Gut Microbiota Composition in Patients with Neurodegenerative Disorders (Parkinson’s and Alzheimer’s) and Healthy Controls: A Systematic Review
Abstract
:1. Introduction
2. Methodology
2.1. Searching Strategy
2.2. Selection Criteria and Screening
2.3. Data Extraction
2.4. Risk of Bias Assessment
2.5. Evaluation of Findings
3. Results
3.1. Study Selection
3.2. Risk of Bias Assessment
3.3. General Characteristics of Included Studies
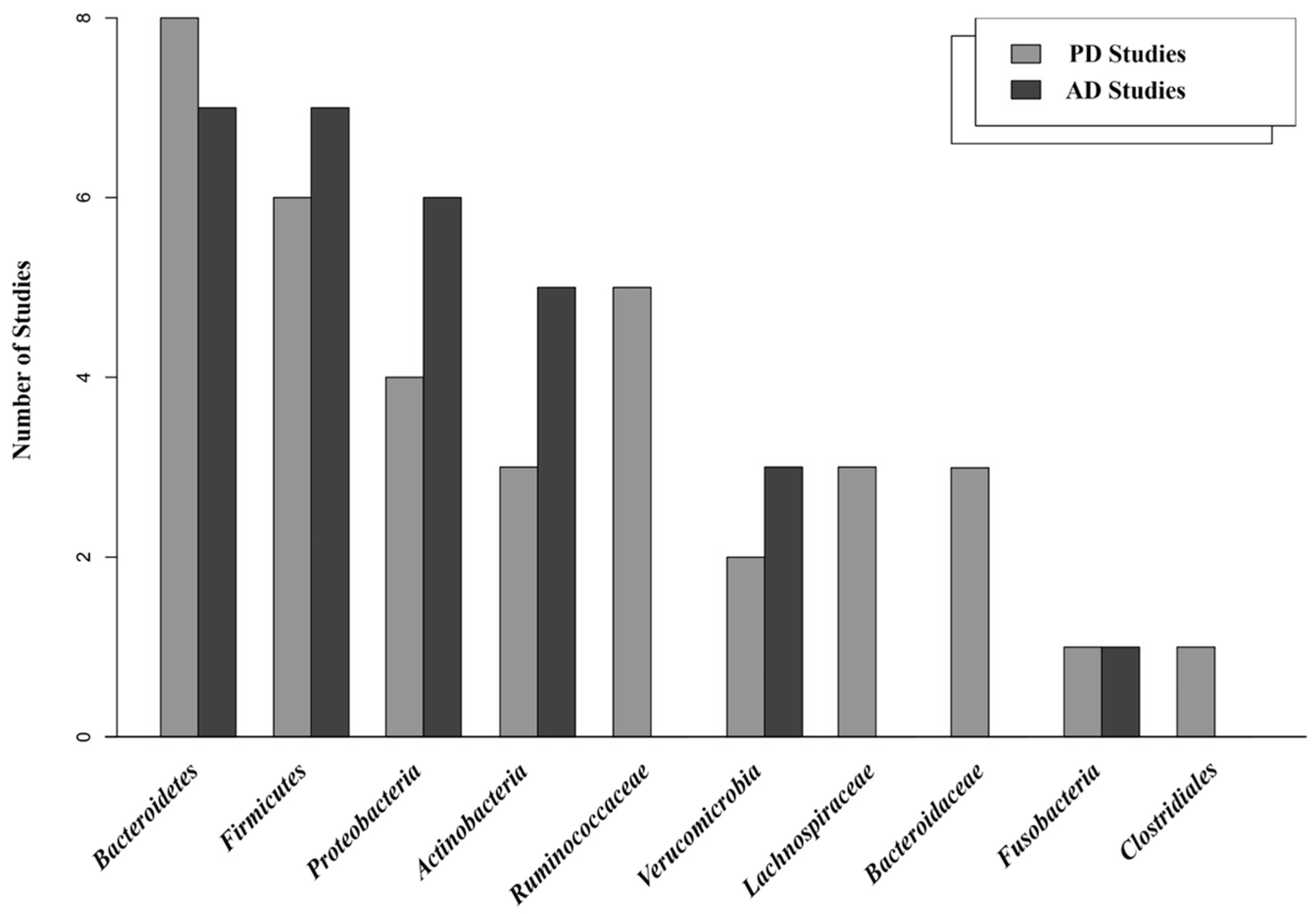
3.4. Gut Microbiota Comparison between Cases and Controls
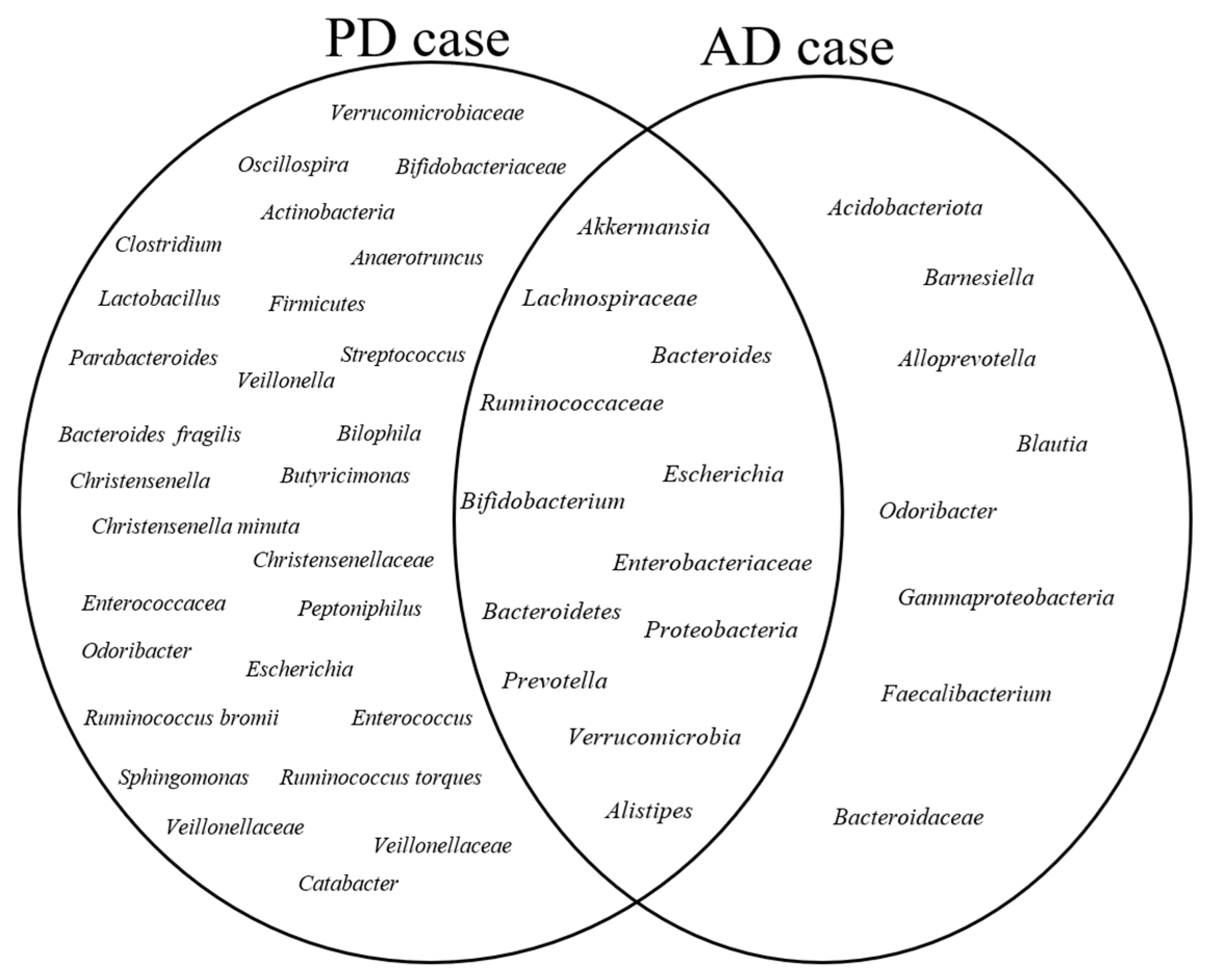
3.5. Core Gut Microbiota
3.6. Microbial Communities in the Healthy Group
3.7. Microbial Communities in the Case Group
3.8. Bacterial Diversity in PD and AD Studies (Alpha and Beta Diversity)
3.9. Gut Microbiota Associated with PD and AD (Differential Abundance Analysis (DAA))
4. Discussion
4.1. Limitation Described in Included Studies as Claimed by the Authors
- Lack of covariate consideration: Important covariates such as diet, exercise, smoking, drug treatment, and comorbidities were not adequately addressed in some studies (13 studies).
