D-Allulose Ameliorates Dysregulated Macrophage Function and Mitochondrial NADH Homeostasis, Mitigating Obesity-Induced Insulin Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diet
2.2. mRNA Sequencing and Data Processing
2.3. GSEA Pre-Ranked Analysis
2.4. Patient Data Acquisition and Processing
2.5. Pathway Analysis and Data Visualization
3. Results
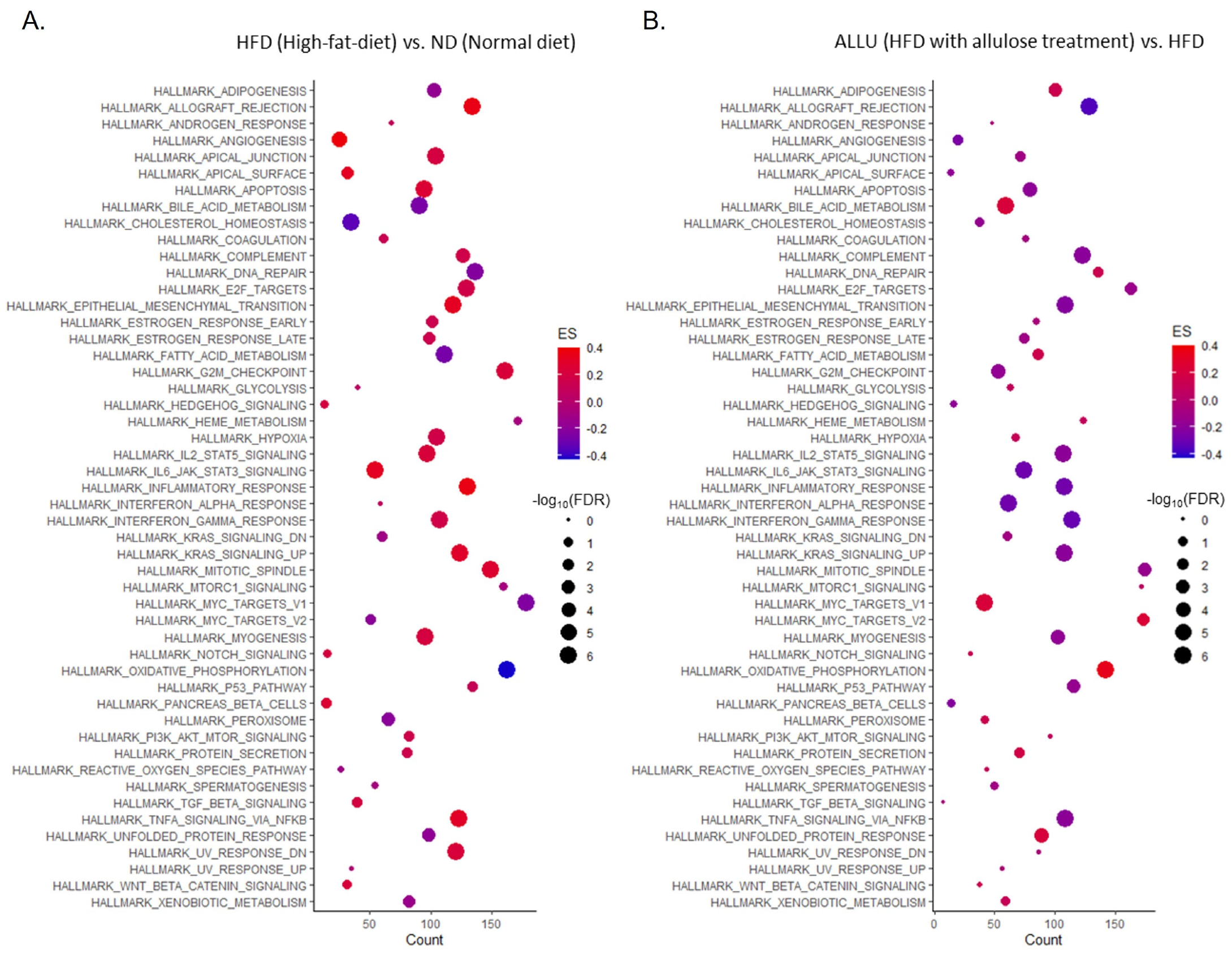
3.1. Time-Dependent Hallmark Gene Set Analysis in the Liver and eWAT from HFD Mice
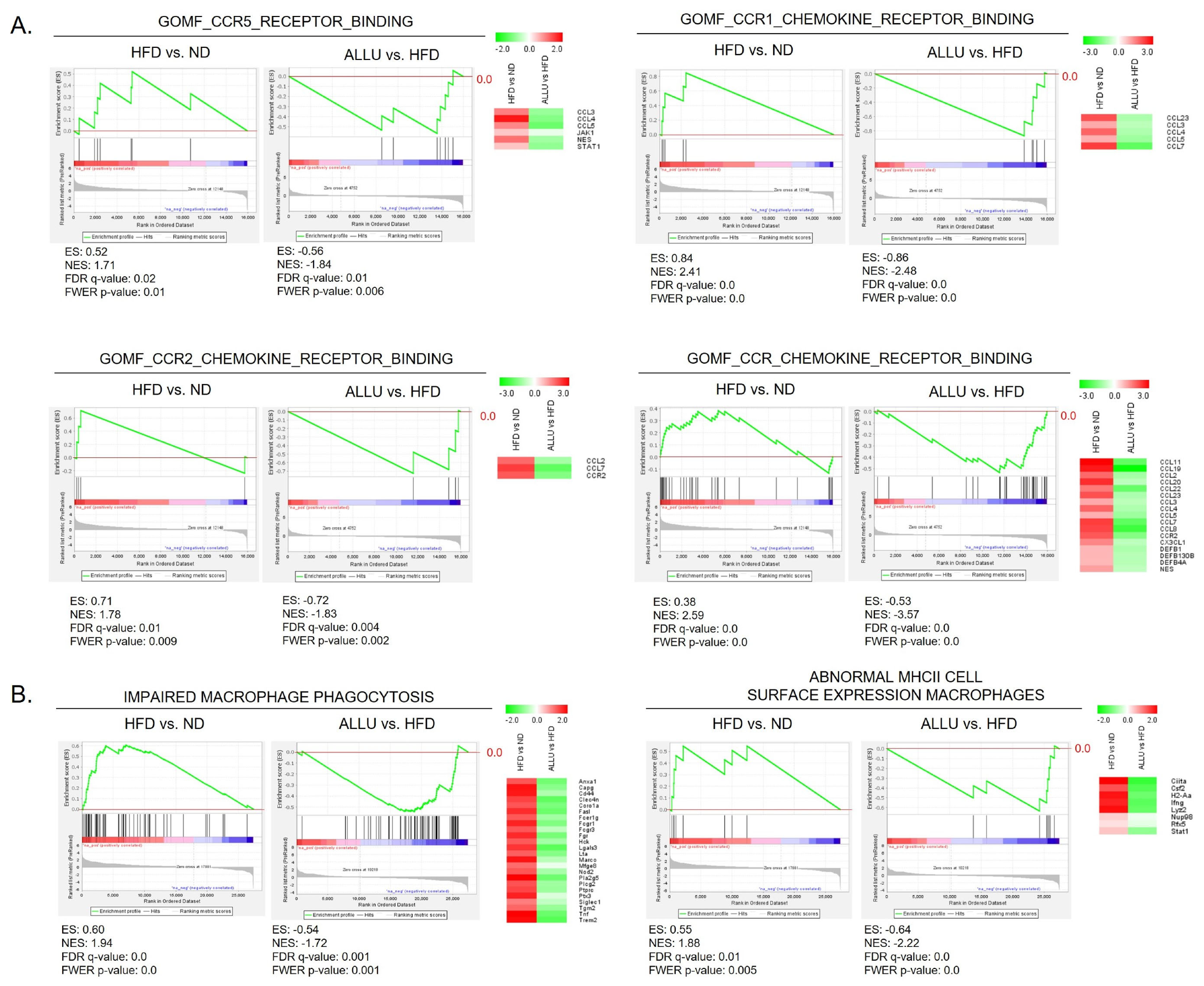
3.2. Suppression of Chemokine Expression and Dysregulated Macrophage Function by Allulose in the Liver and eWAT from HFD Mice
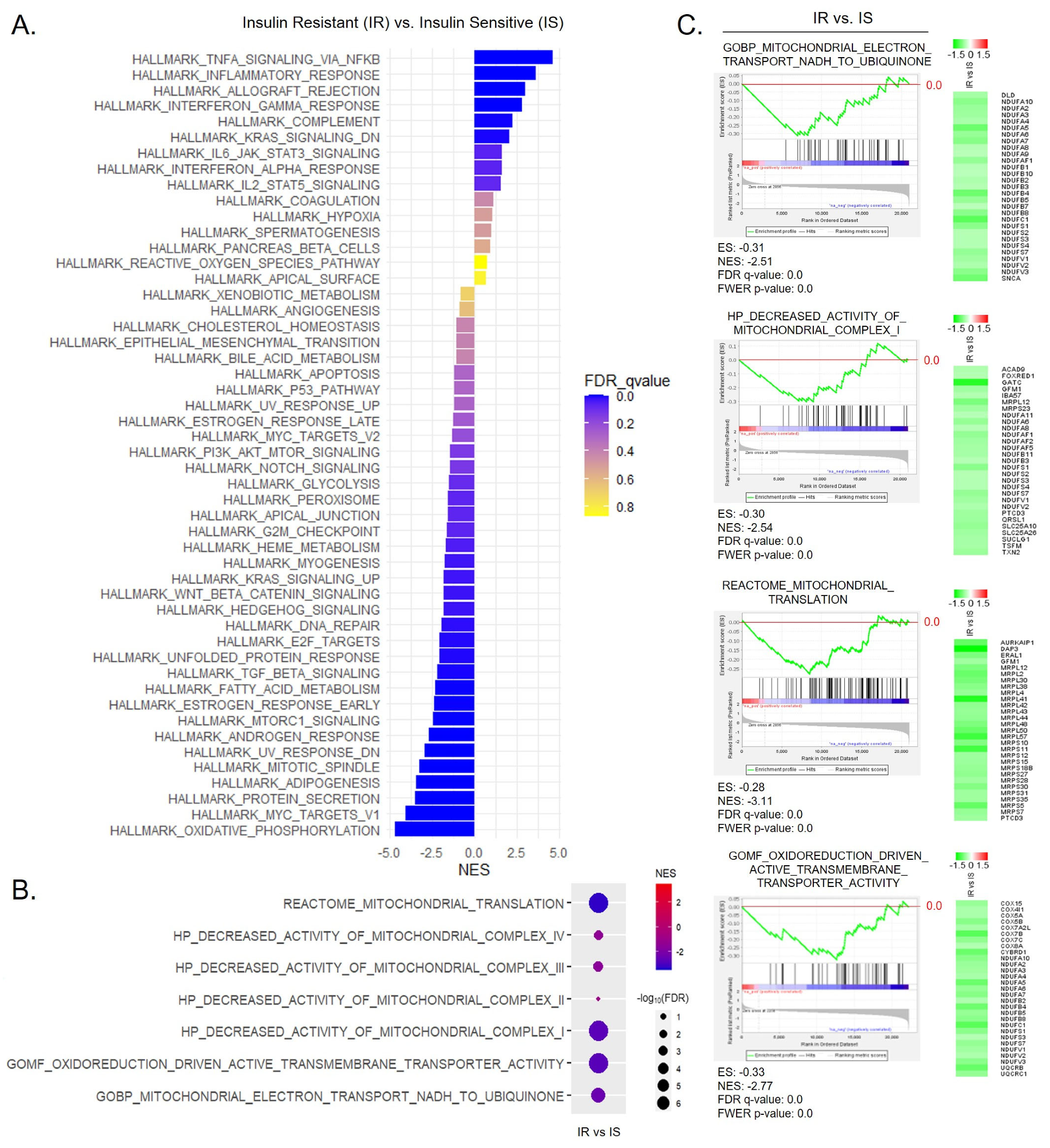
3.3. Correlation between Mitochondrial Energy Expenditure and mRNA Translation Processes in the PBMCs from T2D Patients
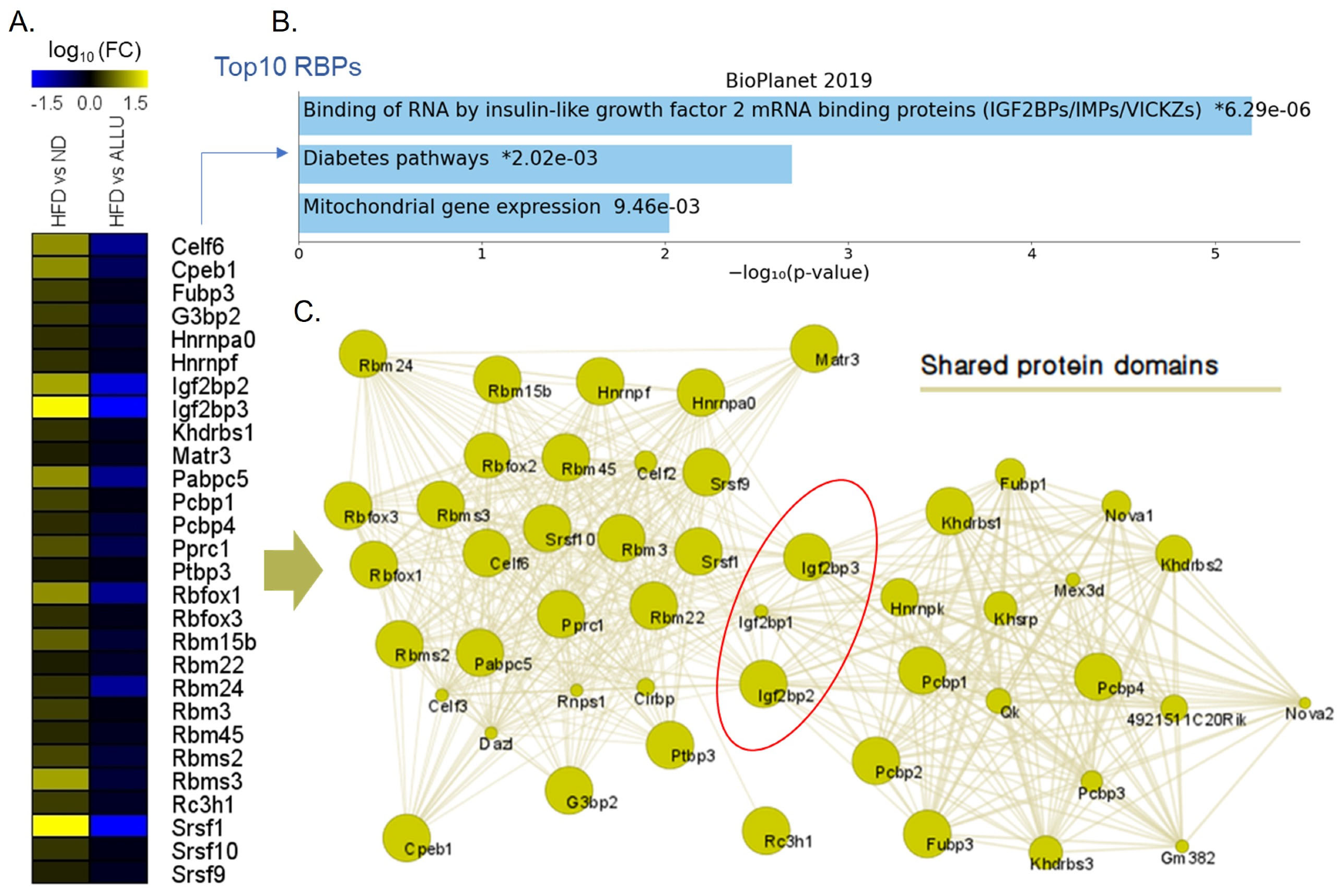
3.4. Effect of Allulose on RBPs and Its Potential Implications for Hepatic Insulin Resistance Triggered by HFD
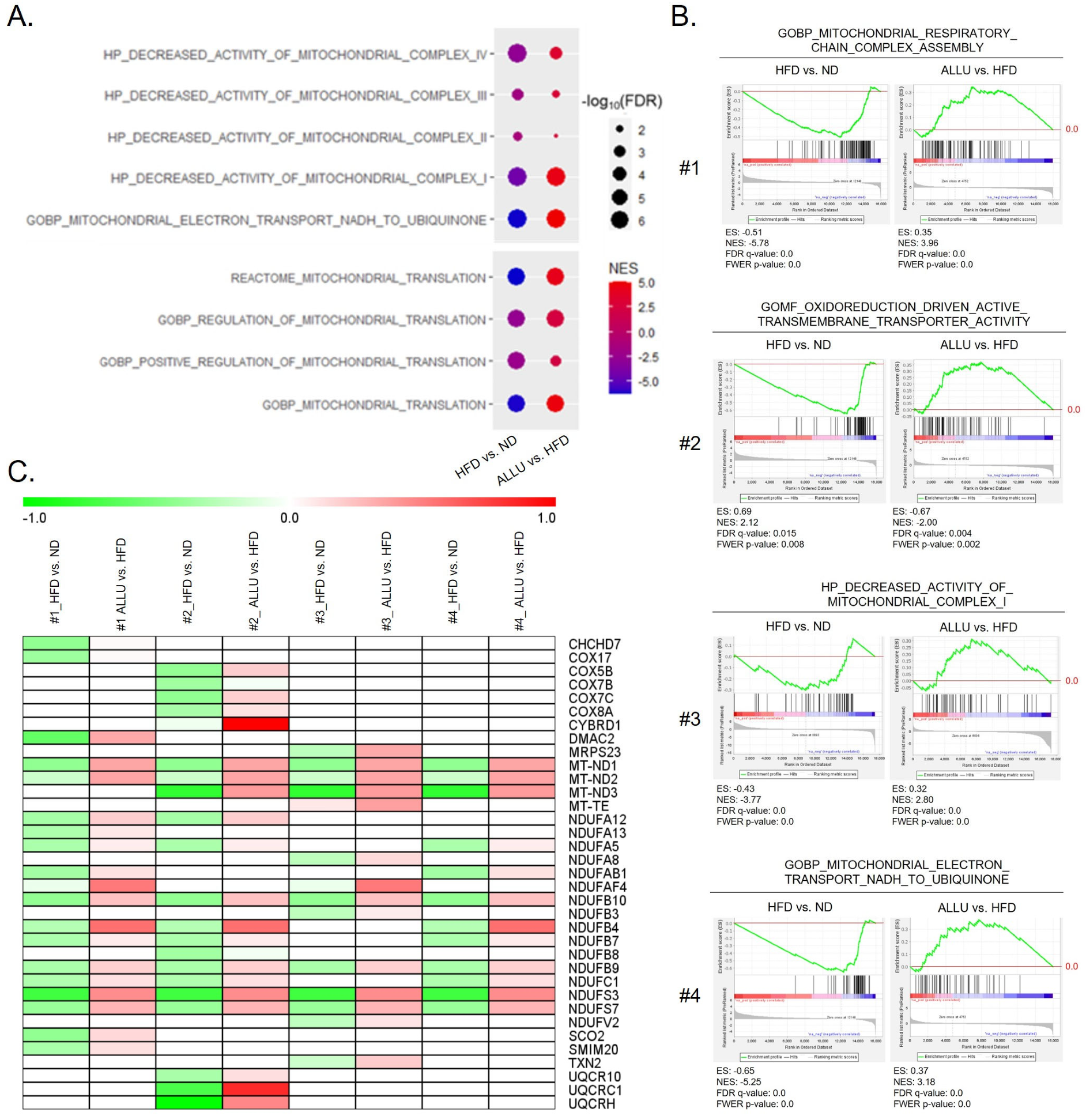
3.5. Effect of Allulose on Altered Mitochondrial NADH Homeostasis and Mitochondrial Translation in the Liver and eWAT from HFD Mice
3.6. Alteration of Mitochondrial NADH Homeostasis and Mitochondrial Translation in the Omental Adipose Tissue Obtained from Human Obese Subjects with Insulin Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, K.; Kim, H.J.; Oh, D.K.; Cha, S.S.; Rhee, S. Crystal structure of D-psicose 3-epimerase from Agrobacterium tumefaciens and its