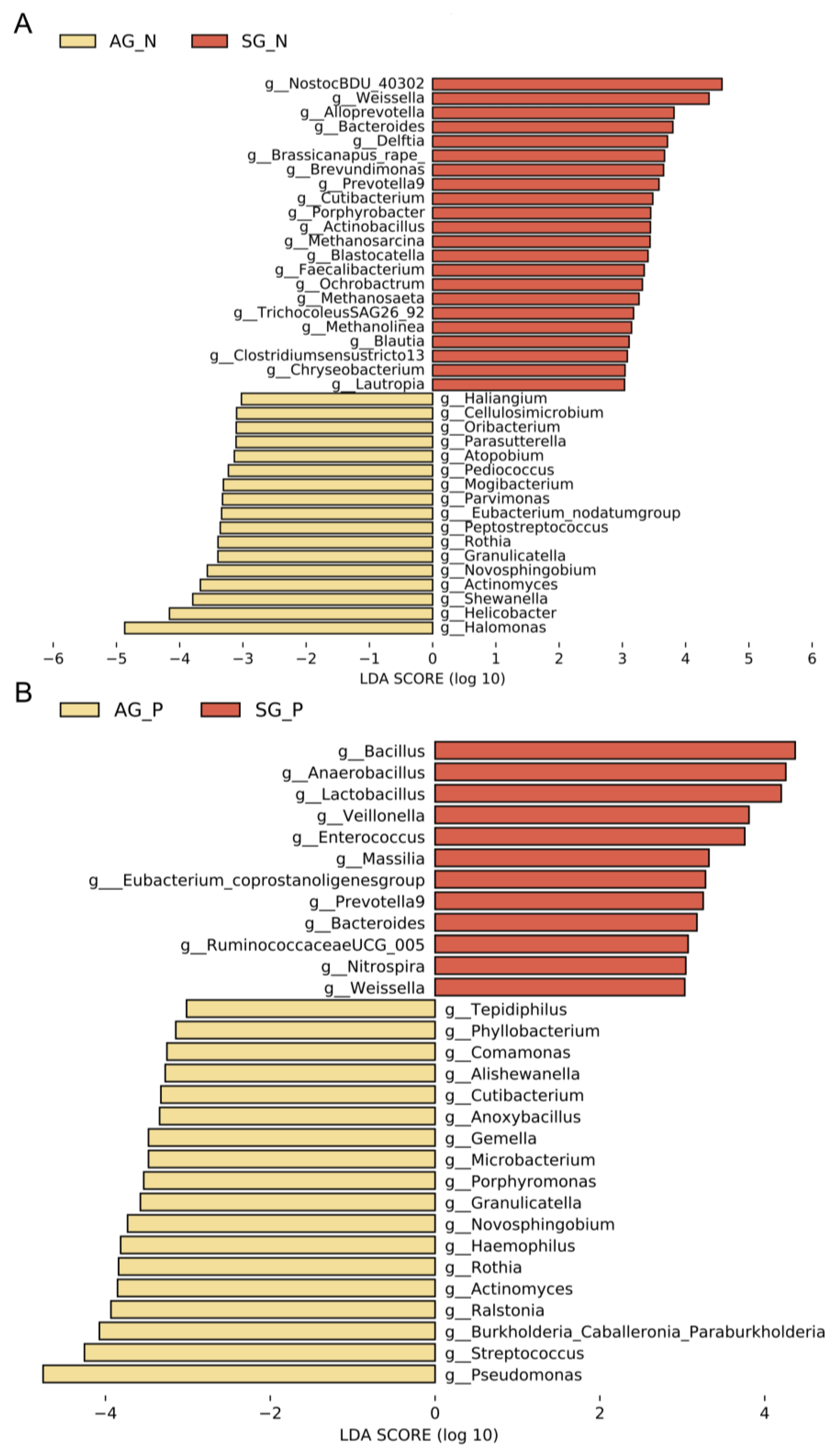
3.3. Gastric Microbial Biomarkers Related to AG
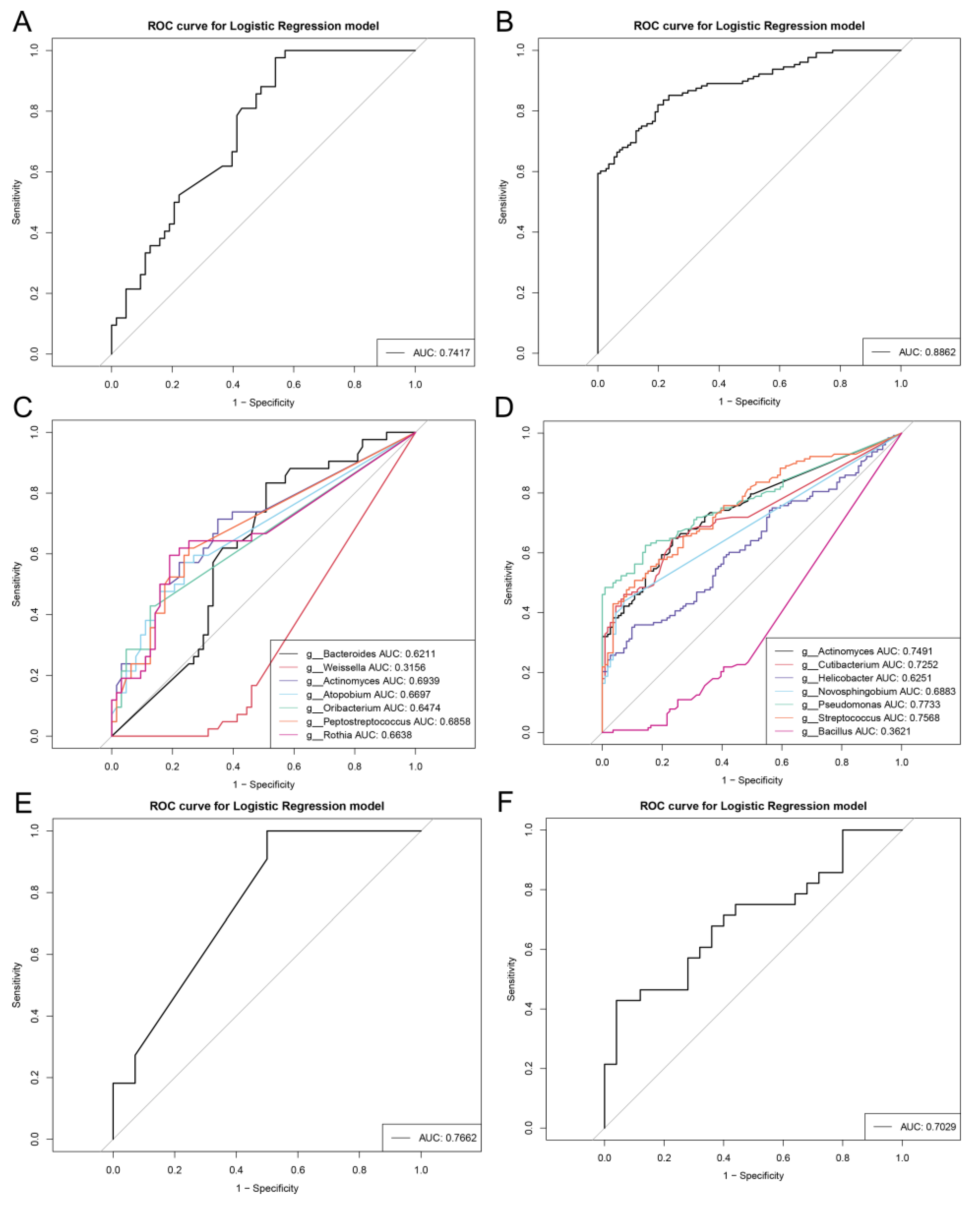
3.4. Repeatable Gastric Microbial Markers for AG
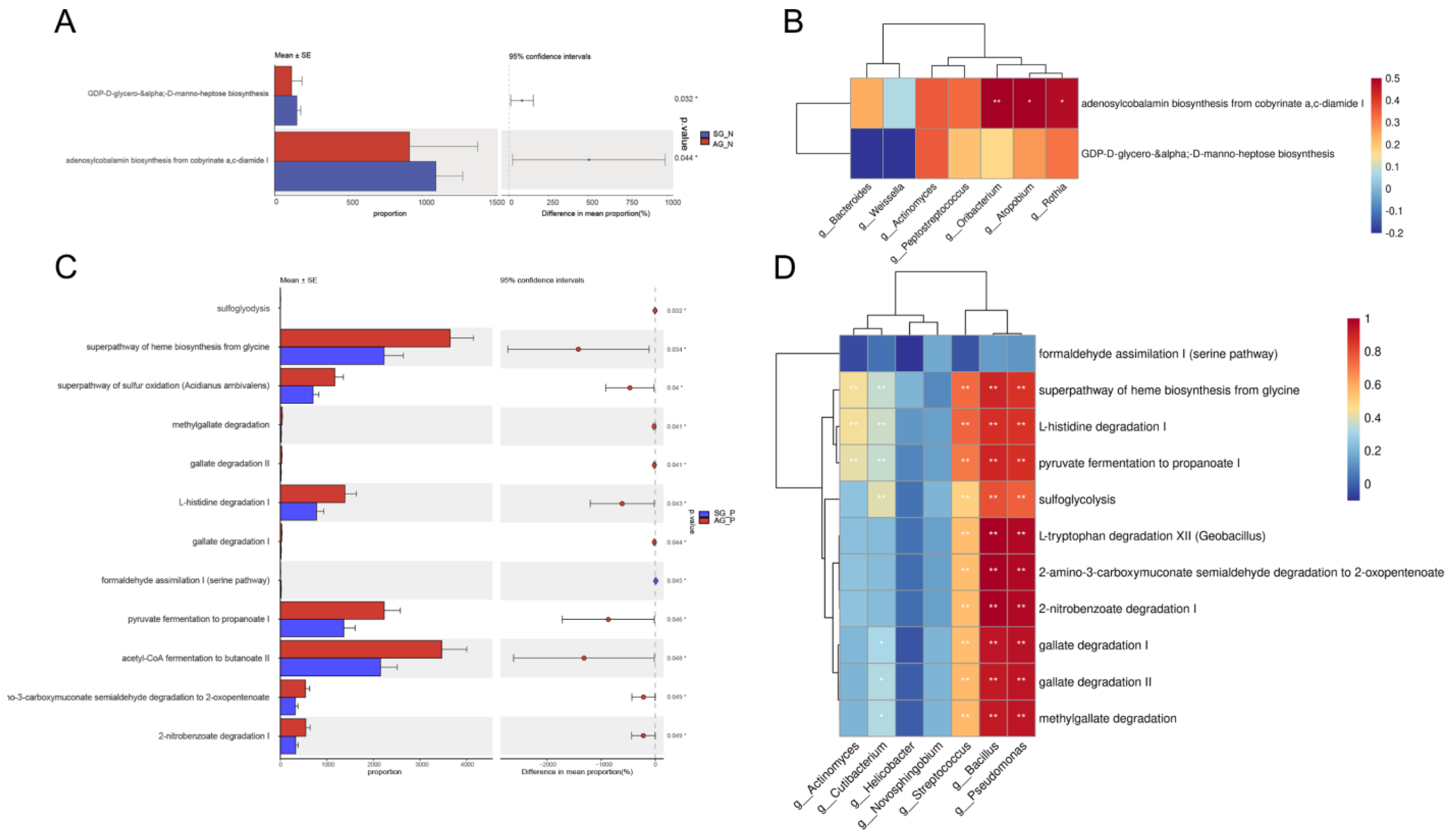
3.5. Crucial Bacteria and Microbial Functions Related to AG
3.6. Dietary-Based Interventions for AG
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, D.; Chen, S.; Gou, Y.; Yu, W.; Zhou, H.; Zhang, R.; Wang, J.; Ye, F.; Liu, Y.; Sun, B.; et al. Gastrointestinal Microbiota Changes in Patients with Gastric Precancerous Lesions. Front. Cell Infect. Microbiol. 2021, 11, 749207. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, T.; Lu, Y.; Wang, C.; Wu, Y.; Li, J.; Tao, Y.; Deng, L.; Zhang, X.; Ma, J. Helicobacter pylori infection affects the human gastric microbiome, as revealed by metagenomic sequencing. FEBS Open Bio 2022, 12, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.-J.; Zhang, Y.; Gerhard, M.; Mejias-Luque, R.; Zhang, L.; Vieth, M.; Ma, J.-L.; Bajbouj, M.; Suchanek, S.; Liu, W.-D.; et al. Association Between Gut Microbiota and Helicobacter pylori-Related Gastric Lesions in a High-Risk Population of Gastric Cancer. Front. Cell Infect. Microbiol. 2018, 8, 202. [Google Scholar] [CrossRef]
- Chen, C.-C.; Liou, J.-M.; Lee, Y.-C.; Hong, T.-C.; El-Omar, E.M.; Wu, M.-S. The interplay between Helicobacter pylori and gastrointestinal microbiota. Gut Microbes 2021, 13, 1909459. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.-K.; Park, J.C.; Kim, K.H.; Yoon, J.; Cho, Y.; Lee, B.; Lee, J.-J.; Jeong, H.; Oh, Y.; Kim, S.-H.; et al. Human gastric microbiota transplantation recapitulates premalignant lesions in germ-free mice. Gut 2022, 71, 1266–1276. [Google Scholar] [CrossRef]
- De Witte, C.; Schulz, C.; Smet, A.; Malfertheiner, P.; Haesebrouck, F. Other Helicobacters and gastric microbiota. Helicobacter 2016, 21, 62–68. [Google Scholar] [CrossRef]
