Enterococcus faecium HDRsEf1 Promotes Systemic Th1 Responses and Enhances Resistance to SalmonellaTyphimurium Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Animals
2.3. Feeding Probiotic
2.4. Histological Tissue Analysis
2.5. Flow Cytometry Analysis
2.6. In Vitro Splenocyte Stimulation
2.7. Coculture of CD4+ T and Bone Marrow-Derived Dendritic Cells (BMDCs)
2.8. Quantitative Expression Analysis by RT-PCR
2.9. Challenging with S. Typhimurium
2.10. Investigation of Bacterial Colonization in the Liver and Spleen
2.11. Statistical Analysis
3. Results
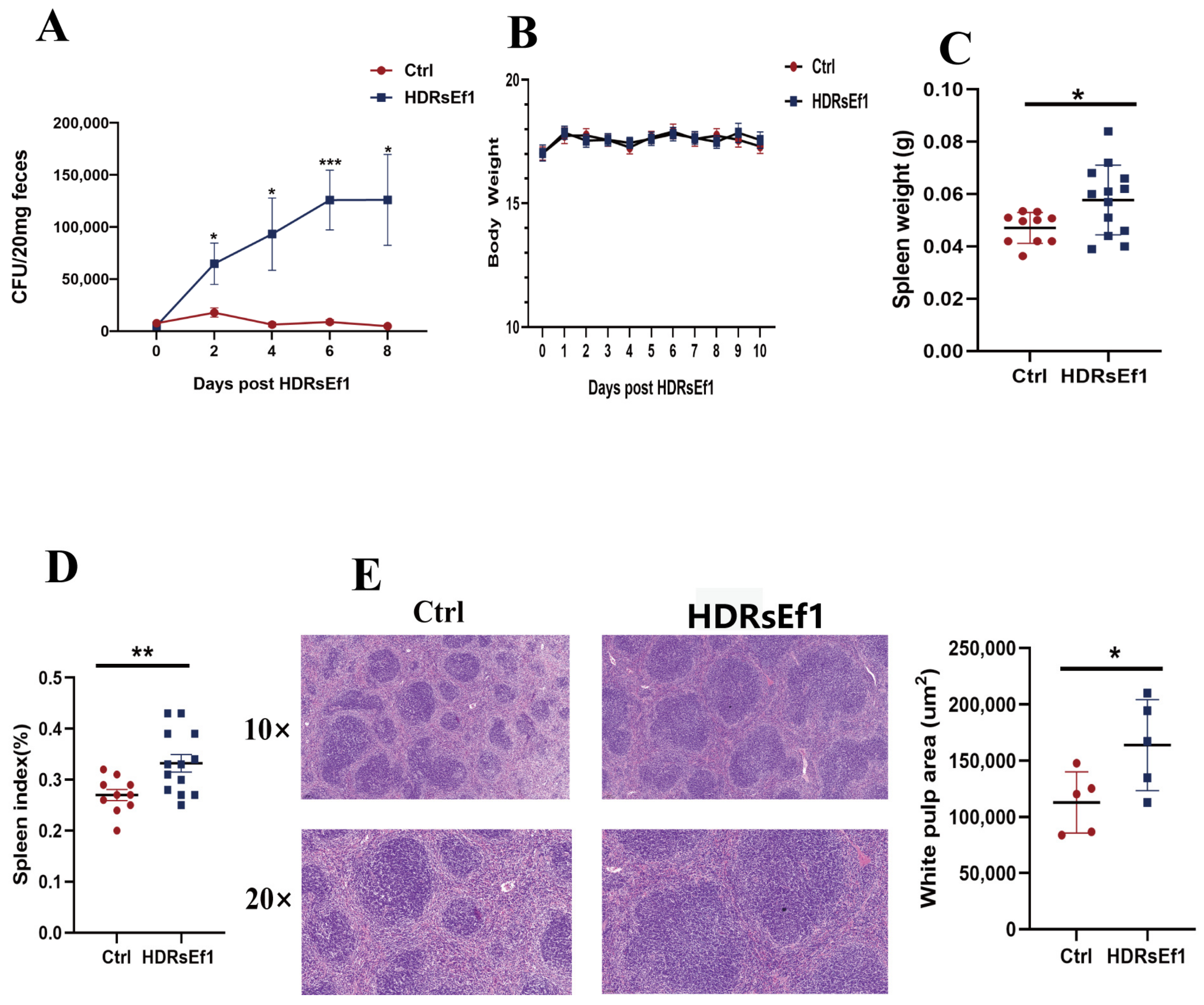
3.1. HDRsEf1 Promotes Spleen Development
3.2. HDRsEf1 Promotes Th1 Differentiation and Function in the Spleen
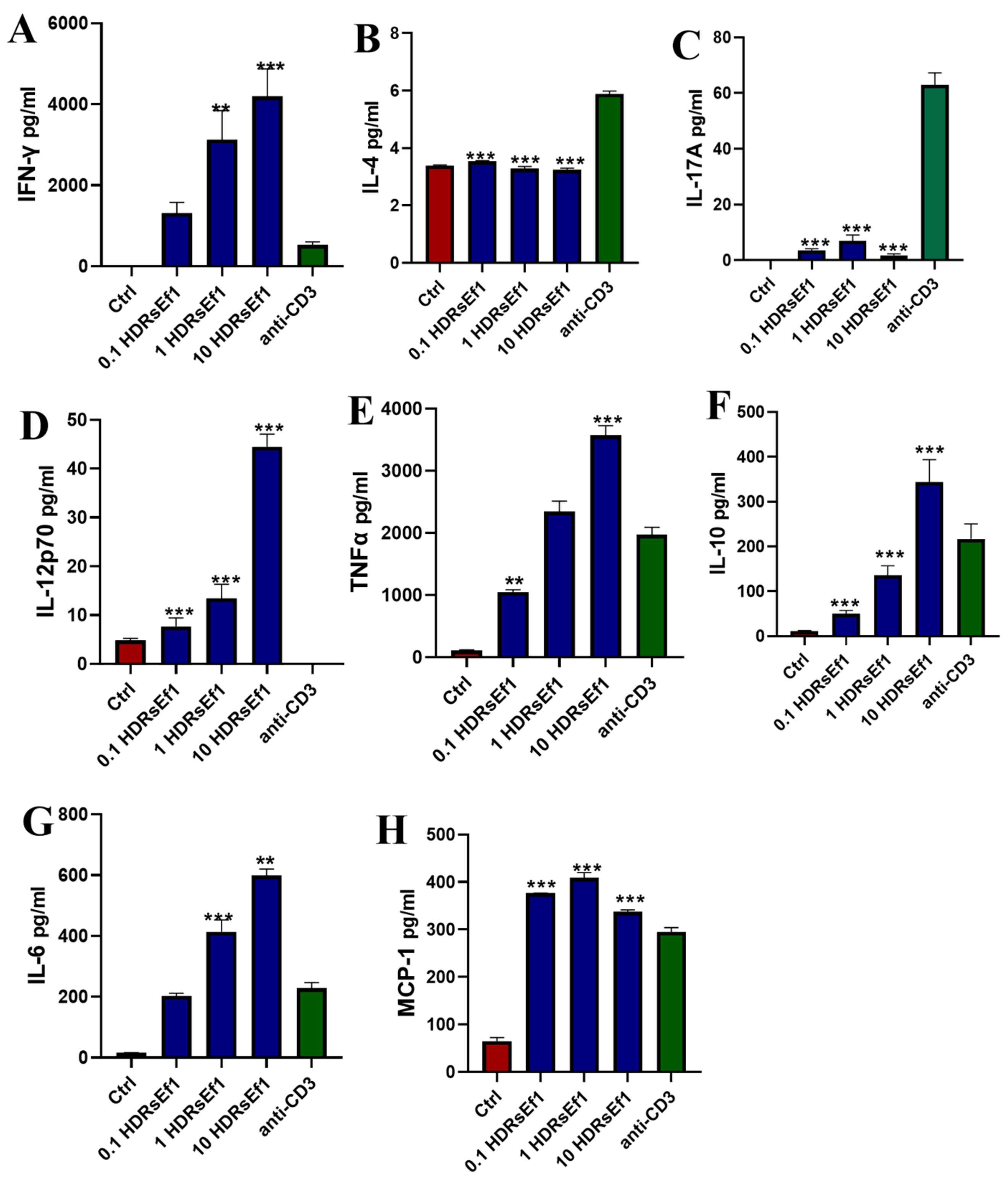
3.3. HDRsEf1 Strongly Promotes IFN-γ Production in Splenocytes In Vitro
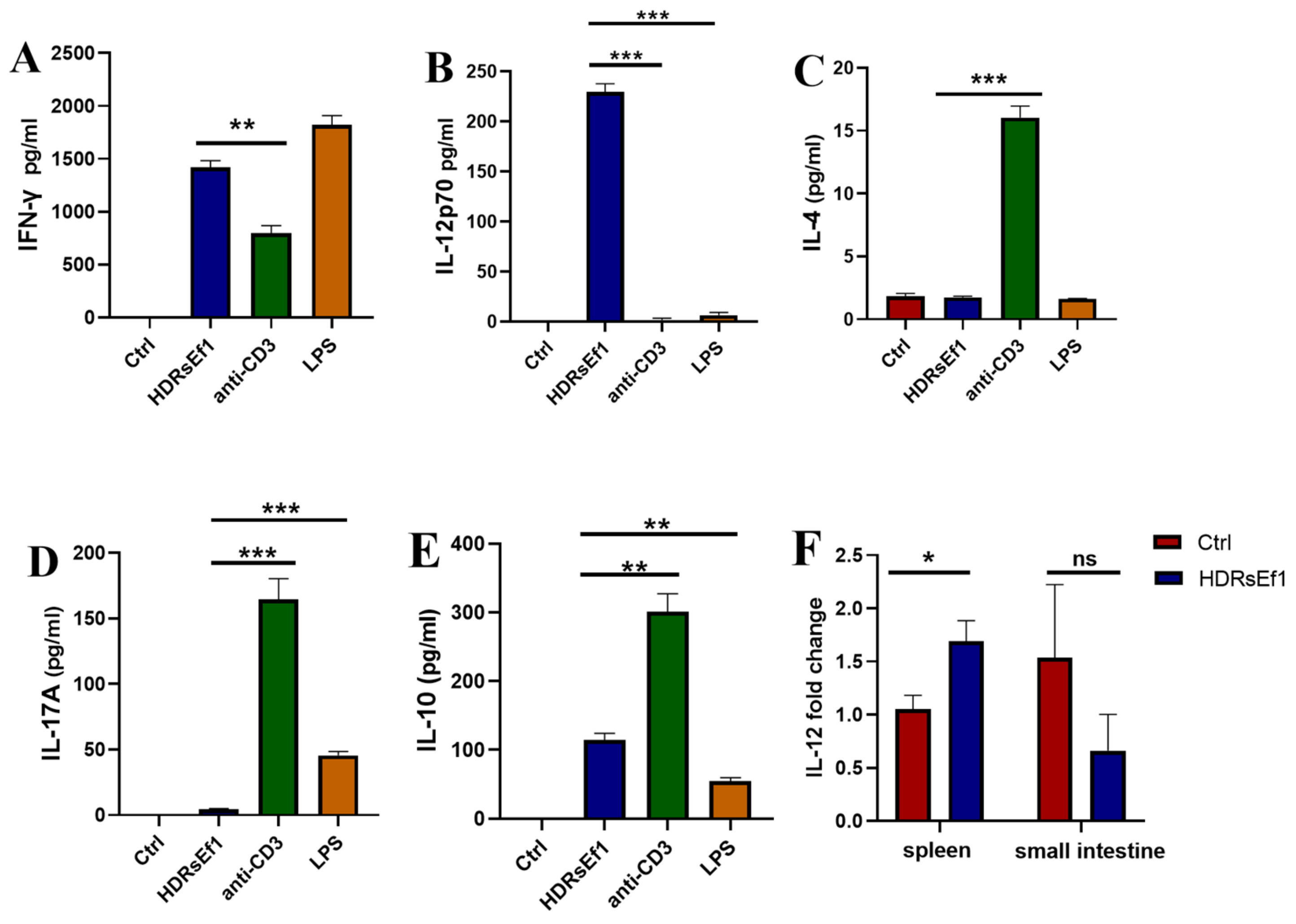
3.4. HDRsEf1 Induces IFN-γ in a Strain-Specific Manner
3.5. HDRsEf1 Promotes IL-12p70 in BMDCs and Spleen Tissue
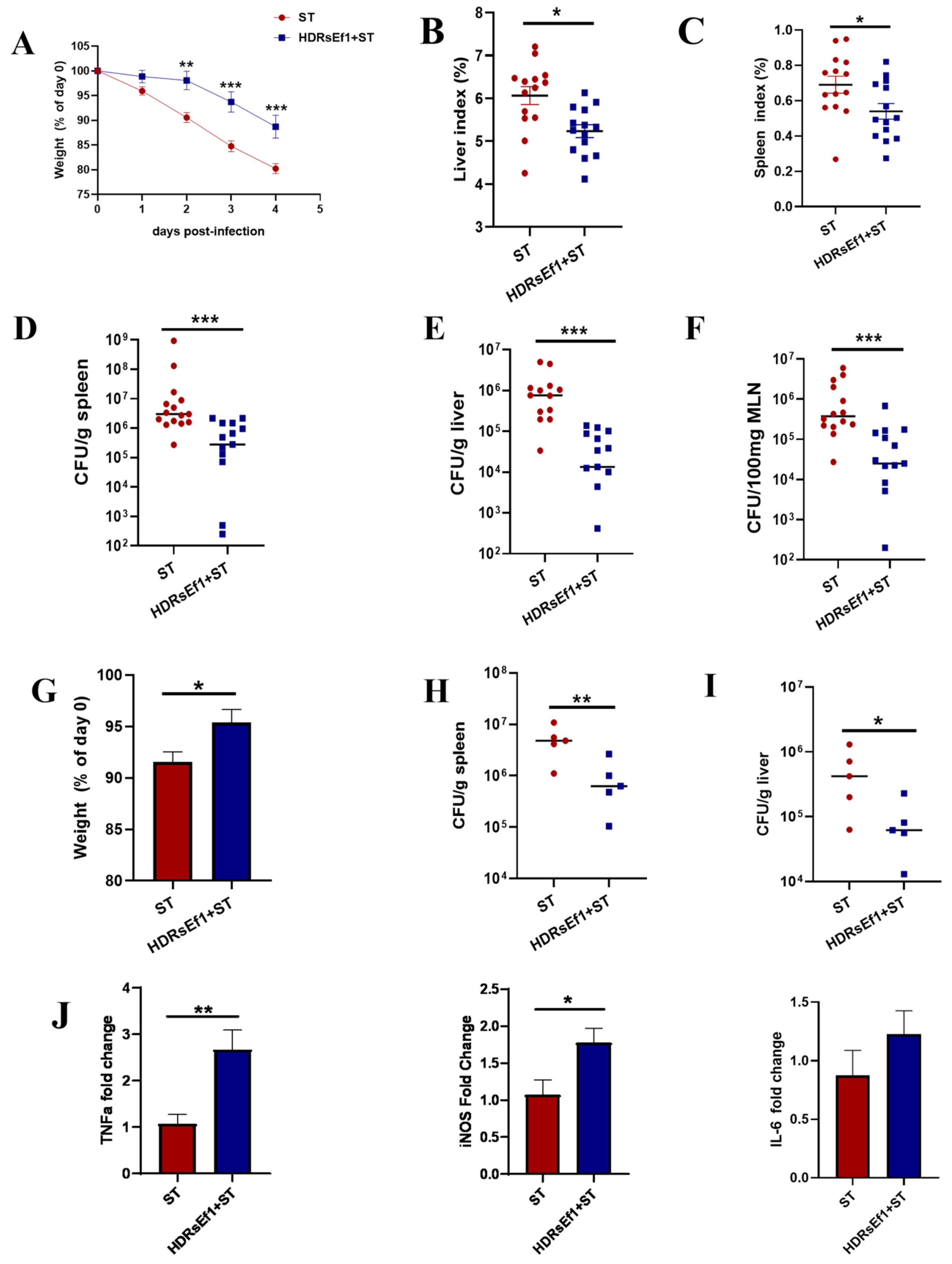
3.6. HDRsEf1 Enhances Resistance to Salmonella Typhimurium Infection in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Geuking, M.B.; Burkhard, R. Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunol. 2020, 13, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.H.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Thiemann, S.; Smit, N.; Roy, U.; Lesker, T.R.; Galvez, E.J.C.; Helmecke, J.; Basic, M.; Bleich, A.; Goodman, A.L.; Kalinke, U.; et al. Enhancement of IFNγ Production by Distinct Commensals Ameliorates Salmonella-Induced Disease. Cell Host Microbe 2017, 21, 682–694.e685. [Google Scholar] [CrossRef]