complex with true substrate D-fructose: A pivotal role of metal in catalysis, an active site for the non-phosphorylated substrate, and its conformational changes. J. Mol. Biol. 2006, 361, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Suzuki, H.; Hashiguchi, M.; Izumori, K. D-psicose is a rare sugar that provides no energy to growing rats. J. Nutr. Sci. Vitaminol. 2002, 48, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Xiao, W.; Zhu, X.; Yang, P.; Zheng, Z.; Lu, S.; Jiang, S.; Zhang, G.; Liu, J. Review on D-Allulose: In vivo Metabolism, Catalytic Mechanism, Engineering Strain Construction, Bio-Production Technology. Front. Bioeng. Biotechnol. 2020, 8, 26. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, X.D.; Li, Z. Recent Advances Regarding the Physiological Functions and Biosynthesis of D-Allulose. Front. Microbiol. 2022, 13, 881037. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Han, H.J.; Kim, A.H.; Choi, J.Y.; Cho, S.J.; Park, Y.B.; Jung, U.J.; Choi, M.S. D-Allulose supplementation normalized the body weight and fat-pad mass in diet-induced obese mice via the regulation of lipid metabolism under isocaloric fed condition. Mol. Nutr. Food Res. 2016, 60, 1695–1706. [Google Scholar] [CrossRef]
- Lee, D.; Han, Y.; Kwon, E.Y.; Choi, M.S. D-allulose Ameliorates Metabolic Dysfunction in C57BL/KsJ-db/db Mice. Molecules 2020, 25, 3656. [Google Scholar] [CrossRef]
