Dys-R Questionnaire: A Novel Screening Tool for Dysbiosis Linked to Impaired Gut Microbiota Richness
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. Participants’ Characterizing Data
2.3. Evaluation of the Gut Microbiota (GM)
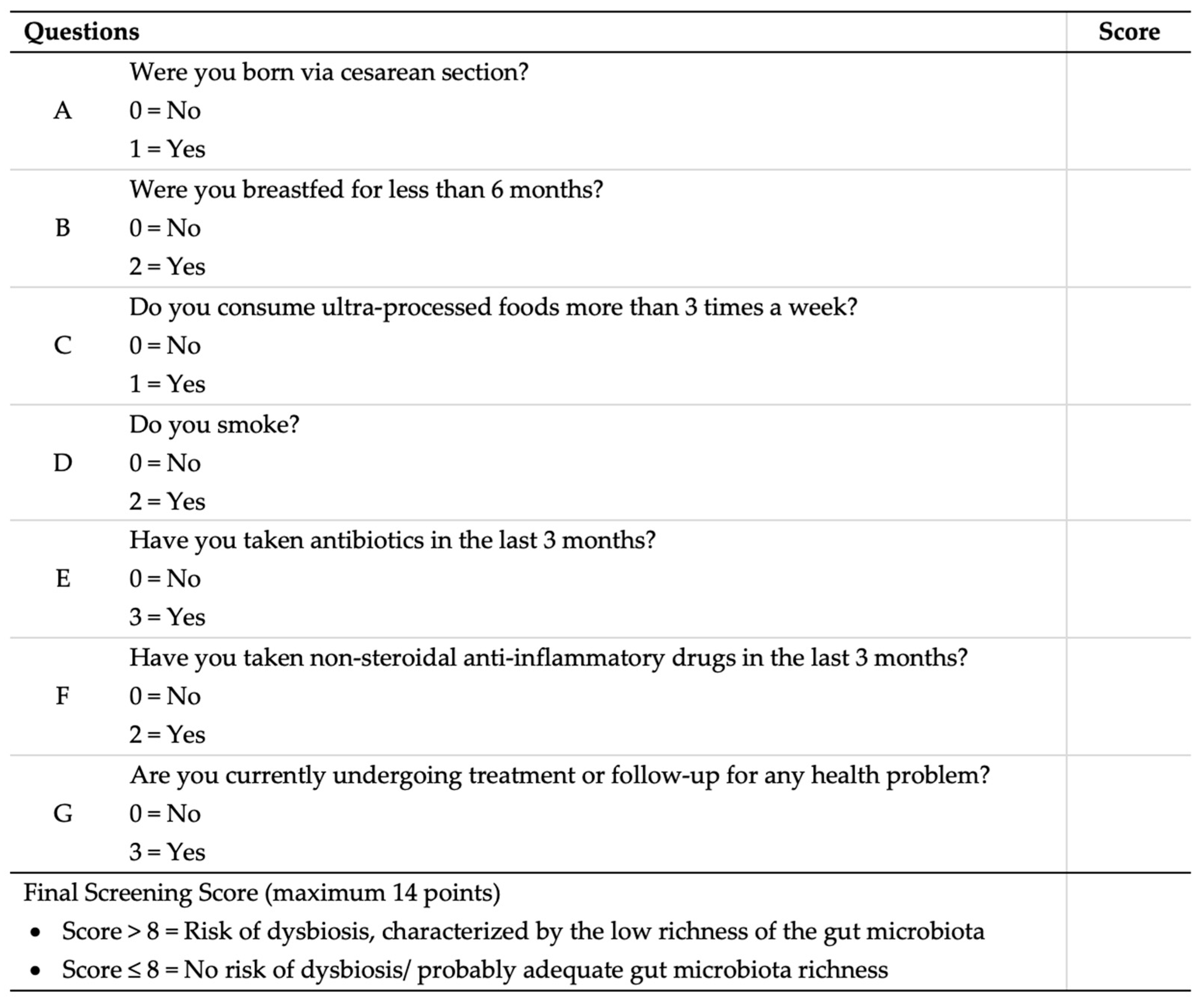
2.4. Development of the Dysbiosis Risk Questionnaire (Dys-R Questionnaire)
2.5. Statistical Analysis
3. Results
3.1. Patient’s Descriptive Data
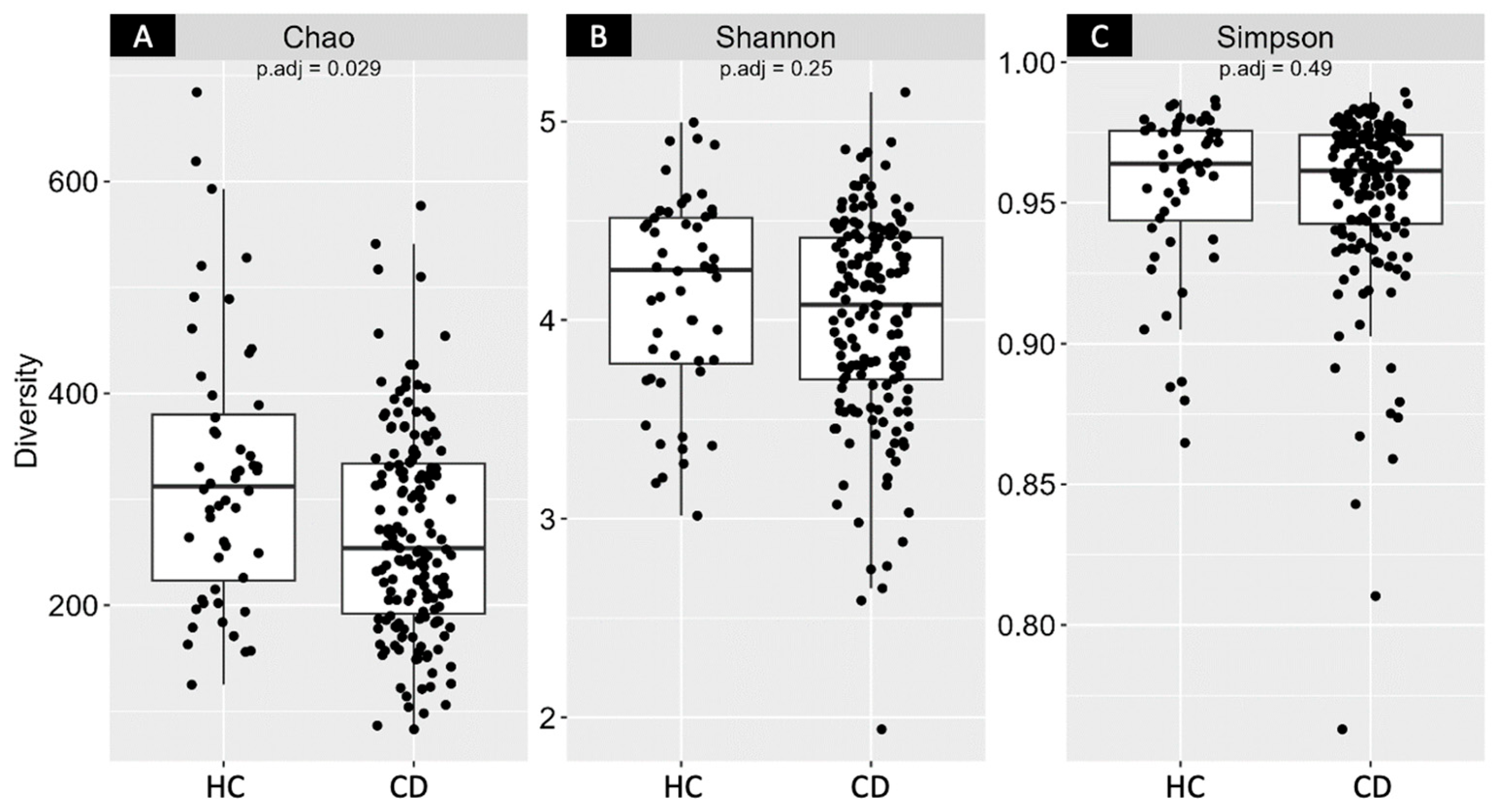
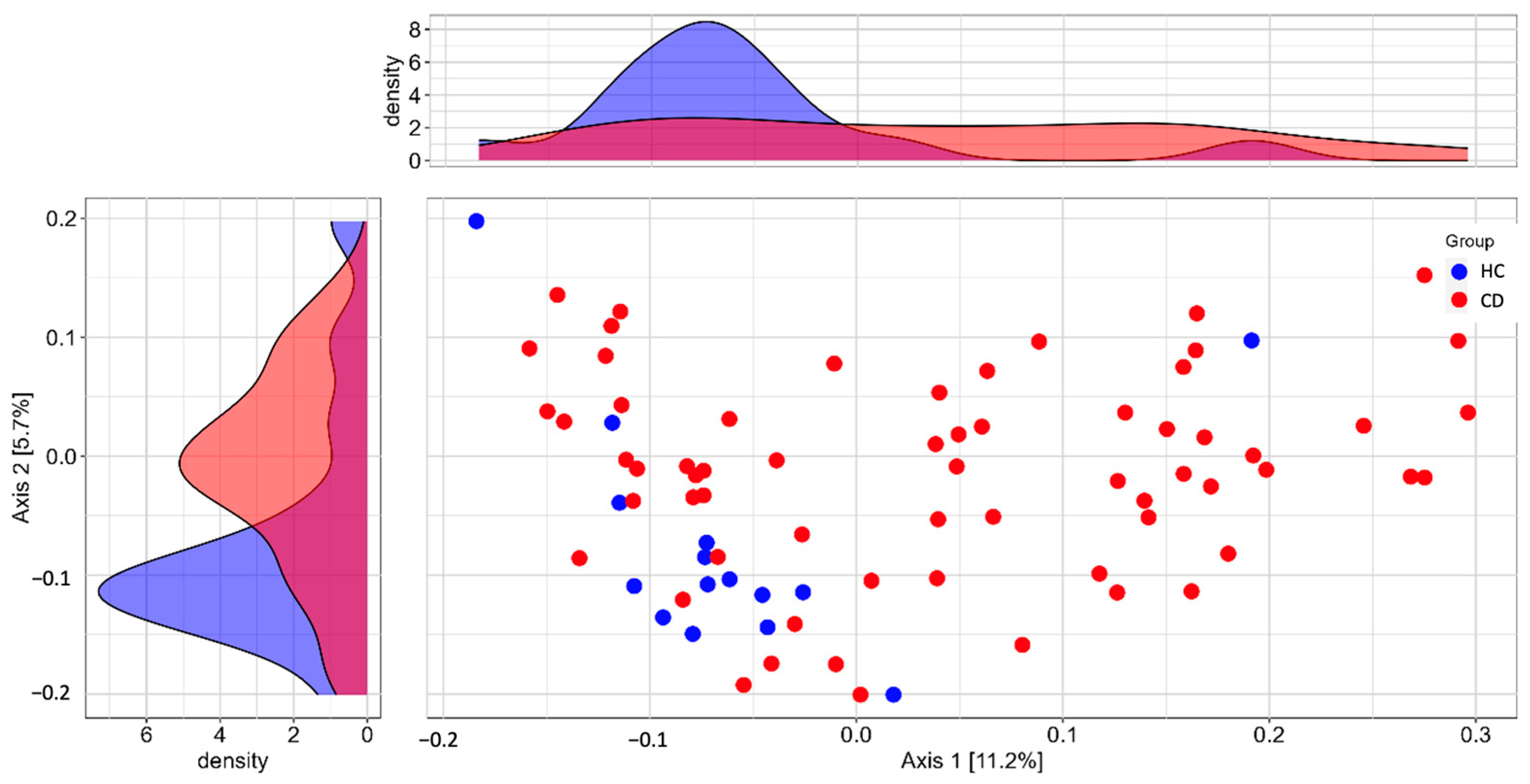
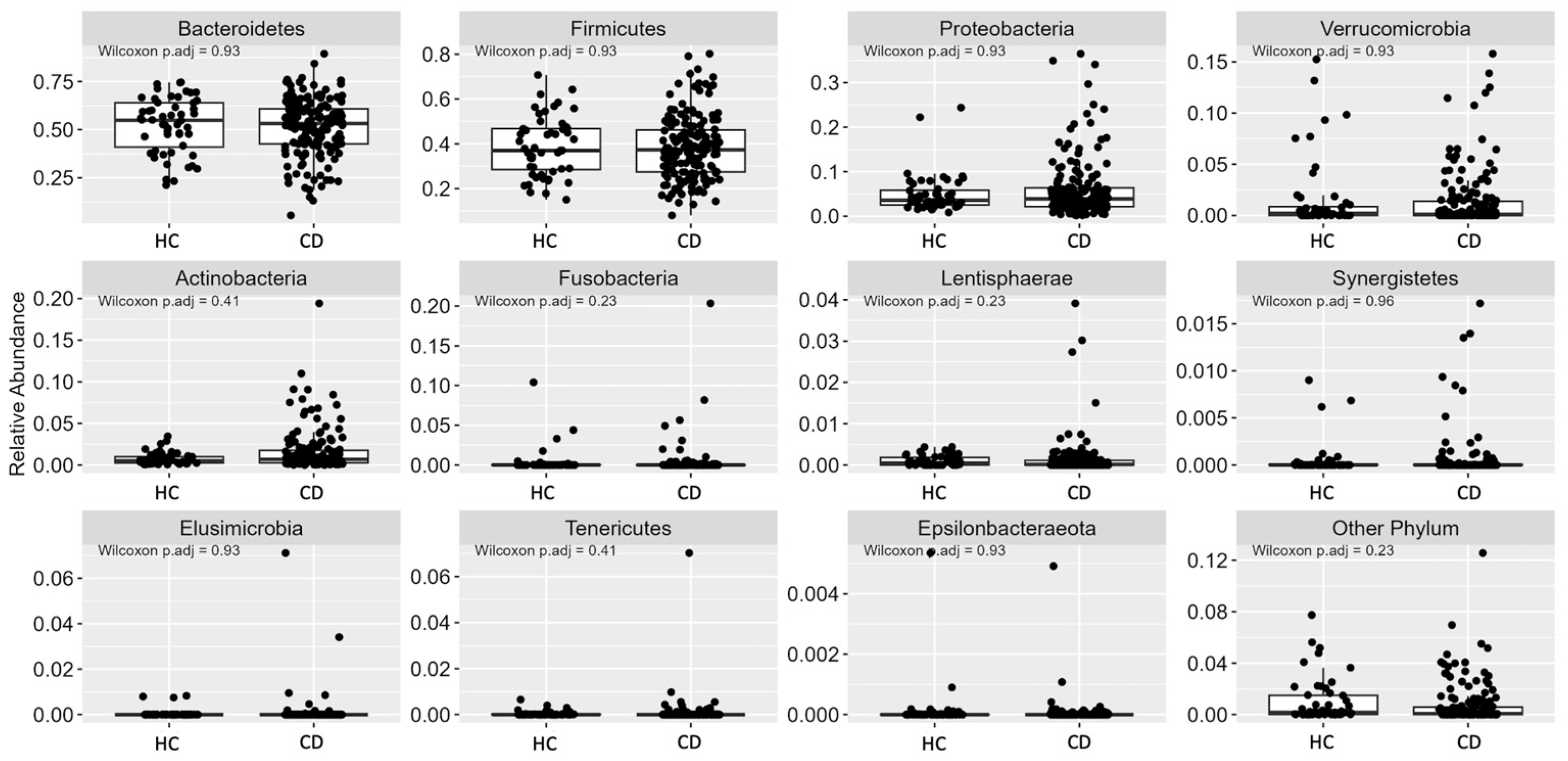
3.2. Gut Microbiota (GM)
3.3. DYS/FQM Performance
3.4. Dys-R Construction
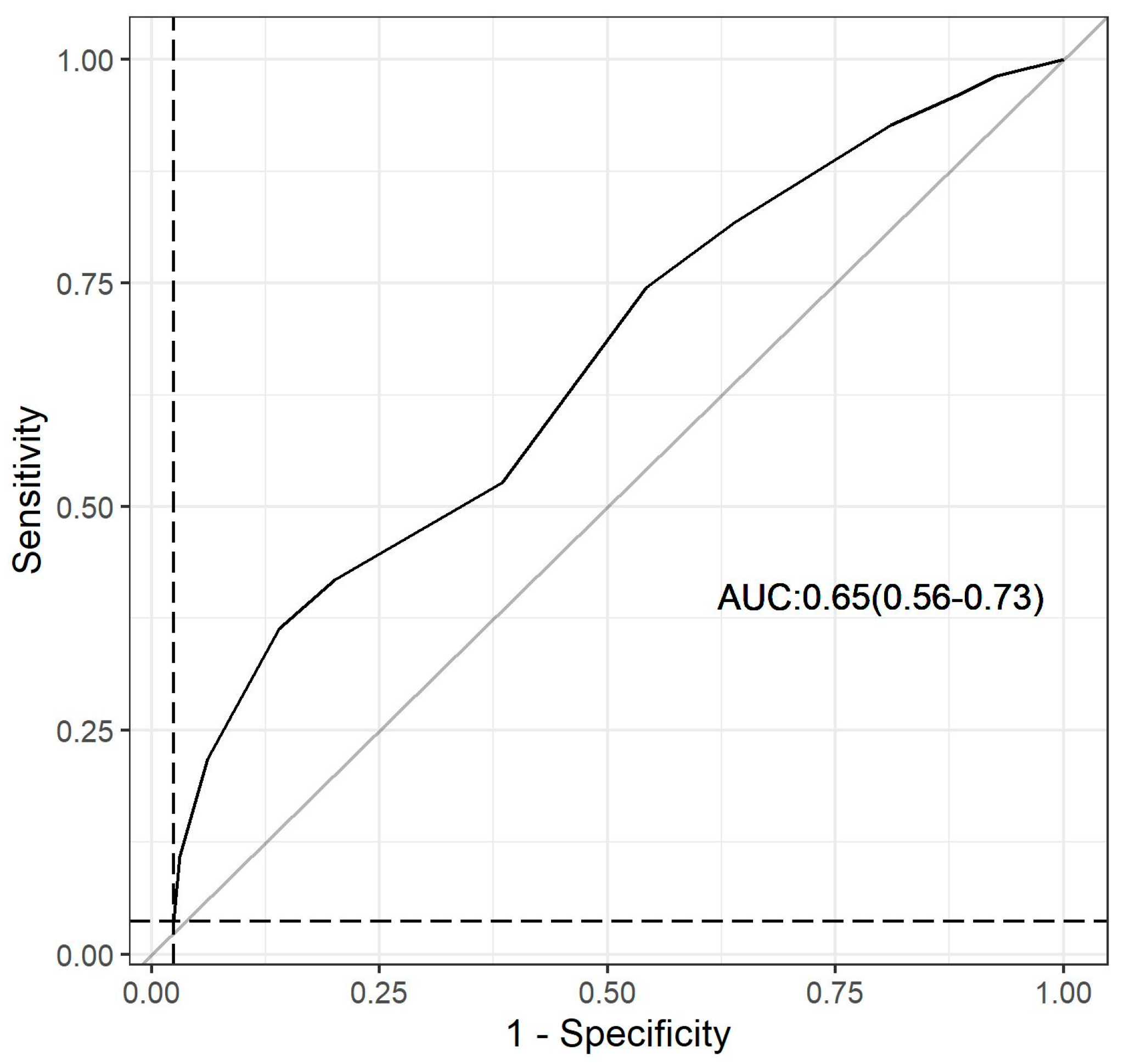
3.5. Dys-R Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Mousa, W.K.; Chehadeh, F.; Husband, S. Recent Advances in Understanding the Structure and Function of the Human Microbiome. Front. Microbiol. 2022, 13, 825338. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulou, K.; Adamopoulos, P.G.; Scorilas, A. Unveiling the Human Gastrointestinal Tract Micro-biome: The Past, Present, and Future of Metagenomics. Biomedicines 2023, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Panwar, R.B.; Sequeira, R.P.; Clarke, T.B. Microbiota-mediated Protection Against Antibiotic-Resistant Pathogens. Genes. Immun. 2021, 22, 255–267. [Google Scholar] [CrossRef]