- Lack of longitudinal data: Several studies lacked longer follow-up periods to capture the microbial community changes during disease progression. Also, the cross-sectional design of many the studies limited their ability to establish causal relationships between the gut microbiota and neurodegenerative disorders (nine studies).
- Small sample size: Several studies reported small sample sizes, which may have limited the statistical power and generalizability of the findings (six studies).
- Lack of host–microbiome interaction consideration: The studies often did not consider the interactions between the host metabolism and the gut microbiota, which could provide a more comprehensive understanding of the mechanisms underlying neurodegenerative disorders (six studies).
- Lack of species/strain resolution: The use of the 16S rRNA sequencing method limited the ability to analyze microbial composition at the species or strain level, which is crucial for identifying the specific microorganisms associated with the diseases (five studies).
- Lack of mucosal microbiota analysis: Although nearly all of the included studies only utilized stool samples, only two studies acknowledged the need for the analysis of mucosal microbiota composition using gastrointestinal biopsies. Such analysis provides a deeper understanding of the local host–microbiota interaction (two studies).
4.2. Strength and Limitation of This Systematic Review
4.3. Future Recommendations
- 1:
- Considering important covariates such as diet, exercise, smoking, comorbidities, and drug treatment and their potential influence on the gut microbiota as a fundamental step in the study design would provide a more comprehensive insight into the role of gut microbiota in AD and PD diseases.
- 2:
- The design of longitudinal studies with longer follow-up periods to capture microbial community changes during disease progression is needed. Also, well-designed intervention studies, such as probiotic or prebiotic trials, can help to determine the therapeutic potential of modulating the gut microbiota in relation to disease symptoms and progression.
- 3:
- Large-scale cohort studies involving diverse populations with larger sample sizes and frequent sampling to capture variations in gut microbiota composition associated with different backgrounds, geographical locations, and lifestyles are essential. This will help to identify potential factors influencing gut microbiota and allow for personalized approaches to managing diseases using gut microbiota markers. Furthermore, considering the heterogeneity among ethnic groups, which is reflected in the wide variation in microbiota, categorizing research studies according to their geographical location and subsequently comparing outcomes between regions could provide a valuable basis for a deeper understanding of gut microbiota profile in diverse human populations.
- 4:
- Investigating the functional analysis of gut microbiota by exploring metabolomic and metagenomic approaches can provide insights into specific mechanisms underlying disease pathogenesis. Also, utilizing advanced sequencing techniques such as shotgun metagenomics allows species- and strain-level resolution, and other omics approaches, such as metratranscriptmics and metabolomics, allow the understanding of the mechanistic insight into host–microbiota interactions.
- 5:
- Additionally, incorporating sigmoid mucosal biopsies and detailed characterization of microbial functions would enhance the understanding of host–microbiota interactions.
- 6:
- Establishing standardized microbiota protocols from sample collection to data analysis to enhance the reliability and comparability of microbiota findings would lead to a better understanding of the dynamic relationship between the host and the gut microbiota [84].
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dorsey, E.; Sherer, T.; Okun, M.S.; Bloem, B.R. The emerging evidence of the Parkinson pandemic. J. Park. Dis. 2018, 8, S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Monica Moore, M.; Díaz-Santos, M.; Vossel, K. Alzheimer’s association 2021 facts and figures report. Alzheimer’s Assoc. 2021, 17. [Google Scholar]
- Nichols, E.; Steinmetz, J.D.; Vollset, S.E.; Fukutaki, K.; Chalek, J.; Abd-Allah, F.; Abdoli, A.; Abualhasan, A.; Abu-Gharbieh, E.; Akram, T.T.; et al. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: An analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105-25. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef]
- Tysnes, O.-B.; Storstein, A. Epidemiology of Parkinson’s disease. J. Neural Transm. 2017, 124, 901–905. [Google Scholar] [CrossRef]
- Ballatore, C.; Lee, V.M.-Y.; Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 2007, 8, 663–672. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Mahony, S.M. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef]
- Appleton, J. The gut-brain axis: Influence of microbiota on mood and mental health. Integr. Med. A Clin. J. 2018, 17, 28. [Google Scholar]
- Abd Mutalib, N.; Syed Mohamad, S.A.; Jusril, N.A.; Hasbullah, N.I.; Mohd Amin, M.C.I.; Ismail, N.H. Lactic Acid Bacteria (LAB) and Neuroprotection, What Is New? An Up-To-Date Systematic Review. Pharmaceuticals 2023, 16, 712. [Google Scholar] [CrossRef]
- Ghezzi, L.; Cantoni, C.; Rotondo, E.; Galimberti, D. The Gut Microbiome–Brain Crosstalk in Neurodegenerative Diseases. Biomedicines 2022, 10, 1486. [Google Scholar] [CrossRef]
- Sharon, G.; Sampson, T.R.; Geschwind, D.H.; Mazmanian, S.K. The central nervous system and the gut microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.M.; Spiegel, J.; Dillmann, K.-U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.-H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Gorecki, A.M.; Preskey, L.; Bakeberg, M.C.; Kenna, J.E.; Gildenhuys, C.; MacDougall, G.; Dunlop, S.A.; Mastaglia, F.L.; Akkari, P.A.; Koengten, F. Altered gut microbiome in Parkinson’s disease and the influence of lipopolysaccharide in a human α-synuclein over-expressing mouse model. Front. Neurosci. 2019, 13, 839. [Google Scholar] [CrossRef]
- Haran, J.P.; Bhattarai, S.K.; Foley, S.E.; Dutta, P.; Ward, D.V.; Bucci, V.; McCormick, B.A. Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-glycoprotein pathway. mBio 2019, 10, e00632-19. [Google Scholar] [CrossRef]