- Han, Y.; Kwon, E.Y.; Choi, M.S. Anti-Diabetic Effects of Allulose in Diet-Induced Obese Mice via Regulation of mRNA Expression and Alteration of the Microbiome Composition. Nutrients 2020, 12, 2113. [Google Scholar] [CrossRef] [PubMed]
- Yuma, T.; Tokuda, M.; Nishimoto, N.; Yokoi, H.; Izumori, K. Allulose for the attenuation of postprandial blood glucose levels in healthy humans: A systematic review and meta-analysis. PLoS ONE 2023, 18, e0281150. [Google Scholar] [CrossRef]
- Fukunaga, K.; Yoshimura, T.; Imachi, H.; Kobayashi, T.; Saheki, T.; Sato, S.; Saheki, N.; Jiang, W.; Murao, K. A Pilot Study on the Efficacy of a Diabetic Diet Containing the Rare Sugar D-Allulose in Patients with Type 2 Diabetes Mellitus: A Prospective, Randomized, Single-Blind, Crossover Study. Nutrients 2023, 15, 2802. [Google Scholar] [CrossRef]
- Hossain, A.; Yamaguchi, F.; Matsuo, T.; Tsukamoto, I.; Toyoda, Y.; Ogawa, M.; Nagata, Y.; Tokuda, M. Rare sugar D-allulose: Potential role and therapeutic monitoring in maintaining obesity and type 2 diabetes mellitus. Pharmacol. Ther. 2015, 155, 49–59. [Google Scholar] [CrossRef]
- Franchi, F.; Yaranov, D.M.; Rollini, F.; Rivas, A.; Rivas Rios, J.; Been, L.; Tani, Y.; Tokuda, M.; Iida, T.; Hayashi, N.; et al. Effects of D-allulose on glucose tolerance and insulin response to a standard oral sucrose load: Results of a prospective, randomized, crossover study. BMJ Open Diabetes Res Care 2021, 9, e001939. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.; Sendo, M.; Dezaki, K.; Hira, T.; Sato, T.; Nakata, M.; Goswami, C.; Aoki, R.; Arai, T.; Kumari, P.; et al. GLP-1 release and vagal afferent activation mediate the beneficial metabolic and chronotherapeutic effects of D-allulose. Nat. Commun. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, M.; Hiraiwa, T.; Wada, Y.; Kuwayama, H.; Shibata, T. Polar pattern formation induced by contact following locomotion in a multicellular system. eLife 2020, 9, e53609. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Yamada, T.; Hayashi, N.; Iida, T.; Nagata, Y.; Ozaki, N.; Toyoda, Y. Rare Sugar Syrup Containing d-Allulose but Not High-Fructose Corn Syrup Maintains Glucose Tolerance and Insulin Sensitivity Partly via Hepatic Glucokinase Translocation in Wistar Rats. J. Agric. Food Chem. 2017, 65, 2888–2894. [Google Scholar] [CrossRef] [PubMed]
- Yermek, R.; Wang, L.; Kaneko, K.; Han, W.; Seino, Y.; Yabe, D.; Yada, T. D-Allulose cooperates with glucagon-like peptide-1 and activates proopiomelanocortin neurons in the arcuate nucleus and central injection inhibits feeding in mice. Biochem. Biophys. Res. Commun. 2022, 613, 159–165. [Google Scholar] [CrossRef]
- Khanna, D.; Khanna, S.; Khanna, P.; Kahar, P.; Patel, B.M. Obesity: A Chronic Low-Grade Inflammation and Its Markers. Cureus 2022, 14, e22711. [Google Scholar] [CrossRef]
- Rohm, T.V.; Meier, D.T.; Olefsky, J.M.; Donath, M.Y. Inflammation in obesity, diabetes, and related disorders. Immunity 2022, 55, 31–55. [Google Scholar] [CrossRef]