- Khosravi, A.; Yanez, A.; Price, J.G.; Chow, A.; Merad, M.; Goodridge, H.S.; Mazmanian, S.K. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014, 15, 374–381. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Nunez, G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef]
- Zegarra-Ruiz, D.F.; Kim, D.V.; Norwood, K.; Kim, M.; Wu, W.H.; Saldana-Morales, F.B.; Hill, A.A.; Majumdar, S.; Orozco, S.; Bell, R.; et al. Thymic development of gut-microbiota-specific T cells. Nature 2021, 594, 413–417. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, J.; Xu, X.; Wang, H.; Qiao, Y.; Chu, W.C.; Xu, S.; Chai, L.; Cottier, F.; Pavelka, N.; et al. Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 2019, 4, 766–773. [Google Scholar] [CrossRef]
- Schlechte, J.; Skalosky, I.; Geuking, M.B.; McDonald, B. Long-distance relationships—Regulation of systemic host defense against infections by the gut microbiota. Mucosal Immunol. 2022, 15, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Deriu, E.; Liu, J.Z.; Pezeshki, M.; Edwards, R.A.; Ochoa, R.J.; Contreras, H.; Libby, S.J.; Fang, F.C.; Raffatellu, M. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 2013, 14, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Li, M.; Liu, Z.; Xu, R.; Qiao, F.; Du, Z.Y.; Zhang, M.L. Pediococcus pentosaceus Enhances Host Resistance Against Pathogen by Increasing IL-1β Production: Understanding Probiotic Effectiveness and Administration Duration. Front. Immunol. 2021, 12, 766401. [Google Scholar] [CrossRef] [PubMed]
- Lemme-Dumit, J.M.; Cazorla, S.I.; Perdigon, G.D.V.; Maldonado-Galdeano, C. Probiotic Bacteria and Their Cell Walls Induce Th1-Type Immunity Against Salmonella Typhimurium Challenge. Front. Immunol. 2021, 12, 660854. [Google Scholar] [CrossRef] [PubMed]