- Olm, M.R.; Dahan, D.; Carter, M.M.; Merrill, B.D.; Yu, F.B.; Jain, S.; Meng, X.; Tripathi, S.; Wastyk, H.; Neff, N.; et al. Robust variation in infant gut microbiome assembly across a spectrum of lifestyles. Science 2022, 376, 1220–1223. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Fang, J.; Du, Y.Q.; Liu, W.Z.; Ren, J.L.; Li, Y.Q.; Chen, X.Y.; Lv, N.H.; Chen, Y.X.; Lv, B. Chinese consensus on chronic gastritis (2017, Shanghai). J. Dig. Dis. 2018, 19, 182–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, J.; Xin, Y.; Geng, C.; Tian, Z.; Yu, X.; Dong, Q. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur. J. Gastroenterol. Hepatol. 2016, 28, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.S.; Kim, B.K.; Han, D.S.; Kim, S.Y.; Kim, K.M.; Choi, B.Y.; Song, K.S.; Kim, Y.S.; Kim, J.F. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014, 19, 407–416. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Cao, W.; Zhang, Z. Alterations of Gastric Microbiota in Gastric Cancer and Precancerous Stages. Front. Cell Infect. Microbiol. 2021, 11, 559148. [Google Scholar] [CrossRef] [PubMed]
- Seekatz, A.M.; Schnizlein, M.K.; Koenigsknecht, M.J.; Baker, J.R.; Hasler, W.L.; Bleske, B.E.; Young, V.B.; Sun, D. Spatial and Temporal Analysis of the Stomach and Small-Intestinal Microbiota in Fasted Healthy Humans. mSphere 2019, 4, e00126-19. [Google Scholar] [CrossRef]
- Park, C.H.; Lee, A.; Lee, Y.; Eun, C.S.; Kil Lee, S.; Han, D.S. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter 2019, 24, e12547. [Google Scholar] [CrossRef]
- Filardo, S.; Scalese, G.; Virili, C.; Pontone, S.; Di Pietro, M.; Covelli, A.; Bedetti, G.; Marinelli, P.; Bruno, G.; Stramazzo, I.; et al. The Potential Role of Hypochlorhydria in the Development of Duodenal Dysbiosis: A Preliminary Report. Front. Cell Infect. Microbiol. 2022, 12, 854904. [Google Scholar] [CrossRef] [PubMed]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037-15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, X.; Zeng, R.; Wu, Q.; Sun, H.; Wu, W.; Zhang, X.; Sun, G.; Yan, B.; Wu, L.; et al. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front. Microbiol. 2020, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Woltemate, S.; Piazuelo, M.B.; Bravo, L.E.; Yepez, M.C.; Romero-Gallo, J.; Delgado, A.G.; Wilson, K.T.; Peek, R.M.; Correa, P.; et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci. Rep. 2016, 6, 18594. [Google Scholar] [CrossRef]