- Birchenough, G.M.H.; Schroeder, B.O.; Sharba, S.; Arike, L.; Recktenwald, C.V.; Puértolas-Balint, F.; Subramani, M.V.; Hansson, K.T.; Yilmaz, B.; Lindén, S.K.; et al. Muc2-dependent Microbial Colonization of the Jejunal Mucus Layer is Diet Sensitive and Confers Local Resistance to Enteric Pathogen Infection. Cell Rep. 2023, 42, 112084. [Google Scholar] [CrossRef]
- Zong, X.; Fu, J.; Xu, B.; Wang, Y.; Jin, M. Interplay Between Gut Microbiota and Antimicrobial Peptides. Anim. Nutr. 2020, 6, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Talapko, J.; Včev, A.; Meštrović, T.; Pustijanac, E.; Jukić, M.; Škrlec, I. Homeostasis and Dysbiosis of the Intestinal Microbiota: Comparing Hallmarks of a Healthy State with Changes in Inflammatory Bowel Disease. Microorganisms 2022, 10, 2405. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the Immune System. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Do, M.H.; Lee, E.; Oh, M.J.; Kim, Y.; Park, H.Y. High-Glucose or -Fructose Diet Cause Changes of the Gut Microbiota and Metabolic Disorders in Mice without Body Weight Change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a Gut Pathobiont Drives Autoim-munity in Mice and Humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef]
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, A.; et al. Targeted Suppression of Human IBD-associated Gut Microbiota Commensals by Phage Consortia for Treatment of Intestinal Inflammation. Cell 2022, 185, 2879–2898.e24. [Google Scholar] [CrossRef]
