Decreased Fatty Acid Oxidation Gene Expression in Pre-Eclampsia According to the Onset and Presence of Intrauterine Growth Restriction
Abstract
:1. Introduction
1.1. Etiopathogenesis of Pre-Eclampsia
1.2. Placental Lipid Metabolism
1.3. Purpose of the Study
2. Materials and Methods
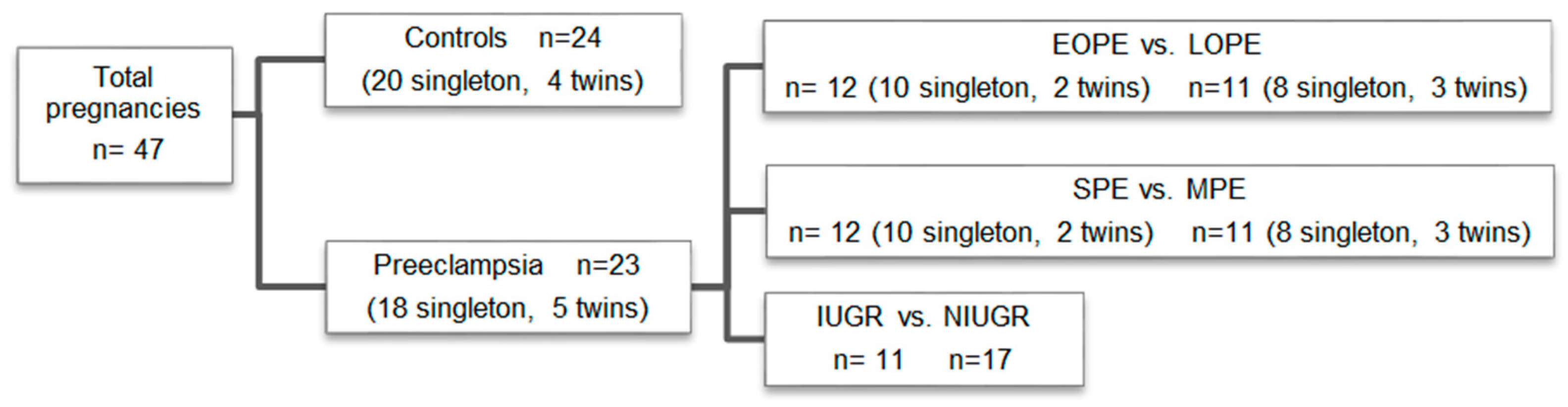