- Helmy, Y.A.; Kassem, I.I.; Rajashekara, G. Immuno-modulatory effect of probiotic E. coli Nissle 1917 in polarized human colonic cells against Campylobacter jejuni infection. Gut Microbes 2021, 13, 1857514. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Bednorz, C.; Guenther, S.; Oelgeschlager, K.; Kinnemann, B.; Pieper, R.; Hartmann, S.; Tedin, K.; Semmler, T.; Neumann, K.; Schierack, P. Feeding the Probiotic Enterococcus faecium Strain NCIMB 10415 to Piglets Specifically Reduces the Number of Escherichia coli Pathotypes That Adhere to the Gut Mucosa. Appl. Environ. Microbiol. 2013, 79, 7896–7904. [Google Scholar] [CrossRef]
- Kumar, M.; Srivastava, S. Antilisterial activity of a broad-spectrum bacteriocin, enterocin LR/6 from Enterococcus faecium LR/6. Appl. Biochem. Biotechnol. 2010, 162, 698–706. [Google Scholar] [CrossRef]
- Pedicord, V.A.; Lockhart, A.A.K.; Rangan, K.J.; Craig, J.W.; Loschko, J.; Rogoz, A.; Hang, H.C.; Mucida, D. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol. 2016, 1, eaai7732. [Google Scholar] [CrossRef]
- Shi, D.; Xiao, Y.; Bi, D.; Xiong, Y.; Wang, X.; Gao, X.; Li, Z.; Zhou, Z.; Liu, M.; Xu, Q. A Beneficial Enterococcus faecium Strain’s Screening and Application. China Patent ZL201,110,452,087.2, 11 November 2013. [Google Scholar]
- Yan, T.; Zhang, F.; He, Y.; Wang, X.A.-O.; Jin, X.; Zhang, P.; Bi, D. Enterococcus faecium HDRsEf1 elevates the intestinal barrier defense against enterotoxigenic Escherichia coli and regulates occludin expression via activation of TLR-2 and PI3K signalling pathways. Lett. Appl. Microbiol. 2018, 67, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Liu, X.; Dai, R.; Xiao, Y.; Wang, X.; Bi, D.; Shi, D. Enterococcus faecium HDRsEf1 Protects the Intestinal Epithelium and Attenuates ETEC-Induced IL-8 Secretion in Enterocytes. Mediat. Inflamm. 