- Wu, H.; Tremaroli, V.; Schmidt, C.; Lundqvist, A.; Olsson, L.M.; Krämer, M.; Gummesson, A.; Perkins, R.; Bergström, G.; Bäckhed, F. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab. 2020, 32, 379–390.e3. [Google Scholar] [CrossRef] [PubMed]
- Bourdeau-Julien, I.; Castonguay-Paradis, S.; Rochefort, G.; Perron, J.; Lamarche, B.; Flamand, N.; Di Marzo, V.; Veilleux, A.; Raymond, F. The Diet Rapidly and Differentially Affects the Gut Microbiota and Host Lipid Mediators in a Healthy Population. Microbiome 2023, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Parkin, K.; Christophersen, C.T.; Verhasselt, V.; Cooper, M.N.; Martino, D. Risk Factors for Gut Dysbiosis in Early Life. Microorganisms 2021, 9, 2066. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal Microbiota Com-position of Breast-Fed Infants is Correlated with Human Milk Oligosaccharides Consumed. J. Pediatr. Gastro-Enterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Shanahan, F.; O’Toole, P.W. The Gut Microbiome as a Modulator of Healthy Ageing. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 565–584. [Google Scholar] [CrossRef]
- Vijay, A.; Valdes, A.M. Role of the Gut Microbiome in Chronic Diseases: A Narrative Review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host Lifestyle Affects Human Microbiota on Daily Timescales. Genome Biol. 2014, 15, R89, Erratum in Genome Biol. 2016, 17, 117. [Google Scholar] [CrossRef]