- Ubeda, C.; Vázquez-Carretero, M.D.; Luque-Tirado, A.; Ríos-Reina, R.; Rubio-Sánchez, R.; Franco-Macías, E.; García-Miranda, P.; Calonge, M.L.; Peral, M.J. Fecal Volatile Organic Compounds and Microbiota Associated with the Progression of Cognitive Impairment in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 24, 707. [Google Scholar]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017, 9, 1–13. [Google Scholar]
- Heintz-Buschart, A.; Pandey, U.; Wicke, T.; Sixel-Döring, F.; Janzen, A.; Sittig-Wiegand, E.; Trenkwalder, C.; Oertel, W.H.; Mollenhauer, B.; Wilmes, P. The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 2018, 33, 88–98. [Google Scholar] [CrossRef]
- Li, F.; Wang, P.; Chen, Z.; Sui, X.; Xie, X.; Zhang, J. Alteration of the fecal microbiota in North-Eastern Han Chinese population with sporadic Parkinson’s disease. Neurosci. Lett. 2019, 707, 134297. [Google Scholar] [CrossRef] [PubMed]
- Cosma-Grigorov, A.; Meixner, H.; Mrochen, A.; Wirtz, S.; Winkler, J.; Marxreiter, F. Changes in gastrointestinal microbiome composition in PD: A pivotal role of covariates. Front. Neurol. 2020, 11, 1041. [Google Scholar] [CrossRef] [PubMed]
- Cilia, R.; Barichella, M.; Severgnini, M.; Cassani, E.; Bolliri, C.; Caronni, S.; Ferri, V.; Cancello, R.; Faierman, S.; Pinelli, G. Unraveling gut microbiota in Parkinson’s disease and atypical parkinsonism. In Proceedings of the Movement Disorders; WILEY: Hoboken, NJ, USA, 2018; p. 1987. [Google Scholar]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Cirstea, M.S.; Kliger, D.; MacLellan, A.D.; Yu, A.C.; Langlois, J.; Fan, M.; Boroomand, S.; Kharazyan, F.; Hsiung, R.G.; MacVicar, B.A. The oral and fecal microbiota in a Canadian cohort of Alzheimer’s disease. J. Alzheimer’s Dis. 2022, 87, 247–258. [Google Scholar] [CrossRef]
- Lin, A.; Zheng, W.; He, Y.; Tang, W.; Wei, X.; He, R.; Huang, W.; Su, Y.; Huang, Y.; Zhou, H. Gut microbiota in patients with Parkinson’s disease in southern China. Park. Relat. Disord. 2018, 53, 82–88. [Google Scholar] [CrossRef]
- Liu, P.; Wu, L.; Peng, G.; Han, Y.; Tang, R.; Ge, J.; Zhang, L.; Jia, L.; Yue, S.; Zhou, K. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav. Immun. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Kleine Bardenhorst, S.; Cereda, E.; Severgnini, M.; Barichella, M.; Pezzoli, G.; Keshavarzian, A.; Desideri, A.; Pietrucci, D.; Aho, V.T.; Scheperjans, F. Gut microbiota dysbiosis in Parkinson disease: A systematic review and pooled analysis. Eur. J. Neurol. 2023, 30, 3581–3594. [Google Scholar] [CrossRef]
- Angoorani, P.; Ejtahed, H.-S.; Siadat, S.D.; Sharifi, F.; Larijani, B. Is there any link between cognitive impairment and gut microbiota? A systematic review. Gerontology 2022, 68, 1201–1213. [Google Scholar] [CrossRef]
- Hung, C.-C.; Chang, C.-C.; Huang, C.-W.; Nouchi, R.; Cheng, C.-H. Gut microbiota in patients with Alzheimer’s disease spectrum: A systematic review and meta-analysis. Aging 2022, 14, 477. [Google Scholar] [CrossRef]
- Solfrizzi, V.; Custodero, C.; Lozupone, M.; Imbimbo, B.P.; Valiani, V.; Agosti, P.; Schilardi, A.; D’Introno, A.; La Montagna, M.; Calvani, M. Relationships of dietary patterns, foods, and micro-and macronutrients with Alzheimer’s disease and late-life cognitive disorders: A systematic review. J. Alzheimer’s Dis. 2017, 59, 815–849. [Google Scholar] [CrossRef]
- Trichopoulou, A.; Kyrozis, A.; Rossi, M.; Katsoulis, M.; Trichopoulos, D.; La Vecchia, C.; Lagiou, P. Mediterranean diet and cognitive decline over time in an elderly Mediterranean population. Eur. J. Nutr. 2015, 54, 1311–1321. [Google Scholar] [CrossRef]
- Fiala, M.; Kooij, G.; Wagner, K.; Hammock, B.; Pellegrini, M. Modulation of innate immunity of patients with Alzheimer’s disease by omega-3 fatty acids. FASEB J. 2017, 31, 3229. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Tangney, C.C.; Wang, Y.; Sacks, F.M.; Barnes, L.L.; Bennett, D.A.; Aggarwal, N.T. MIND diet slows cognitive decline with aging. Alzheimer’s Dement. 2015, 11, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, L.A.; Higgins, J.P. Risk-of-bias VISualization (robvis): An R package and Shiny web app for visualizing risk-of-bias assessments. Res. Synth. Methods 2021, 12, 55–61. [Google Scholar] [CrossRef]
- Aho, V.T.; Pereira, P.A.; Voutilainen, S.; Paulin, L.; Pekkonen, E.; Auvinen, P.; Scheperjans, F. Gut microbiota in Parkinson’s disease: Temporal stability and relations to disease progression. EBioMedicine 2019, 44, 691–707. [Google Scholar] [CrossRef]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 1–21. [Google Scholar] [CrossRef]
- Chen, L.; Xu, X.; Wu, X.; Cao, H.; Li, X.; Hou, Z.; Wang, B.; Liu, J.; Ji, X.; Zhang, P. A comparison of the composition and functions of the oral and gut microbiotas in Alzheimer’s patients. Front. Cell. Infect. Microbiol. 2022, 12, 942460. [Google Scholar] [CrossRef]
- Hopfner, F.; Künstner, A.; Müller, S.H.; Künzel, S.; Zeuner, K.E.; Margraf, N.G.; Deuschl, G.; Baines, J.F.; Kuhlenbäumer, G. Gut microbiota in Parkinson disease in a northern German cohort. Brain Res. 2017, 1667, 41–45. [Google Scholar] [CrossRef]
- Sheng, C.; Lin, L.; Lin, H.; Wang, X.; Han, Y.; Liu, S.-L. Altered gut microbiota in adults with subjective cognitive decline: The SILCODE study. J. Alzheimer’s Dis. 2021, 82, 513–526. [Google Scholar]
- Weis, S.; Schwiertz, A.; Unger, M.M.; Becker, A.; Faßbender, K.; Ratering, S.; Kohl, M.; Schnell, S.; Schäfer, K.-H.; Egert, M. Effect of Parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. NPJ Park. Dis. 2019, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Cirstea, M.S.; Yu, A.C.; Golz, E.; Sundvick, K.; Kliger, D.; Radisavljevic, N.; Foulger, L.H.; Mackenzie, M.; Huan, T.; Finlay, B.B. Microbiota composition and metabolism are associated with gut function in Parkinson’s disease. Mov. Disord. 2020, 35, 1208–1217. [Google Scholar] [CrossRef]
- Guo, M.; Peng, J.; Huang, X.; Xiao, L.; Huang, F.; Zuo, Z. Gut microbiome features of Chinese patients newly diagnosed with Alzheimer’s disease or mild cognitive impairment. J. Alzheimer’s Dis. 2021, 80, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Kaiyrlykyzy, A.; Kozhakhmetov, S.; Babenko, D.; Zholdasbekova, G.; Alzhanova, D.; Olzhayev, F.; Baibulatova, A.; Kushugulova, A.R.; Askarova, S. Study of gut microbiota alterations in Alzheimer’s dementia patients from Kazakhstan. Sci. Rep. 2022, 12, 15115. [Google Scholar] [CrossRef]