- Sung, J.; Kim, N.; Kim, J.; Jo, H.J.; Park, J.H.; Nam, R.H.; Seok, Y.-J.; Kim, Y.-R.; Lee, D.H.; Jung, H.C. Comparison of Gastric Microbiota Between Gastric Juice and Mucosa by Next Generation Sequencing Method. J. Cancer Prev. 2016, 21, 60–65. [Google Scholar] [CrossRef]
- Dong, Q.; Xin, Y.; Wang, L.; Meng, X.; Yu, X.; Lu, L.; Xuan, S. Characterization of Gastric Microbiota in Twins. Curr. Microbiol. 2017, 74, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhou, X.; Liu, X.; Ling, Z.; Ji, F. Role of the Gastric Microbiome in Gastric Cancer: From Carcinogenesis to Treatment. Front. Microbiol. 2021, 12, 641322. [Google Scholar] [CrossRef] [PubMed]
- Ndegwa, N.; Ploner, A.; Andersson, A.F.; Zagai, U.; Andreasson, A.; Vieth, M.; Talley, N.J.; Agreus, L.; Ye, W. Gastric Microbiota in a Low–Helicobacter pylori Prevalence General Population and Their Associations With Gastric Lesions. Clin. Transl. Gastroenterol. 2020, 11, e00191. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, R.; Chen, S.; Sun, B.; Zhang, K. Analysis of gastric microbiome reveals three distinctive microbial communities associated with the occurrence of gastric cancer. BMC Microbiol. 2022, 22, 184. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, L.; Li, Z.; Liu, Y.; Xiao, B.; Ding, Y.; Wen, H. Differences in gastric microbiota and mucosal function between patients with chronic superficial gastritis and intestinal metaplasia. Front. Microbiol. 2022, 13, 950325. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-L.; Pang, W.; Huang, Y.; Zhang, Y.; Zhang, C.-J. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Front. Cell Infect. Microbiol. 2018, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, J. Benchmarking differential abundance analysis methods for correlated microbiome sequencing data. Briefings Bioinform. 2023, 24, bbac607. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, Y.; Zheng, X.; Li, F.; Bo, X. RevEcoR: An R package for the reverse ecology analysis of microbiomes. BMC Bioinform. 2016, 17, 294. [Google Scholar] [CrossRef]