- Patangia, D.V.; Anthony Ryan, C.; Dempsey, E.; Paul Ross, R.; Stanton, C. Impact of Antibiotics on the Human Microbiome and Consequences for Host Health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. The Microbiome-Gut-Brain Axis in Health and Disease. Gastroenterol. Clin. N. Am. 2017, 46, 77–89. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Wu, C. Modulation of Gut Microbiota and Immune System by Probiotics, Pre-biotics, and Post-biotics. Front. Nutr. 2022, 8, 634897. [Google Scholar] [CrossRef]
- Yen, S.; Johnson, J.S. Metagenomics: A Path to Understanding the Gut Microbiome. Mamm. Genome 2021, 32, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, D.; Kanitkar, T.; Narouz, M.; Azadian, B.S.; Moore, L.S.P.; Mughal, N. Clinical Utility and Cost-effectiveness of Bacterial 16S rRNA and Targeted PCR Based Diagnostic Testing in a UK Microbiology Laboratory Network. Sci. Rep. 2020, 10, 7965. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Oliveira, F.S.; Correa, F.B.; Morais, D.K.; Fernandes, G.R. TAG.ME: Taxonomic Assignment of Genetic Markers for Ecology. BioRxiv 2018, 263293. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Beccati, A.; Gerken, J.; Quast, C.; Yilmaz, P.; Glöckner, F.O. SILVA Tree Viewer: Interactive Web Browsing of the Silva Phylogenetic Guide Trees. BMC Bioinform. 2017, 18, 433. [Google Scholar] [CrossRef]
- Wei, S.; Bahl, M.I.; Baunwall, S.M.D.; Hvas, C.L.; Licht, T.R. Determining Gut Microbial Dysbiosis: A Review of Applied Indexes for Assessment of Intestinal Microbiota Imbalances. Appl. Environ. Microbiol. 2021, 87, e00395-21. [Google Scholar] [CrossRef]