- Li, C.; Cui, L.; Yang, Y.; Miao, J.; Zhao, X.; Zhang, J.; Cui, G.; Zhang, Y. Gut microbiota differs between Parkinson’s disease patients and healthy controls in Northeast China. Front. Mol. Neurosci. 2019, 12, 171. [Google Scholar] [CrossRef]
- Li, W.; Wu, X.; Hu, X.; Wang, T.; Liang, S.; Duan, Y.; Jin, F.; Qin, B. Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Sci. China Life Sci. 2017, 60, 1223–1233. [Google Scholar] [CrossRef]
- Lin, C.-H.; Chen, C.-C.; Chiang, H.-L.; Liou, J.-M.; Chang, C.-M.; Lu, T.-P.; Chuang, E.Y.; Tai, Y.-C.; Cheng, C.; Lin, H.-Y. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflamm. 2019, 16, 1–9. [Google Scholar] [CrossRef]
- Petrov, V.; Saltykova, I.; Zhukova, I.; Alifirova, V.; Zhukova, N.; Dorofeeva, Y.B.; Tyakht, A.; Kovarsky, B.; Alekseev, D.; Kostryukova, E. Analysis of gut microbiota in patients with Parkinson’s disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef]
- Pietrucci, D.; Cerroni, R.; Unida, V.; Farcomeni, A.; Pierantozzi, M.; Mercuri, N.B.; Biocca, S.; Stefani, A.; Desideri, A. Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Park. Relat. Disord. 2019, 65, 124–130. [Google Scholar] [CrossRef]
- Qian, Y.; Yang, X.; Xu, S.; Huang, P.; Li, B.; Du, J.; He, Y.; Su, B.; Xu, L.-M.; Wang, L. Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain 2020, 143, 2474–2489. [Google Scholar] [CrossRef]
- Qian, Y.; Yang, X.; Xu, S.; Wu, C.; Song, Y.; Qin, N.; Chen, S.-D.; Xiao, Q. Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 2018, 70, 194–202. [Google Scholar] [CrossRef]
- Stadlbauer, V.; Engertsberger, L.; Komarova, I.; Feldbacher, N.; Leber, B.; Pichler, G.; Fink, N.; Scarpatetti, M.; Schippinger, W.; Schmidt, R. Dysbiosis, gut barrier dysfunction and inflammation in dementia: A pilot study. BMC Geriatr. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.H.; Chong, C.W.; Lim, S.Y.; Yap, I.K.S.; Teh, C.S.J.; Loke, M.F.; Song, S.L.; Tan, J.Y.; Ang, B.H.; Tan, Y.Q. Gut microbial ecosystem in Parkinson disease: New clinicobiological insights from multi-omics. Ann. Neurol. 2021, 89, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V. Gut microbiota and metabolome alterations associated with Parkinson’s disease. Msystems 2020, 5, e00561-20. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [PubMed]
- Wanapaisan, P.; Chuansangeam, M.; Nopnipa, S.; Mathuranyanon, R.; Nonthabenjawan, N.; Ngamsombat, C.; Thientunyakit, T.; Muangpaisan, W. Association between Gut Microbiota with Mild Cognitive Impairment and Alzheimer’s Disease in a Thai Population. Neurodegener. Dis. 2023, 22, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Ding, D.; Zhu, H.; Wang, R.; Su, F.; Wu, W.; Xiao, Z.; Liang, X.; Zhao, Q.; Hong, Z. Disturbed microbial ecology in Alzheimer’s disease: Evidence from the gut microbiota and fecal metabolome. BMC Microbiol. 2021, 21, 1–13. [Google Scholar] [CrossRef]
- Yan, Z.; Yang, F.; Cao, J.; Ding, W.; Yan, S.; Shi, W.; Wen, S.; Yao, L. Alterations of gut microbiota and metabolome with Parkinson’s disease. Microb. Pathog. 2021, 160, 105187. [Google Scholar] [CrossRef] [PubMed]
- Yıldırım, S.; Nalbantoğlu, Ö.U.; Bayraktar, A.; Ercan, F.B.; Gündoğdu, A.; Velioğlu, H.A.; Göl, M.F.; Soylu, A.E.; Koç, F.; Gülpınar, E.A. Stratification of the gut microbiota composition landscape across the alzheimer’s disease continuum in a Turkish cohort. Msystems 2022, 7, e00004–e00022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yue, L.; Fang, X.; Wang, G.; Li, C.; Sun, X.; Jia, X.; Yang, J.; Song, J.; Zhang, Y. Altered gut microbiota in Parkinson’s disease patients/healthy spouses and its association with clinical features. Park. Relat. Disord. 2020, 81, 84–88. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Quan, M.; Zhao, H.; Jia, J. Gut microbiota changes and their correlation with cognitive and neuropsychiatric symptoms in Alzheimer’s disease. J. Alzheimer’s Dis. 2021, 81, 583–595. [Google Scholar] [CrossRef]
- Zhu, Z.; Ma, X.; Wu, J.; Xiao, Z.; Wu, W.; Ding, S.; Zheng, L.; Liang, X.; Luo, J.; Ding, D. Altered Gut Microbiota and Its Clinical Relevance in Mild Cognitive Impairment and Alzheimer’s Disease: Shanghai Aging Study and Shanghai Memory Study. Nutrients 2022, 14, 3959. [Google Scholar] [CrossRef]
- Zhuang, Z.-Q.; Shen, L.-L.; Li, W.-W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.-L. Gut microbiota is altered in patients with Alzheimer’s disease. J. Alzheimer’s Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef]
- Ren, T.; Gao, Y.; Qiu, Y.; Jiang, S.; Zhang, Q.; Zhang, J.; Wang, L.; Zhang, Y.; Wang, L.; Nie, K. Gut microbiota altered in mild cognitive impairment compared with normal cognition in sporadic Parkinson’s disease. Front. Neurol. 2020, 11, 137. [Google Scholar] [CrossRef]
- Borsom, E.M.; Conn, K.; Keefe, C.R.; Herman, C.; Orsini, G.M.; Hirsch, A.H.; Palma Avila, M.; Testo, G.; Jaramillo, S.A.; Bolyen, E. Predicting neurodegenerative disease using Prepathology gut microbiota composition: A longitudinal study in mice modeling Alzheimer’s disease pathologies. Microbiol. Spectr. 2023, 11, e03458-22. [Google Scholar] [CrossRef]
- Ling, Z.; Zhu, M.; Yan, X.; Cheng, Y.; Shao, L.; Liu, X.; Jiang, R.; Wu, S. Structural and functional dysbiosis of fecal microbiota in Chinese patients with Alzheimer’s disease. Front. Cell Dev. Biol. 2021, 8, 634069. [Google Scholar] [CrossRef]
- Lei, W.; Cheng, Y.; Gao, J.; Liu, X.; Shao, L.; Kong, Q.; Zheng, N.; Ling, Z.; Hu, W. Akkermansia muciniphila in neuropsychiatric disorders: Friend or foe? Front. Cell. Infect. Microbiol. 2023, 13, 1224155. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Cattaneo, A.; Cattane, N.; Galluzzi, S.; Provasi, S.; Lopizzo, N.; Festari, C.; Ferrari, C.; Guerra, U.P.; Paghera, B.; Muscio, C. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 2017, 49, 60–68. [Google Scholar] [CrossRef]
- Minter, M.R.; Zhang, C.; Leone, V.; Ringus, D.L.; Zhang, X.; Oyler-Castrillo, P.; Musch, M.W.; Liao, F.; Ward, J.F.; Holtzman, D.M. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 2016, 6, 30028. [Google Scholar] [CrossRef]