- Valour, F.; Sénéchal, A.; Dupieux, C.; Karsenty, J.; Lustig, S.; Breton, P.; Gleizal, A.; Boussel, L.; Laurent, F.; Braun, E.; et al. Actinomycosis: Etiology, clinical features, diagnosis, treatment, and management. Infect. Drug Resist. 2014, 7, 183–197. [Google Scholar] [CrossRef]
- Teixeira, C.G.; Fusieger, A.; Milião, G.L.; Martins, E.; Drider, D.; Nero, L.A.; de Carvalho, A.F. Weissella: An Emerging Bacterium with Promising Health Benefits. Probiotics Antimicrob. Proteins 2021, 13, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Karunakaran, G.; Yang, Y.; Tremblay, V.; Ning, Z.; Martin, J.; Belaouad, A.; Figeys, D.; Brunzelle, J.S.; Giguere, P.M.; Stintzi, A.; et al. Structural analysis of Atopobium parvulum SufS cysteine desulfurase linked to Crohn′s disease. FEBS Lett. 2022, 596, 898–909. [Google Scholar] [CrossRef]
- Yang, Y.; Jobin, C. Novel insights into microbiome in colitis and colorectal cancer. Curr. Opin. Gastroenterol. 2017, 33, 422. [Google Scholar] [CrossRef] [PubMed]
- Molina-Santiago, C.; Vela-Corcía, D.; Petras, D.; Díaz-Martínez, L.; Pérez-Lorente, A.I.; Sopeña-Torres, S.; Pearson, J.; Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; de Vicente, A.; et al. Chemical interplay and complementary adaptative strategies toggle bacterial antagonism and co-existence. Cell Rep. 2021, 36, 109449. [Google Scholar] [CrossRef]
- Berthelot, J.-M.; Corvec, S.; Hayem, G. SAPHO, autophagy, IL-1, FoxO1, and Propionibacterium ( Cutibacterium ) acnes. Jt. Bone Spine 2018, 85, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Mikami, A.; Ogita, T.; Namai, F.; Shigemori, S.; Sato, T.; Shimosato, T. Oral administration of Flavonifractor plautii attenuates inflammatory responses in obese adipose tissue. Mol. Biol. Rep. 2020, 47, 6717–6725. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.K.; Kazmierczak, B.I. Inflammation: A Double-Edged Sword in the Response to Pseudomonas aeruginosa Infection. J. Innate Immun. 2017, 9, 250–261. [Google Scholar] [CrossRef]