- Balmant, B.D.; Fonseca, D.C.; Prudêncio, A.P.A.; Rocha, I.M.; Callado, L.; Alves, J.T.M.; Torrinhas, R.S.M.M.; Borba, E.F.; Waitzberg, D.L. Megamonas funiformis, Plasma Zonulin, and Sodium Intake Affect C3 Complement Levels in Inactive Systemic Lupus Erythematosus. Nutrients 2023, 15, 1999. [Google Scholar] [CrossRef]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, J.; Liang, H.; Ye, L.; Lan, L.; Lu, F.; Wang, Q.; Lei, T.; Yang, X.; Cui, P.; et al. Differences in Alpha Diversity of Gut Microbiota in Neurological Diseases. Front. Neurosci. 2022, 16, 879318. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.G.D.; Knight, R.; Gilbert, J.A.; Blaser, M.J. Preserving Microbial Diversity. Science 2018, 362, 33–34. [Google Scholar] [CrossRef] [PubMed]
- Pant, A.; Maiti, T.K.; Mahajan, D.; Das, B. Human Gut Microbiota and Drug Metabolism. Microb. Ecol. 2023, 86, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Clauss, M.; Gérard, P.; Mosca, A.; Leclerc, M. Interplay Between Exercise and Gut Microbiome in the Context of Human Health and Performance. Front. Nutr. 2021, 8, 637010. [Google Scholar] [CrossRef]
- Putignani, L.; Del Chierico, F.; Vernocchi, P.; Cicala, M.; Cucchiara, S.; Dallapiccola, B.; Dysbiotrack Study Group. Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Child-hood-Adulthood Transition. Inflamm. Bowel Dis. 2016, 22, 487–504. [Google Scholar] [CrossRef]