- Laudani, S.; Torrisi, S.A.; Alboni, S.; Bastiaanssen, T.F.; Benatti, C.; Rivi, V.; Moloney, R.D.; Fuochi, V.; Furneri, P.M.; Drago, F. Gut microbiota alterations promote traumatic stress susceptibility associated with p-cresol-induced dopaminergic dysfunctions. Brain Behav. Immun. 2023, 107, 385–396. [Google Scholar] [CrossRef]
- Hamamah, S.; Aghazarian, A.; Nazaryan, A.; Hajnal, A.; Covasa, M. Role of microbiota-gut-brain axis in regulating dopaminergic signaling. Biomedicines 2022, 10, 436. [Google Scholar] [CrossRef]
- Ojha, S.; Patil, N.; Jain, M.; Kole, C.; Kaushik, P. Probiotics for Neurodegenerative Diseases: A Systemic Review. Microorganisms 2023, 11, 1083. [Google Scholar] [CrossRef]
- Bisaglia, M. Mediterranean Diet and Parkinson’s Disease. Int. J. Mol. Sci. 2022, 24, 42. [Google Scholar] [CrossRef]
- Baert, F.; Matthys, C.; Maselyne, J.; Van Poucke, C.; Van Coillie, E.; Bergmans, B.; Vlaemynck, G. Parkinson’s disease patients’ short chain fatty acids production capacity after in vitro fecal fiber fermentation. NPJ Park. Dis. 2021, 7, 72. [Google Scholar] [CrossRef]
- Vervier, K.; Moss, S.; Kumar, N.; Adoum, A.; Barne, M.; Browne, H.; Kaser, A.; Kiely, C.J.; Neville, B.A.; Powell, N. Two microbiota subtypes identified in irritable bowel syndrome with distinct responses to the low FODMAP diet. Gut 2022, 71, 1821–1830. [Google Scholar] [CrossRef]
- Minerbi, A.; Gonzalez, E.; Brereton, N.J.; Anjarkouchian, A.; Dewar, K.; Fitzcharles, M.-A.; Chevalier, S.; Shir, Y. Altered microbiome composition in individuals with fibromyalgia. Pain 2019, 160, 2589–2602. [Google Scholar] [CrossRef] [PubMed]
- Shade, A. Diversity is the question, not the answer. ISME J. 2017, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mirzayi, C.; Renson, A.; Genomic Standards Consortium; Massive Analysis and Quality Control Society; Cesare, F.; Susanna-Assunta, S.; Zohra, F.; Elsafoury, S.; Zohra, F.; Elsafoury, S.; et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nat. Med. 2021, 27, 1885–1892. [Google Scholar] [CrossRef] [PubMed]



| Study | Year | Location | Population Size | Age | Gender | Sample Type | Matching Factor | Sample Preservative | Extraction Method | Sequencing Platform | Sequenced Regions | Forward Primer | Reverse Primer | Data Availability | Main Microbiota Finding |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [38] | 2019 | Finland | 128 (64 case, 64 control) | case: 65.2 ± 5.52/control: 64.45 ± 6.9; (years, mean ±SD) | Female: case: 48.6%/control: 50.0% | stool | age, sex | DNA stabilizer PSP (Spin Stool DNA Plus Kit, STRATEC Molecular) | PSP Spin Stool DNA Plus Kit (STRATEC Molecular) | Illumina MiSeq | V3–V4 | 341F1–4 (5′ CCTACGGGNGGCWGCAG 3′) | 785R1–4 (5′ GACTACHVGGGTATCTAATCC 3′) | PRJEB27564 | Significant differences in gut microbiota between cases and controls. Disease progression did not influence gut microbiota. No difference in Firmicutes/Bacteroidetes ratio between cases and controls. |
| [39] | 2020 | Ireland | 309 (147 case, 162 control) | case: 69.3 ± 8.6/control: 63.3 ± 8.3; (mean ± SD) | Female: case: 31.5%/control: 35.8% | stool | age, sex | MNIgene.GUT® kit | Chemagic DNA blood protocol | Illumina MiSeq | V3–V4 | NR | NR | Available on request | Composition of the gut microbiome could potentially serve as a marker of disease severity in PD. Bilophila and Paraprevotella abundance were significantly associated with disease severity. |
| [23] | 2018 | Italy | 350 (237 cases, 113 control) | cases: 67.6 (9.7)/control: 65.9 (9.9); (y, mean (SD)) | cases: 115 (59.6)/control 47 (41.6): Male gender, n (%) | stool | age, nutritional status, geographical area | No preservative | QIAamp DNA Stool Mini Kit | Illumina MiSeq | V3–V4 | NR | NR | NR | Low Lachnospiraceae in cases. |
| [19] | 2017 | Germany | 59 (31 case, 28 control) | case: 64.8 ± 9.5/control 65.6 ± 10.4; (years, mean ± SD) | Male: all case and control | stool | age | NR | NR | Illumina Hiseq4000 | NR | NR | NR | ERP019674 | Case and controls had different gut microbiota composition, characterized by increased levels of Verrucomicrobiaceae (Akkermansia muciniphila) and unclassified Firmicutes, and decreased levels of Prevotellaceae (Prevotella copri) and Erysipelotrichaceae (Eubacterium biforme) in cases. |
| [44] | 2020 | Canada | 300 (197 case, 103 control) | case:66 (59–71)/control 66 (58–71); (years) | Female: case: 38.1%/control: 51.5% | feces | age | DNA stabilizer buffer | OMNIgeneGUT Kit | Illumina MiSeq | V4 | 515F | 806R | On publisher’s website | Increased abundance of Akkermansia and Bifidobacterium and decreased abundance of Faecalibacterium, Lachnospiraceae, and SCFA-producing bacteria in cases. |
| [22] | 2020 | Germany | 101 (71 case, 30 control) | case: 65.3 ± 10.2/controls: 64.3 ± 8.9; (years) | Female: case 45.7%/controls: 45.2% | feces | MS score, NMS constipation item, Wexner Constipation, coffee consumption | DNA stabilizing solution | PSP R Spin Stool DNA Plus Kit | Illumina MiSeq | V3–V4 | 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) | 806R (5′-GGACTAC HVGGGTWTCTAAT-3′) | Available on request | High abundance of Faecalibacterium, Ruminococcus, Clostridia, Lachnospiraceae, Oscillospira, Betaproteaobacteria, Burkholderiales, Alcaligenaceae, and Sutterella in cases compared to controls. |
| [20] | 2017 | Germany | 154 (76 case, 78 control) | case: 68.0 6 9.7/control: 68.4 6 6.7; (year) | Male: case 66%/control 59% | stool | diabetes medication | Stool specimen collector (MedAuxil) | Qiagen AllPrep kit | Illumina MiSeq | V4 | 515F | 805R | PRJNA381395 | High abundance of Akkermansia in PD, different gut microbiota between cases and controls. |
| [24] | 2017 | USA | 327 (197 case, 130 control) | Total 69, 68.4, 9.2; (Median, Mean, SD) | Female: 65; 132 (67.0%), Male (% Male) | stool | PD medications, disease duration, spousal relationship, geographic site | Media-free swabs kit with DNA/RNA-free sterile swabs | According to the Earth Microbiome Project Protocol | Illumina MiSeq | V4 | 515F | 806R | ERP016332 | Significantly higher abundance of Bifidobacteriaceae, Christensenellaceae, Lactobacillaceae, Pasteurellaceae, and Verrucomicrobiaceae families in cases compared to controls/association of a high abundance of Akkermansia, Lactobacillus, Bifidobacterium, and reduced Lachnospiraceae (chain fatty acids producer) with disease development. |