- Bae, H.R.; Shin, S.K.; Yoo, J.H.; Kim, S.; Young, H.A.; Kwon, E.Y. Chronic inflammation in high-fat diet-fed mice: Unveiling the early pathogenic connection between liver and adipose tissue. J. Autoimmun. 2023, 139, 103091. [Google Scholar] [CrossRef]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Zyla, J.; Marczyk, M.; Weiner, J.; Polanska, J. Ranking metrics in gene set enrichment analysis: Do they matter? BMC Bioinform. 2017, 18, 256. [Google Scholar] [CrossRef] [PubMed]
- Hardy, O.T.; Perugini, R.A.; Nicoloro, S.M.; Gallagher-Dorval, K.; Puri, V.; Straubhaar, J.; Czech, M.P. Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity. Surg. Obes. Relat. Dis. 2011, 7, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.R.; Choi, M.S.; Kim, S.; Young, H.A.; Gershwin, M.E.; Jeon, S.M.; Kwon, E.Y. IFNgamma is a Key Link between Obesity and Th1-Mediated AutoImmune Diseases. Int. J. Mol. Sci. 2020, 22, 208. [Google Scholar] [CrossRef]
- Bae, H.R.; Leung, P.S.C.; Hodge, D.L.; Fenimore, J.M.; Jeon, S.M.; Thovarai, V.; Dzutsev, A.; Welcher, A.A.; Boedigheimer, M.; Damore, M.A.; et al. Multi-omics: Differential expression of IFN-gamma results in distinctive mechanistic features linking chronic inflammation, gut dysbiosis, and autoimmune diseases. J. Autoimmun. 2020, 111, 102436. [Google Scholar] [CrossRef]