- Monnoyer, R.; Eftedal, I.; Hjelde, A.; Deb, S.; Haugum, K.; Lautridou, J. Functional Profiling Reveals Altered Metabolic Activity in Divers’ Oral Microbiota During Commercial Heliox Saturation Diving. Front. Physiol. 2021, 12, 702634. [Google Scholar] [CrossRef]
- Ortiz, J.P.M.; Read, M.N.; McClure, D.D.; Holmes, A.; Dehghani, F.; Shanahan, E.R. High throughput genome scale modeling predicts microbial vitamin requirements contribute to gut microbiome community structure. Gut Microbes 2022, 14, 2118831. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Zadeh, M.; Mohamadzadeh, M. Vitamin B12 coordinates ileal epithelial cell and microbiota functions to resist Salmonella infection in mice. J. Exp. Med. 2022, 219, e20220057. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Tang, Y.; Tang, W.; Chen, Y. Heptose-containing bacterial natural products: Structures, bioactivities, and biosyntheses. Nat. Prod. Rep. 2021, 38, 1887–1909. [Google Scholar] [CrossRef] [PubMed]
- Klein, V.J.; Irla, M.; Gil López, M.; Brautaset, T.; Brito, L.F. Unravelling Formaldehyde Metabolism in Bacteria: Road towards Synthetic Methylotrophy. Microorganisms 2022, 10, 220. [Google Scholar] [CrossRef]
- Delépine, B.; Gil López, M.; Carnicer, M.; Vicente, C.M.; Wendisch, V.F.; Heux, S. Charting the Metabolic Landscape of the Facultative Methylotroph Bacillus methanolicus. mSystems 2020, 5, e00745-20. [Google Scholar] [CrossRef] [PubMed]
- Gamage, S.M.; Lee, K.T.; Dissabandara, D.L.O.; Lam, A.K.-Y.; Gopalan, V. Dual role of heme iron in cancer; promotor of carcinogenesis and an inducer of tumour suppression. Exp. Mol. Pathol. 2021, 120, 104642. [Google Scholar] [CrossRef]
- Constante, M.; Fragoso, G.; Calvé, A.; Samba-Mondonga, M.; Santos, M.M. Dietary Heme Induces Gut Dysbiosis, Aggravates Colitis, and Potentiates the Development of Adenomas in Mice. Front. Microbiol. 2017, 8, 1809. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Wang, R.; Huang, H.; Hou, Y.; Wang, X.; He, W.; Gong, X.; Hu, J. Construction of double network hydrogels using agarose and gallic acid with antibacterial and anti-inflammatory properties for wound healing. Int. J. Biol. Macromol. 2023, 227, 698–710. [Google Scholar] [CrossRef] [PubMed]