| [41] | 2017 | Germany | 58 (29 case, 29 control) | case: 69.2, 6.5/control: 69.4, 6.7 (years; mean, SD) | case: 23 male, 6 female/control: 13 male, 16 female | stool | age | Shipment with no preservative, at room temp | PowerSoil Kit | Illumina MiSeq | V1–V2 | NR | NR | NR | High abundance of Lactobacillaceae, Barnesiellaceae, and Enterococcacea in cases. |
| [14] | 2015 | USA | 72 (38 case, 34 control) | case: 61.6 ± 9.4/control: 45.1 ± 14.4; (mean) | case: 24/14, control: 18/16; male/female | fecal samples and sigmoid mucosal biopsies | NR | Supporting info | Supporting info | NR | V4 | NR | NR | NR | Anti-inflammatory butyrate- producing bacteria from the genera Blautia, Coprococcus, and Roseburia were significantly more abundant in the feces of controls than PD patients/Faecalibacterium were significantly more abundant in the mucosa of controls than in PD/proinflammatory bacteria such as Proteobacteria were significantly more abundant in the mucosa of PD than controls/the ratio of Firmicutes-to-Bacteroidetes in fecal samples was not significantly different between PD and HC groups/positive correlation of Bacteroidetes and Proteobacteria with PD duration/negative correlation of Firmicutes with PD duration. |
| [47] | 2019 | China | 99 (51 case, 48 control) | case: 62.4 ± 8.2, control: 62.2 ± 9.2 (years, mean ± SD) | case: male: 32, female: 19/control: control: male: 19, female: 29 | stool | age | Sterilized tube | QIAamp Fast DNA Stool Mini Kit | HiSeq2500 PE250 | V4 | 515F | 806R | NR | Low alpha and beta diversity, high abundance of Akkermansia, P. copri Prevotella, Ruminococcaceae, Veillonellaceae Verrucomicrobiaceae, methanobrevibacter smithii, Ruminococcus callidus, Roseburia inulinivorans, Parabacteroides merde, Ruminococcus torques in cases and a high abundance of Bacteroidales, Lactobacillaceae, lactobacillus gasseri in controls. |
| [21] | 2019 | China | 20 (10 case, 10 control) | over 65 | case: male: 7 (70%)/control: male: 5 (50%) | feces | NR | DNA/RNA Shield | PSP SPIN Stool DNA plus kit | Illumina MiSeq | V1–V3 | 341F | 805R | NR | Slightly different gut microbiota between cases and controls, a significant abundance of Bacteroides and Prevotellaceae in healthy controls, a significant abundance of Ruminococcaceae, Verrucomicrobiaceae, Porphyromondaceae, Hydrogenoanaerobacterium, and Lachnospiraceae in PD cases. |
| [27] | 2018 | China | 120 (75 case, 45 control) | case: 60.48 ± 10.72/control; 63.20 ± 6.00 | case: male: 49, female: 26/control: male: 23, female: 22 | stool | age | Without preservation solution at room temperature during shipment | Huirui.® DNA kit | Illumina HiSeq PE250 | V4 | V4T9 (5′-GTGTGYCAGCMG-CCGCGG TAA-3′) | V4R19 (5′-CCGGACTACNVGGGTWTCTAAT-3′) | NR | Significant reductions in Tenericutes, Euryarchaeota, and Firmicutes, Lachnospiraceae in patients with PD. Veillonellaceae and Verrucomicrobiaceae showed marked increases but without statistical significance. Significant differences in alpha diversity (but not beta) between patients with PD who had a disease duration of greater than 5 years compared to those with a disease duration of fewer than 5 years. |
| [49] | 2019 | Taiwan | 157 (80 case, 77 control) | case: 62.3 ± 7.8/control: 61.8 ± 8.3 | case: 62.5% men/control: 60% men | stool | age and sex | Flash-frozen on dry ice, and stored at −80 °C | QIAamp Fast DNA Stool Mini Kit | Illumina MiSeq | V3–V4 | NR | NR | NR | Microbiota from patients with PD dominated by Verrucomicrobia, Mucispirillum, Porphyromonas, Lactobacillus, and Parabacteroides. In contrast, Prevotella was more abundant in controls. The abundances of Bacteroides were more increased in patients with non-tremor PD subtype than patients with tremor subtype. Bacteroides abundance was correlated with motor symptom severity defined by UPDRS part III motor scores. |
| [48] | 2017 | China | 38 (24 case, 14 control) | case: 73.75 ± 6.26/control: 74.64 ± 5.57 | case: 16 men/control: 6 men | stool | age and sex | NR | TIANamp stool DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) | Illumina MiSeq | V3–V5 | 5′-CCTACGGRRBGCASCAGKVRVGAAT-3′ | e 5′-GGACTACNVGGGTWTCTAATCC-3′ | NR | The genera Blautia, Faecalibacterium, and Ruminococcus were significantly decreased in PD compared to healthy controls. The genera Escherichia-Shigella, Streptococcus, Proteus, and Enterococcus were significantly increased in PD subjects. |
| [50] | 2017 | Russia | 155 (89 case, 66 control) | case: 67/control: 63 | NR | stool | age and sex | NR | NR | Illumina MiSeq | V3–V4 | NR | NR | NR | Reduction in taxonomic diversity of gut microbiota in patients with PD. |
| [51] | 2019 | Italy | 152 (80 case, 72 control) | NR | NR | stool | age, sex, loss of weight | NR | PSP Spin Stool DNA Kit Plus (Stratec Molecular) | Illumina MiSeq | V3–V4 | NR | NR | PRJNA510730 | Significantly higher levels of Lactobacillaceae, Enterobacteriaceae, and Enterococcaceae families compared to healthy individuals. On the other hand, the levels of Lachnospiraceae were significantly reduced in PD patients. |
| [52] | 2020 | China | 80 (40 case, 40 control) | case: 66.6 ± 7.1/control: 66.3 ± 8.1 | case: 19 men/control: 21 men | stool | lifestyle, gender, age, and BMI | NR | QIAamp DNA Stool Mini Kit (Qiagen) | Illumina HiSeq X-ten | NR | NR | NR | PRJNA433459 | The diversity and community of gut microbial genes in PD patients differed from those of healthy control subjects. Thirty-six different taxa were enriched in the PD patients, and no taxon was enriched in the healthy controls. |
| [53] | 2018 | China | 90 (45 case, 45 control) | case: 68.1 ± 8.0/control: 67.9 ± 8.0 | case: 22 men/control: 23 men | stool | age, sex, BMI, constipation | NR | QIAamp DNA Stool Mini Kit (Qiagen) | Illumina MiSeq | V3–V4 | 341F | 806R | PRJNA391524 | Some bacteria were correlated with PD clinical characteristics, including disease duration, severity, medication, and non-motor symptoms. |
| [66] | 2020 | China | 40 (27 case, 13 control) | case: 62.1 ± 10.2/control: 63 ± 8.76 | case: 19 men/control: 3 men | stool | age | NR | QIAamp DNA Stool Mini Kit (Qiagen) | Illumina MiSeq | V3–V4 | 515F 5′-GTGCCAGCMGCCGCGGTAA-3′ | 926R 5′-CCGTCAATTCMTTTGAGTTT-3′ | PRJNA561023 | Compared with HC and patients with PD-NC, the gut microbiota of patients with PD-MCI was significantly altered, particularly manifesting in enriched genera from Porphyromonadaceae family and decreased abundance of genera Blautia and Ruminococcus. |