- Chen, L.; Reynolds, C.; David, R.; Peace Brewer, A. Development of an Index Score for Intestinal Inflam-mation-Associated Dysbiosis Using Real-World Stool Test Results. Dig. Dis. Sci. 2020, 65, 1111–1124. [Google Scholar] [CrossRef]
- Hrncir, T. Gut Microbiota Dysbiosis: Triggers, Consequences, Diagnostic and Therapeutic Options. Microorganisms 2022, 10, 578. [Google Scholar] [CrossRef]
- Okipney, A.; Souza, J.R.A.; Campos, A.C.L.; Campos, L.F.; Anjo, P.R.; Abreu, C. Risk Stratification for Intestinal Dysbiosis in Hospitalized Adult Patients According to the National Dysbiosis Survey (INDIS). Braspen J. 2020, 35, 149–159. [Google Scholar] [CrossRef]
- Rodrigues, C.; Catapani, W.R. A Theoretical Exploratory Analysis on the Risk of Dysbiosis in Individuals with Inflammatory Bowel Diseases Using the DYS/FQM questionnaire. Gastroenterol. Hepatol. Open Access 2023, 14, 53–56. [Google Scholar] [CrossRef]
| Variables | CD 1 (n = 167) | HC 2 (n = 52) | p-Value |
|---|---|---|---|
| Age, years | 47.0 ± 13.9 | 40.4 ± 17.9 | <0.001 3 |
| Gender | |||
| Female, n (%) | 111 (66.5) | 23 (44.2) | 0.004 4 |
| Male, n (%) | 56 (33.5) | 29 (55.8) | |
| Ethnicity | |||
| Black, n (%) | 21(12.6) | 3 (5.8) | <0.001 4 |
| Brown, n (%) | 61 (36.5) | 4 (7.7) | |
| White, n (%) | 84 (50.3) | 39 (75.0) | |
| Yellow, n (%) | 0 (0) | 6 (11.5) | |
| Indigenous, n (%) | 1 (0.6) | 0 (0) | |
| Anthropometric | |||
| Weight, kg | 79.8 ± 26.5 | 68.6 ± 13.1 | 0.014 3 |
| Height, cm | 164 ± 0.1 | 167 ± 0.1 | 0.001 3 |
| BMI 5, kg/m2 | 29.6 ± 9.2 | 23.9 ± 2.9 | <0.001 3 |