- Eltayeb, S.; Berg, A.L.; Lassmann, H.; Wallstrom, E.; Nilsson, M.; Olsson, T.; Ericsson-Dahlstrand, A.; Sunnemark, D. Temporal expression and cellular origin of CC chemokine receptors CCR1, CCR2 and CCR5 in the central nervous system: Insight into mechanisms of MOG-induced EAE. J. Neuroinflamm. 2007, 4, 14. [Google Scholar] [CrossRef]
- Kaufmann, A.; Salentin, R.; Gemsa, D.; Sprenger, H. Increase of CCR1 and CCR5 expression and enhanced functional response to MIP-1 alpha during differentiation of human monocytes to macrophages. J. Leukoc. Biol. 2001, 69, 248–252. [Google Scholar] [CrossRef]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef]
- Li, H.; Meng, Y.; He, S.; Tan, X.; Zhang, Y.; Zhang, X.; Wang, L.; Zheng, W. Macrophages, Chronic Inflammation, and Insulin Resistance. Cells 2022, 11, 3001. [Google Scholar] [CrossRef]
- Ieronymaki, E.; Theodorakis, E.M.; Lyroni, K.; Vergadi, E.; Lagoudaki, E.; Al-Qahtani, A.; Aznaourova, M.; Neofotistou-Themeli, E.; Eliopoulos, A.G.; Vaporidi, K.; et al. Insulin Resistance in Macrophages Alters Their Metabolism and Promotes an M2-Like Phenotype. J. Immunol. 2019, 202, 1786–1797. [Google Scholar] [CrossRef]
- Orliaguet, L.; Dalmas, E.; Drareni, K.; Venteclef, N.; Alzaid, F. Mechanisms of Macrophage Polarization in Insulin Signaling and Sensitivity. Front. Endocrinol. 2020, 11, 62. [Google Scholar] [CrossRef]
- Hojlund, K.; Mogensen, M.; Sahlin, K.; Beck-Nielsen, H. Mitochondrial dysfunction in type 2 diabetes and obesity. Endocrinol. Metab. Clin. N. Am. 2008, 37, 713–731. [Google Scholar] [CrossRef] [PubMed]