| [55] | 2021 | Malaysia | 200 (104 case, 96 control) | case: 65.4 ± 8.4/control: 62.4 ± 9.0 | case: 62.5% male/control: 37.5% male | stool | age, sex | Preservatives were not added | QIAamp DNA Stool Mini Kit (Qiagen) | Illumina MiSeq | V3–V4 | 341F (5′-CCTACGGGNGGCWGCAG-3′) | 805R (5′-GACTACHVGGGTATCTAATCC-3′) | PRJNA494620 | Ten bacterial taxa were significantly increased in PD; largest fold changes were observed for B. fragilis, Lactobacillus acidophilus, Megasphaera and Gammaproteobacteria. |
| [56] | 2020 | Italy | 115 (64 case, 51 control) | case: 71.39 ± 10.99/51.67 ± 12.42 | case: 44 men/control: 31 men | stool | sex, age, BMI, coffee consumption, smoking | NR | QIAamp DNA Stool Mini Kit (Qiagen) | Illumina MiSeq | V3–V4 | NR | NR | PRJEB30401 | The most significant changes within the PD group highlighted a reduction in bacterial taxa, which are linked to anti-inflammatory/neuroprotective effects, particularly in the Lachnospiraceae family and key members, such as Butyrivibrio, Pseudobutyrivibrio, Coprococcus, and Blautia. |
| [43] | 2019 | Germany | 59 (34 case, 25 control) | case: 67.9 ± 8.6/63.9 ± 5.8 | case: 23 men/control: 11 men | stool | age | Sterile containers | FastDNA SPIN kit | Illumina MiSeq | V4–V5 | 520 F (5′-AYTGGGYDTAAAGNG-3′) | 926 R (5′-CCGTCAATTCMTTTRAGTTT-3′) | PRJEB30615 | PD patients exhibit alterations in their gut microbiota composition, characterized by a decrease in beneficial bacteria and an increase in certain bacterial groups. A potential link between the gut microbiome and PD development, as well as the influence of PD medications on the gut microbiota. |
| [60] | 2021 | China | 40 (20 case, 20 control) | case: 63.65 ± 5.64/61.95 ± 4.73 | case: 10 men/control: 10 men | stool | age, sex, BMI | NR | DNeasy PowerSoil Kit (Qiagen) | Illumina MiSeq | V3–V4 | 343 F: 5′-TACGGRAGGCAGCAG-3′ | 798 R: 5′-AGGGTATCTAATCCT-3′ | NR | A greater abundance of Alistipes, Rikenellaceae_RC9_gut_group, Bifidobacterium, Parabacteroides, while Faecalibacterium was decreased in fecal samples from PD patients. |
| [62] | 2020 | China | 126 (63 case, 63 control) | case: 64.0 ± 7.4/63.9 ± 7.9 | case: 40 men/control: 23 men | stool | age | NR | QIAamp Fast DNA Stool Mini Kit (Qiagen) | Illumina HiSeq PE250 | V4 | NR | NR | CRA001938 | There were markedly different microbial compositions among PD, HS, and HP samples by alpha/beta diversity. They also found differential microbial compositions among Hoehn and Yahr stage/disease duration. |
| [25] | 2014 | Finland | 144 (72 case, 72 control) | case: 65.3 ± 6 5.5/64.5 ± 6.9 | case: 51.4% male/control: 50% men | stool | age, sex | NR | NR | NR | V1–V3 | NR | NR | NR | Reduced abundance of Prevotellaceae in PD patients and the positive association of Enterobacteriaceae abundance with PIGD symptoms. |
| [40] | 2022 | China | 172 (132 case (mild: 43, moderate: 89), 40 control) | 60 to 90 | M/F case: 15/28 (mild AD), 33/56 (moderate AD)/control: 16/24 | feces | age, sex | Special cytoprotective agent | E.Z.N.A. Soil DNA Kit | Illumina MiSeq | V3–V4 | CCTACGGGNGGCWGCAG | GACTACHVGGGTATCTAATCC | PRJNA855571 | Elevated abundance of certain bacteria in cases (moderate vs. control), including Proteobacteria, Verrucomicrobia, Actinobacteria, and Synergistetes. Reduced levels of Firmicutes and Bacteroidetes in cases. Controls, when compared to mild and moderate cases, showed higher levels of Firmicutes, Erysipelotrichia, Acidaminococcaceae, Ruminococcaceae, and Bacteroidetes. |
| [26] | 2022 | Canada | 99 (45 case, 54 control) | case: 74: (65–78)/control: 70 (66–74); (year) | Female: case 33.3%/control 33.3% | feces | age | OMNIgene® GUT | QIAamp PowerFecal DNA Kits | MiSeq | V4 | 515F (GTGCCAGC MGCCGCGGTAA) | 806R (GGACTACHVHHHTWTCTAAT) | PRJNA770746 | No significant different between AD patients and controls (beta diversity), lower alpha diversity in cases, higher abundance of Erysipelotrichaceae in cases. |
| [45] | 2021 | China | 64 (28 cases (18 AD), 18 control) | case: 63.5 (4.7)/control: 64.5 (4.5); (SD) | Male (%): case 2 (11)/control: 4 (22) | feces | age, sex | NR | PowerSoil DNA Isolation Kit | Illumina Miseq/Microseq | V3–V4 | NR | NR | NR | AD cases exhibited increased microbial diversity, decreased levels of Bacteroides, Lachnospira, and Ruminiclostridium, and increased Prevotella. |
| [17] | 2019 | USA | 108 | case: 84.7 (8.1)/control: 83 (10.2); (mean [SD]) (year) | Male: case: 4 (16.7)/control: 8 (15.7) | stool | age, sex | NR | PowerMag soil DNA isolation kit | NextSeq 500 | NR | NR | NR | Upon request | Significant increase in Bacteroides, Alistipes, Odoribacter, Barnesiella, Osplanchnicus, Odoribacter spp., K. pneumoniae, B. fragilis, E. lenta, and Desulfovibrio AD (sulfate-reducing bacteria). Conversely, there were significant decreases in bacteria, including Lachnoclostridium, Butyrivibrio, B. proteoclasticus, B. hungatei, Eubacterium, E. eligens, E. hallii, E. rectale, Clostridium sp. SY8519, R. hominis, and F. prausnitzii./significantly different beta diversity between controls and AD cases. |
| [46] | 2022 | Kazakhstan | 84 (41 case, 43 control) | case: 68 (62–74)/control 68 (61–75); (median (IQR)) | Female, n (%): case 30 (73.2%)/control 35 (81.4%) | feces | NR | special kit | QIAamp DNA stool Mini Kit | Illumina MiSeq | NR | NR | NR | PRJNA811324 | Increased abundance of Acidobacteriota, Verrucomicrobiota, Planctomycetota, and Synergistota and decreased abundance of Bifidobacterium, Clostridia bacterium, Castellaniella, Erysipelotrichaceae UCG-003, Roseburia, Tuzzerella, Lactobacillaceae and Monoglobus in AD patients. |
| [28] | 2019 | China | 97 (33 case (32 aMCI), 32 control) | case: 74.85 ± 11.37 (AD), 70.53 ± 11.0 (aMCI)/control: 76.88 ± 9.35; (years, mean ± SD) | Male: case: 14 (AD; 42.42%), 18 (56.25%) aMCI/control 16 (50%) | feces | age, sex | Sterile collection containers | QIAGEN | Illumina MiSeq | V3–V4 | 5′-CAAGCAGAAG ACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGA TCT-3′ | 5′-AATGATACGGCGACCACCGAGATC TACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ | PRJNA496408 | Decreased diversity in AD patients compared to controls. Different gut microbiota between healthy and cases/reduced abundance of Firmicutes, increased abundance of Proteobacteria in cases/a significant correlation between AD severity and the abundance of altered microbiomes/association of Enterobacteriaceae with AD. |