| Body composition | |||
| LMP 6, % | 61.4 ± 11.3 | 72.4 ± 7.5 | <0.001 3 |
| FMP 7, % | 38.6 ± 11.3 | 27.6 ± 7.5 | <0.001 3 |
| Variables, n (%) | CD 1 (n = 167) | HC 2 (n = 52) | p-Value 3 |
|---|---|---|---|
| Age ≥ 60 years | 39 (23.4) | 9 (17.3) | 0.358 |
| Born by cesarean section | 30 (18.0) | 25 (48.1) | <0.001 |
| Breastfed for less than 6 months | 104 (62.3) | 32 (61.5) | 0.924 |
| Consumption of less than 5 daily servings of fruits, vegetables, legumes, and/or whole grains | 155 (92.8) | 44 (84.6) | 0.073 |
| Consumption of refined sugar or artificial sweeteners more than once a day | 156 (93.4) | 40 (77.0) | <0.001 |
| Consumption of ultraprocessed foods more than 3 times a week | 75 (44.9) | 19 (36.5) | 0.287 |
| Consumption of greater than 4 doses of alcoholic beverage per week | 2 (1.2) | 3 (5.8) | 0.054 |
| Less than 150 min of physical activity per week | 135 (80.8) | 30 (57.7) | <0.001 |
| High self-reported psychological stress | 56 (33.5) | 11 (21.2) | 0.091 |
| Smoking | 16 (9.6) | 1 (1.9) | 0.072 |
| Use of antibiotics in the last 3 months | 45 (27.0) | 5 (9.6) | 0.009 |
| Use of nonsteroidal anti-inflammatory drugs in the last 3 months | 38 (22.8) | 7 (13.5) | 0.148 |
| Continuous consumption of more than 3 medications | 74 (44.3) | 0 (0) | <0.001 |
| Current treatment or monitoring for any health condition | 167 (100) | 52 (100) | <0.001 |