- Rugge, M.; Meggio, A.; Pennelli, G.; Piscioli, F.; Giacomelli, L.; De Pretis, G.; Graham, D.Y. Gastritis staging in clinical practice: The OLGA staging system. Gut 2007, 56, 631–636. [Google Scholar] [CrossRef]
- Wurm, P.; Dörner, E.; Kremer, C.; Spranger, J.; Maddox, C.; Halwachs, B.; Harrison, U.; Blanchard, T.; Haas, R.; Högenauer, C.; et al. Qualitative and Quantitative DNA- and RNA-Based Analysis of the Bacterial Stomach Microbiota in Humans, Mice, and Gerbils. mSystems 2018, 3, e00262-18. [Google Scholar] [CrossRef]
- Mailhe, M.; Ricaboni, D.; Vitton, V.; Gonzalez, J.-M.; Bachar, D.; Dubourg, G.; Cadoret, F.; Robert, C.; Delerce, J.; Levasseur, A.; et al. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol. 2018, 18, 157. [Google Scholar] [CrossRef]
- Kavitake, D.; Devi, P.B.; Shetty, P.H. Overview of exopolysaccharides produced by Weissella genus—A review. Int. J. Biol. Macromol. 2020, 164, 2964–2973. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Qin, S.; Shen, Y. Probiotic potential of Weissella strains isolated from horse feces. Microb. Pathog. 2019, 132, 117–123. [Google Scholar] [CrossRef] [PubMed]

| Reference | Dataset Geographic Regions (Data Source) | Sequence Accession Codes | Samples Used in This Study | Age (Average ± s.d.) | Sex F(%)/M(%) |
|---|---|---|---|---|---|
| Wang et al., 2016 [12] | Qingdao (China) | SRP060550 | SG (6) | 55.8 ± 13.5 | 39.7/60.3 |
| Eun et al., 2014 [13] | Hanyang (Korea) | SRP038955 | SG (10) | 50.4 ± 11.5 | 50/50 |
| Zhang et al., 2021 [14] | Nanjing (China) | PRJNA634837 | SG (17) | 56 ± 10.6 | 47.1/52.9 |
| Seekatz et al., 2019 [15] | Michigan (USA) | PRJNA495320 | HC (49) | 37.7 ± 10.2 | 34.7/65.3 |
| Ferreira et al., 2017 [3] | Porto (Portugal) | PRJNA413125 | AG (81) | 43.6 ± 7 | 2.5/97.5 |
| Park et al., 2019 [16] | Hanyang (Korea) | PRJNA389357 | SG (62) | 32.1 ± 10.5 | 58.1/41.9 |