| [42] | 2021 | China | 105 (case: 53 (SCD) 14 (CI: MCI, n = 8; mild AD dementia, n = 6))/38 control) | case: 66.68 ± 6.32 (SCD), 73.21 ± 7.89 (CI)/control: 66.79 ± 5.13; (year) | M/F: case: 10/43 (SCD), 4/10 (CI)/control: 15/23 | feces | age, sex, educational years, other potential factors | Cytoprotective agents | QIAamp DNA Stool Mini Kit | Illumina Miseq PE250 | V3–V4 | 341F (CCTAC GGGRSGCAGCAG) | 806R (GGACTACVVGGGTATCTAATC) | NR | Decreasing abundance of Firmicutes, Clostridia, Clostridiales, Ruminococcaceae, and Faecalibacterium in cases. |
| [54] | 2020 | Austria | 41 (23 case, 18 control) | case: 88 (73, 85)/control: 75 (74, 76); (years) | F/M: case: 15/18, control: 11/7 | stool | age, sex | Collection tubes | MagnaPure LC DNA Isolation Kit III | Illumina MiSeq | V1–V2 | 27F (AGAGTT TGATCCTGGCTCAG) | R357 (CTGCTGCCTYCCGTA) | PRJNA608281 | Decreased abundance of Lachnospiraceae in cases, clear clustering of case and controls at different stages of dementia according to beta diversity, AD association with a reduction in bacteria producing short-chain fatty acids (SCFA) and increased biomarkers of gut permeability and inflammation. Increased abundance of C. clostridioforme and Eisenbergiella is associated with cognitive impairment. |
| [18] | 2022 | Spain | 22 (12 case, 10 control) | 60 to 70 | case: 2 men/control: 6 men | stool | NR | NR | QIAamp PowerFecal Pro DNA isolation kit (Qiagen, Madrid, Spain) | Illumina MiSeq | NR | 515F-Y (50 GTG YCA GCM GCC GCG GTA A 30 | 806R (50 GGA CTA CNV GGG TWT CTA AT 30 | NR | At a more advanced stage of AD, the gut microbiota and volatiles shifted towards a profile with increases in Ruminococcus and Blautia. |
| [58] | 2022 | Thailand | 40 (20 case, 20 control) | case: 72.8 ± 5.6/control: 69.4 ± 6.2 | case: 45.5% male/control: 38.5% male | stool | NR | Preservation System (Norgen Biotek Corp., Thorold, ON, Canada) | QIAamp Stool Mini kit (Qiagen, USA) | Illumina MiSeq | V3–V4 | NR | NR | NR | A significant difference at the operational taxonomic unit level. The altered gut microbiome could be potentially targeted for the early diagnosis of dementia and the reduction in AD risk. |
| [59] | 2021 | China | 65 (21 case, 44 control) | case: 76.2 ± 9.9/control: 78.4 ± 6.6 | case: 13 men/control: 20 men | stool | NR | Sterile fecal collection containers | E.Z.N.A.@ Stool DNA Kit (Omega Bio-Tek, Norcross, GA, USA) | Illumina Miseq | V3–V4 | 338 forward (5′-ACTCCTACGGGAGGCAGCAG-3′) | 806 reverse (5′-GGACTACHVGGGTWTCTAAT-3′) | SRP252374 | Gut microbial alterations and related metabolic output changes may be associated with pathogenesis of AD. Fecal markers might be used as a non-invasive examination to assist screening and diagnosis of AD. |
| [61] | 2022 | Turkey | 98 (47 case, 51 control) | case: 71.4 ± 5.1/control: 67 ± 5.3 | case: 24 men/control: 28 men | stool | NR | NR | QiaAmp DNA stool minikit (Qiagen, Germany) | Illumina MiSeq | V3–V4 | 341 F (59-CCTACGGGNGGCWGCAG-39) | 805 R (59-GACTACHVGGGTATCTAATCC-39) | NCBI BioProject database | A different gut microbiota composition in AD cases marked primarily by Prevotella and Bacteroides, but also subnetworks of other taxa exist in the community. |
| [63] | 2021 | China | 92 (60 case, 32 control) | case: 72.82 ± 7.25/control: 71.06 ± 5.92 | case: 24 men/control: 14 men | stool | age | NR | QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany) | Illumina MiSeq | V3–V4 | 5-CCTACGGGNGGCWGCAG-3 | 5-GACTACHVGGGTATCTAATCC-3 | NR | AD patients had gut microbiota alterations related to cognition, and differential taxa between AD patients with and without NPS associated differently with NPS domains. |
| [65] | 2018 | China | 86 (43 case, 43 control) | case: 70.12 ± 8.76/control: 69.72 ± 9.24 | case: 23 men/control: 23 men | stool | age, sex | NR | QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany) | Illumina MiSeq | V3–V4 | 338F | 806R | NR | Altered gut microbiota composition and diversity AD cases compared to cognitively normal controls. Several bacterial taxa, including Actinobacteria, Bacteroidales, Ruminococcaceae, Selenomonadales, and Lachnoclostridium, were found to contribute to these differences. |
| [64] | 2022 | China | 302 (125 MCI, 83 AD case, 94 control) | case: 71.8 ± 8.3/control: 74.3 ± 10.6 | case: 53 men/control: 58 men | stool | NR | Sterile containers | E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) | Illumina MiSeq | V3–V4 | 338F (50 -ACTCCTACGGGAGGCAGCAG-30) | 806R (50-GGACTACHV GGGTWTCTAAT-30) | NR | No significant difference in the alpha and beta diversity among groups. Patients with AD or MCI had increased bacterial taxa including Erysipelatoclostridiaceae, Erysipelotrichales, Patescibacteria, Saccharimonadales, and Saccharimonadia, compared with NC group. |
| [57] | 2017 | USA | 50 (25 case, 25 control) | case: 71.3 ± 7.3/control: 69.3 ± 7.5 | case: 8 men/control: 7 men | stool | age, sex | NR | NR | Illumina MiSeq | V4 | NR | NR | NR | Decreased microbial diversity in AD cases and compositionally distinct from controls. |
| PD Studies | AD Studies | |
|---|---|---|
| Alpha Diversity (case vs. control) |
|
|
| Beta Diversity (case vs. control) |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heravi, F.S.; Naseri, K.; Hu, H. Gut Microbiota Composition in Patients with Neurodegenerative Disorders (Parkinson’s and Alzheimer’s) and Healthy Controls: A Systematic Review. Nutrients 2023, 15, 4365. https://doi.org/10.3390/nu15204365
Heravi FS, Naseri K, Hu H. Gut Microbiota Composition in Patients with Neurodegenerative Disorders (Parkinson’s and Alzheimer’s) and Healthy Controls: A Systematic Review. Nutrients. 2023; 15(20):4365. https://doi.org/10.3390/nu15204365
Chicago/Turabian StyleHeravi, Fatemah Sadeghpour, Kaveh Naseri, and Honghua Hu. 2023. "Gut Microbiota Composition in Patients with Neurodegenerative Disorders (Parkinson’s and Alzheimer’s) and Healthy Controls: A Systematic Review" Nutrients 15, no. 20: 4365. https://doi.org/10.3390/nu15204365
APA StyleHeravi, F. S., Naseri, K., & Hu, H. (2023). Gut Microbiota Composition in Patients with Neurodegenerative Disorders (Parkinson’s and Alzheimer’s) and Healthy Controls: A Systematic Review. Nutrients, 15(20), 4365. https://doi.org/10.3390/nu15204365