| Three or more liquid bowel movements per day or difficult bowel movements, hard stools, and/or less than 3 bowel movements per week | 44 (26.4) | 5 (9.6) | 0.011 |
| Medium or major surgical procedure in the last 60 days or bariatric surgery at any time in life | 2 (1.2) | 1 (1.9) | 0.694 |
| Treatment with chemotherapy or radiotherapy | NA 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balmant, B.D.; Fonseca, D.C.; Rocha, I.M.; Callado, L.; Torrinhas, R.S.M.d.M.; Waitzberg, D.L. Dys-R Questionnaire: A Novel Screening Tool for Dysbiosis Linked to Impaired Gut Microbiota Richness. Nutrients 2023, 15, 4261. https://doi.org/10.3390/nu15194261
Balmant BD, Fonseca DC, Rocha IM, Callado L, Torrinhas RSMdM, Waitzberg DL. Dys-R Questionnaire: A Novel Screening Tool for Dysbiosis Linked to Impaired Gut Microbiota Richness. Nutrients. 2023; 15(19):4261. https://doi.org/10.3390/nu15194261
Chicago/Turabian StyleBalmant, Bianca Depieri, Danielle Cristina Fonseca, Ilanna Marques Rocha, Letícia Callado, Raquel Susana Matos de Miranda Torrinhas, and Dan Linetzky Waitzberg. 2023. "Dys-R Questionnaire: A Novel Screening Tool for Dysbiosis Linked to Impaired Gut Microbiota Richness" Nutrients 15, no. 19: 4261. https://doi.org/10.3390/nu15194261
APA StyleBalmant, B. D., Fonseca, D. C., Rocha, I. M., Callado, L., Torrinhas, R. S. M. d. M., & Waitzberg, D. L. (2023). Dys-R Questionnaire: A Novel Screening Tool for Dysbiosis Linked to Impaired Gut Microbiota Richness. Nutrients, 15(19), 4261. https://doi.org/10.3390/nu15194261