2016, 2016, 7474306. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; He, Y.; Wang, Z.; Zhao, H.; Jin, X.; Shi, D.; Wang, X. Enterococcus faecium HDRsEf1 inhibits Lipopolysaccharide-induced downregulation of zona occludens-1 expression via toll-like receptor 2/4-mediated c-Jun N-terminal kinase/activator protein-1 signalling pathways. J. Appl. Microbiol. 2022, 132, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Xiao, Y.; HE, X.; Xu, W.; Wang, X.; Bi, D.; Shi, D. Study on probiotic agents substituting for antibiotics in weaned piglet diets. Prog. Vet. Med. 2014, 35, 53–58. [Google Scholar] [CrossRef]
- Mueller-Ortiz, S.L.; Shivshankar, P.; Wetsel, R.A. The Second Receptor for C5a, C5aR2, Is Detrimental to Mice during Systemic Infection with Listeria monocytogenes. J. Immunol. 2019, 203, 2701–2711. [Google Scholar] [CrossRef]
- Bastos, K.R.; Barboza, R.; Sardinha, L.; Russo, M.; Alvarez, J.M.; Lima, M.R. Role of endogenous IFN-γ in macrophage programming induced by IL-12 and IL-18. J. Interferon Cytokine Res. 2007, 27, 399–410. [Google Scholar] [CrossRef]
- Wu, C.; Xue, Y.; Wang, P.; Lin, L.; Liu, Q.; Li, N.; Xu, J.; Cao, X. IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b. J. Immunol. 2014, 193, 3036–3044. [Google Scholar] [CrossRef]
- Salcedo, S.P.; Noursadeghi, M.; Cohen, J.; Holden, D.W. Intracellular replication of Salmonella Typhimurium strains in specific subsets of splenic macrophages in vivo. Cell Microbiol. 2001, 3, 587–597. [Google Scholar] [CrossRef]
- Son, Y.I.; Egawa, S.I.; Tatsumi, T.; Redlinger, R.E., Jr.; Kalinski, P.; Kanto, T. A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J. Immunol. Methods 2002, 262, 145–157. [Google Scholar] [CrossRef]