- Krako Jakovljevic, N.; Pavlovic, K.; Jotic, A.; Lalic, K.; Stoiljkovic, M.; Lukic, L.; Milicic, T.; Macesic, M.; Stanarcic Gajovic, J.; Lalic, N.M. Targeting Mitochondria in Diabetes. Int. J. Mol. Sci. 2021, 22, 6642. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.; Apostolova, N.; Diaz-Rua, R.; Muntane, J.; Victor, V.M. Mitochondria and T2D: Role of Autophagy, ER Stress, and Inflammasome. Trends Endocrinol. Metab. 2020, 31, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, A.; Torrecilla-Parra, M.; Fernandez-de Frutos, M.; Martin-Martin, Y.; Pardo-Marques, V.; Ramirez, C.M. Posttranscriptional Regulation of Insulin Resistance: Implications for Metabolic Diseases. Biomolecules 2022, 12, 208. [Google Scholar] [CrossRef]
- Jannig, P.R.; Ruas, J.L. Targeting mitochondrial mRNA translation to tackle obesity-induced insulin resistance: Thumbs up for exercise. Acta Physiol. 2017, 219, 14–16. [Google Scholar] [CrossRef][Green Version]
- Nutter, C.A.; Kuyumcu-Martinez, M.N. Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wiley Interdiscip. Rev. RNA 2018, 9, e1459. [Google Scholar] [CrossRef]
- Chen, X.; Wu, J.; Li, Z.; Han, J.; Xia, P.; Shen, Y.; Ma, J.; Liu, X.; Zhang, J.; Yu, P. Advances in the study of RNA-binding proteins in diabetic complications. Mol. Metab. 2022, 62, 101515. [Google Scholar] [CrossRef]
- Martinez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Wu, J.; Jin, Z.; Zheng, H.; Yan, L.J. Sources and implications of NADH/NAD(+) redox imbalance in diabetes and its complications. Diabetes Metab. Syndr. Obes. 2016, 9, 145–153. [Google Scholar] [CrossRef]