| Coker et al., 2018 [10] | Inner Mongolia (China) | PRJNA375772 | SG (52) | 52.8 ± 15 | 50/50 |
| AG (62) | 56 ± 12.3 | 50/50 | |||
| Coker et al., 2018 [10] | Xian (China) | PRJNA375772 | SG (20) | 47.1 ± 11.7 | 50/50 |
| AG (16) | 50.4 ± 11.7 | 56.2/43.8 | |||
| Filardo et al., 2022 [17] | Rome (Italy) | PRJNA795512 | SG (18) | 50.4 ± 11 | 77.8/22.2 |
| Bassis et al., 2015 [18] | Michigan (USA) | PRJNA263948 | HC (34) | 41.3 ± 16.2 | 67.6/32.4 |
| Wang et al., 2020 [19] | Beijing (China) | PRJEB26931 | HC (51) | 44.8 ± 13.6 | 53/47 |
| Yang et al., 2016 [20] | Tumaco (Columbia) | PRJEB11763 | SG (12) | 49 ± 6.7 | 66.7/33.3 |
| AG (6) | 46.7 ± 2.1 | 66.7/33.3 | |||
| Yang et al., 2016 [20] | Túquerres (Columbia) | PRJEB11763 | SG (10) | 47.1 ± 4.4 | 60/40 |
| AG (8) | 48.4 ± 7.5 | 75/25 | |||
| Sung et al., 2016 [21] | Seoul (Korea) | GSE61493 | HC (31) | 58 ± 11.6 | 44.9/55.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Yin, P.; Ren, Y.; Yu, L.; Tian, F.; Zhao, J.; Chen, W.; Xue, Y.; Zhai, Q. Predicting Personalized Diets Based on Microbial Characteristics between Patients with Superficial Gastritis and Atrophic Gastritis. Nutrients 2023, 15, 4738. https://doi.org/10.3390/nu15224738
Gao X, Yin P, Ren Y, Yu L, Tian F, Zhao J, Chen W, Xue Y, Zhai Q. Predicting Personalized Diets Based on Microbial Characteristics between Patients with Superficial Gastritis and Atrophic Gastritis. Nutrients. 2023; 15(22):4738. https://doi.org/10.3390/nu15224738
Chicago/Turabian StyleGao, Xiaoxiang, Pingping Yin, Yilin Ren, Leilei Yu, Fengwei Tian, Jianxin Zhao, Wei Chen, Yuzheng Xue, and Qixiao Zhai. 2023. "Predicting Personalized Diets Based on Microbial Characteristics between Patients with Superficial Gastritis and Atrophic Gastritis" Nutrients 15, no. 22: 4738. https://doi.org/10.3390/nu15224738
APA StyleGao, X., Yin, P., Ren, Y., Yu, L., Tian, F., Zhao, J., Chen, W., Xue, Y., & Zhai, Q. (2023). Predicting Personalized Diets Based on Microbial Characteristics between Patients with Superficial Gastritis and Atrophic Gastritis. Nutrients, 15(22), 4738. https://doi.org/10.3390/nu15224738