- Barberi, C.; Campana, S.; De Pasquale, C.; Khorasgani, M.R.; Ferlazzo, G.; Bonaccorsi, I. T cell polarizing properties of probiotic bacteria. Immunol. Lett. 2015, 168, 337–342. [Google Scholar] [CrossRef]
- Makino, S.; Sato, A.; Goto, A.; Nakamura, M.; Ogawa, M.; Chiba, Y.; Hemmi, J.; Kano, H.; Takeda, K.; Okumura, K.; et al. Enhanced natural killer cell activation by exopolysaccharides derived from yogurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J. Dairy Sci. 2016, 99, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Taha-Abdelaziz, K.; Astill, J.; Kulkarni, R.R.; Read, L.R.; Najarian, A.; Farber, J.M.; Sharif, S. In vitro assessment of immunomodulatory and anti-Campylobacter activities of probiotic lactobacilli. Sci. Rep. 2019, 9, 17903. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.; McCoy, K.D.; Macpherson, A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007, 19, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.G.; Sefik, E.; Geva-Zatorsky, N.; Kua, L.; Naskar, D.; Teng, F.; Pasman, L.; Ortiz-Lopez, A.; Jupp, R.; Wu, H.J.; et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E8141–E8150. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Ju, D.B.; Kye, Y.C.; Ju, Y.J.; Kim, C.G.; Lee, I.K.; Park, S.M.; Choi, I.S.; Cho, K.K.; Lee, S.H.; et al. Galectin-9 Induced by Dietary Probiotic Mixture Regulates Immune Balance to Reduce Atopic Dermatitis Symptoms in Mice. Front. Immunol. 2019, 10, 3063. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, R.; Shah, N.P. Immune system stimulation by probiotic microorganisms. Crit. Rev. Food Sci. Nutr. 2014, 54, 938–956. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C., Jr. Innate immune recognition: Mechanisms and pathways. Immunol. Rev. 2000, 173, 89–97. [Google Scholar] [CrossRef]