- Titov, D.V.; Cracan, V.; Goodman, R.P.; Peng, J.; Grabarek, Z.; Mootha, V.K. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 2016, 352, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, H.R.; Shin, S.-K.; Han, Y.; Yoo, J.-H.; Kim, S.; Young, H.A.; Kwon, E.-Y. D-Allulose Ameliorates Dysregulated Macrophage Function and Mitochondrial NADH Homeostasis, Mitigating Obesity-Induced Insulin Resistance. Nutrients 2023, 15, 4218. https://doi.org/10.3390/nu15194218
Bae HR, Shin S-K, Han Y, Yoo J-H, Kim S, Young HA, Kwon E-Y. D-Allulose Ameliorates Dysregulated Macrophage Function and Mitochondrial NADH Homeostasis, Mitigating Obesity-Induced Insulin Resistance. Nutrients. 2023; 15(19):4218. https://doi.org/10.3390/nu15194218
Chicago/Turabian StyleBae, Heekyong R., Su-Kyung Shin, Youngji Han, Ji-Hyeon Yoo, Suntae Kim, Howard A. Young, and Eun-Young Kwon. 2023. "D-Allulose Ameliorates Dysregulated Macrophage Function and Mitochondrial NADH Homeostasis, Mitigating Obesity-Induced Insulin Resistance" Nutrients 15, no. 19: 4218. https://doi.org/10.3390/nu15194218
APA StyleBae, H. R., Shin, S.-K., Han, Y., Yoo, J.-H., Kim, S., Young, H. A., & Kwon, E.-Y. (2023). D-Allulose Ameliorates Dysregulated Macrophage Function and Mitochondrial NADH Homeostasis, Mitigating Obesity-Induced Insulin Resistance. Nutrients, 15(19), 4218. https://doi.org/10.3390/nu15194218