- Morinobu, A.; Gadina, M.; Strober, W.; Visconti, R.; Fornace, A.; Montagna, C.; Feldman, G.M.; Nishikomori, R.; O’Shea, J. STAT4 serine phosphorylation is critical for IL-12-induced IFN-γ production but not for cell proliferation. Proc. Natl. Acad. Sci. USA 2002, 99, 12281–12286. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Chiba, Y.; Shida, K.; Nagata, S.; Wada, M.; Bian, L.; Wang, C.; Shimizu, T.; Yamashiro, Y.; Kiyoshima-Shibata, J.; Nanno, M.; et al. Well-controlled proinflammatory cytokine responses of Peyer’s patch cells to probiotic Lactobacillus casei. Immunology 2010, 130, 352–362. [Google Scholar] [CrossRef]
- Clarke, T.B.; Davis, K.M.; Lysenko, E.S.; Zhou, A.Y.; Yu, Y.; Weiser, J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010, 16, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, M.; Gautam, U.K.; Makki, K.; Lambert, A.; Brabec, T.; Joly, A.; Srutkova, D.; Poinsot, P.; Novotna, T.; Geoffroy, S.; et al. Microbe-mediated intestinal NOD2 stimulation improves linear growth of undernourished infant mice. Science 2023, 379, 826–833. [Google Scholar] [CrossRef]
- Coburn, B.; Grassl, G.A.; Finlay, B.B. Salmonella, the host and disease: A brief review. Immunol. Cell Biol. 2007, 85, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Eisele, N.A.; Ruby, T.; Jacobson, A.; Manzanillo, P.S.; Cox, J.S.; Lam, L.; Mukundan, L.; Chawla, A.; Monack, D.M. Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 2013, 14, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Monack, D.M.; Bouley, D.M.; Falkow, S. Salmonella Typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J. Exp. Med. 2004, 199, 231–241. [Google Scholar] [CrossRef]
- Khan, N.; Vidyarthi, A.; Nadeem, S.; Negi, S.; Nair, G.; Agrewala, J.N. Alteration in the Gut Microbiota Provokes Susceptibility to Tuberculosis. Front. Immunol. 2016, 7, 529. [Google Scholar] [CrossRef]
- Braverman, J.; Stanley, S.A.-O. Nitric Oxide Modulates Macrophage Responses to Mycobacterium tuberculosis Infection through Activation of HIF-1α and Repression of NF-κB. J. Immunol. 2017, 199, 1805–1816. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Wang, T.; Fan, L.; Xiao, H.; Ji, H.; Zhou, N.; Zhou, Z.; Liu, H.; Akhtar, M.; Xiao, Y.; et al. Enterococcus faecium HDRsEf1 Promotes Systemic Th1 Responses and Enhances Resistance to SalmonellaTyphimurium Infection. Nutrients 2023, 15, 4241. https://doi.org/10.3390/nu15194241
Zhou J, Wang T, Fan L, Xiao H, Ji H, Zhou N, Zhou Z, Liu H, Akhtar M, Xiao Y, et al. Enterococcus faecium HDRsEf1 Promotes Systemic Th1 Responses and Enhances Resistance to SalmonellaTyphimurium Infection. Nutrients. 2023; 15(19):4241. https://doi.org/10.3390/nu15194241
Chicago/Turabian StyleZhou, Jin, Tingyang Wang, Lele Fan, Hongde Xiao, Hui Ji, Naiji Zhou, Zutao Zhou, Huazhen Liu, Muhammad Akhtar, Yuncai Xiao, and et al. 2023. "Enterococcus faecium HDRsEf1 Promotes Systemic Th1 Responses and Enhances Resistance to SalmonellaTyphimurium Infection" Nutrients 15, no. 19: 4241. https://doi.org/10.3390/nu15194241
APA StyleZhou, J., Wang, T., Fan, L., Xiao, H., Ji, H., Zhou, N., Zhou, Z., Liu, H., Akhtar, M., Xiao, Y., & Shi, D. (2023). Enterococcus faecium HDRsEf1 Promotes Systemic Th1 Responses and Enhances Resistance to SalmonellaTyphimurium Infection. Nutrients, 15(19), 4241. https://doi.org/10.3390/nu15194241