2.1. Study Participants
2.2. Sample Collection
2.3. Quantification of Placental mRNA Levels
2.4. Statistical Analysis
3. Results
3.1. Maternal, Obstetric, and Perinatal Outcomes
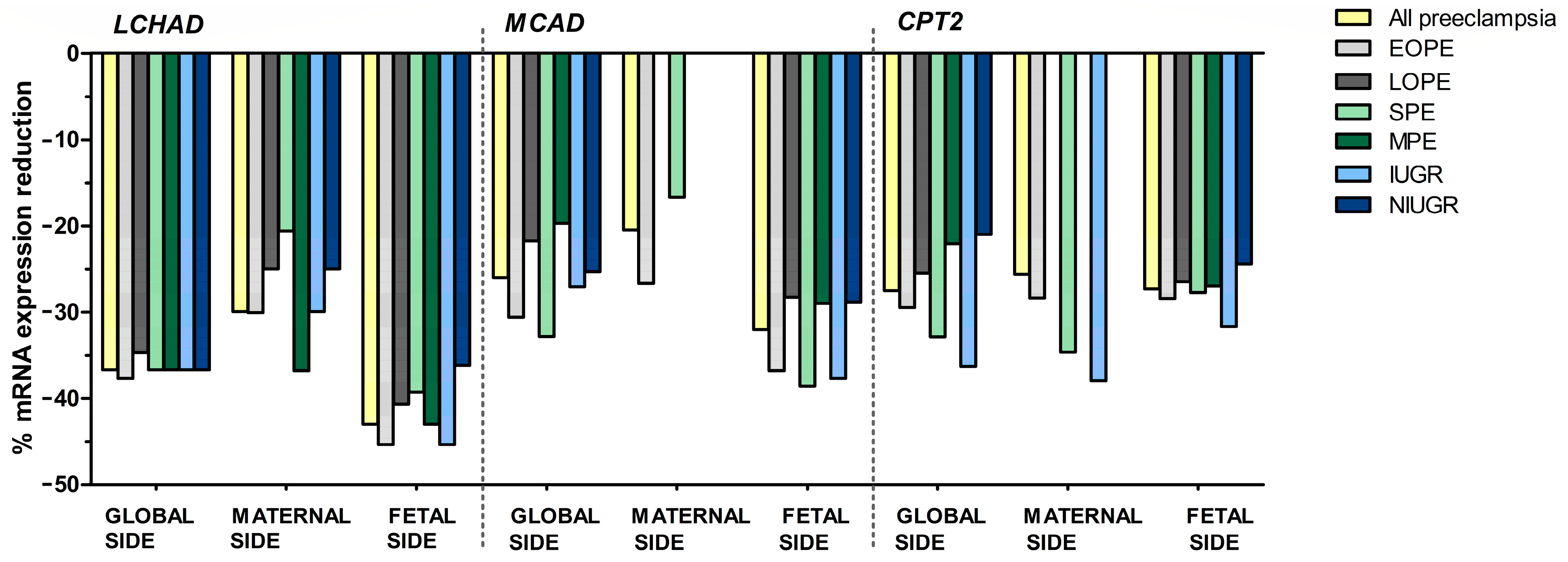
3.2. Analysis of the Placental mRNA Levels
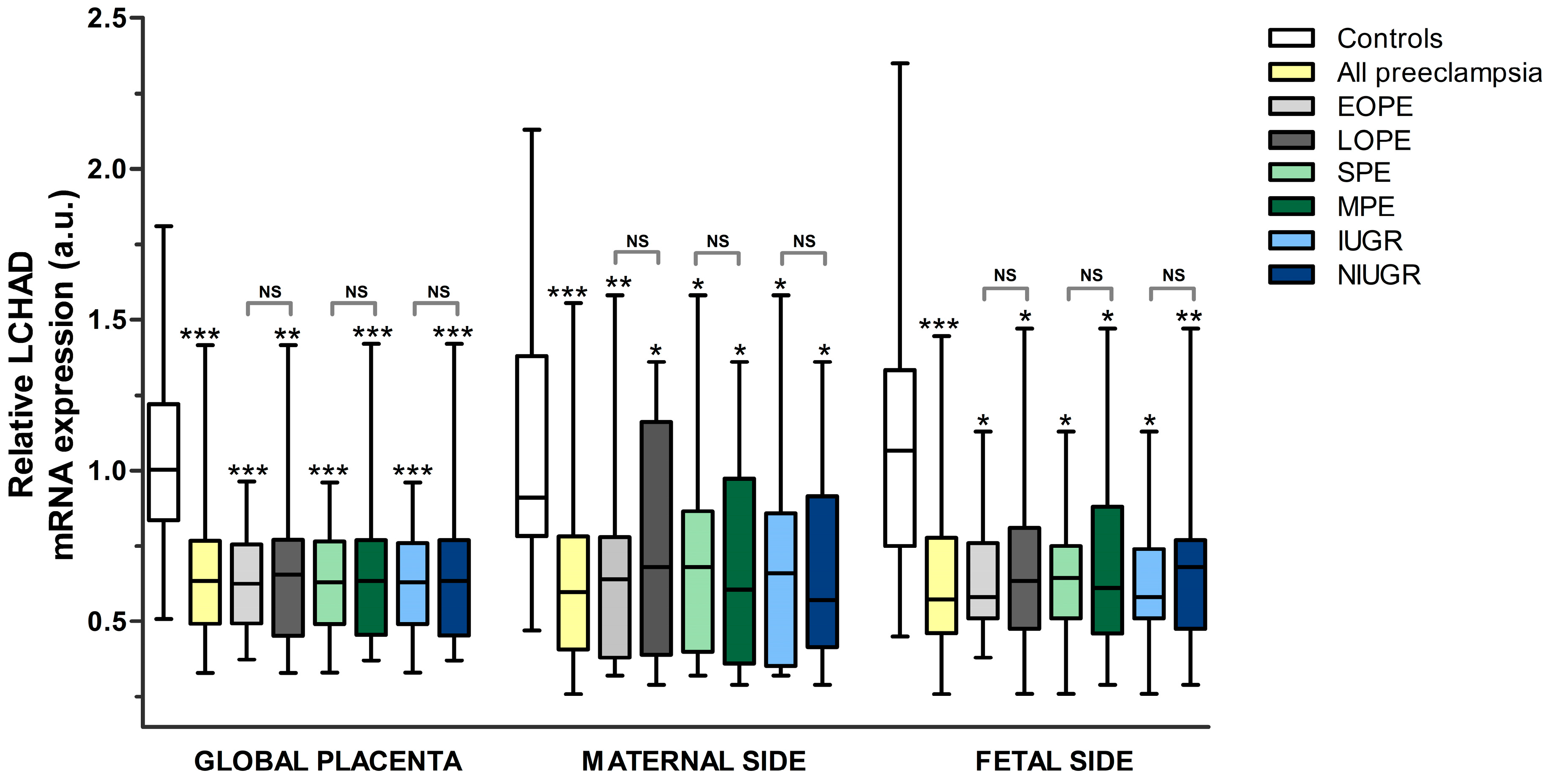
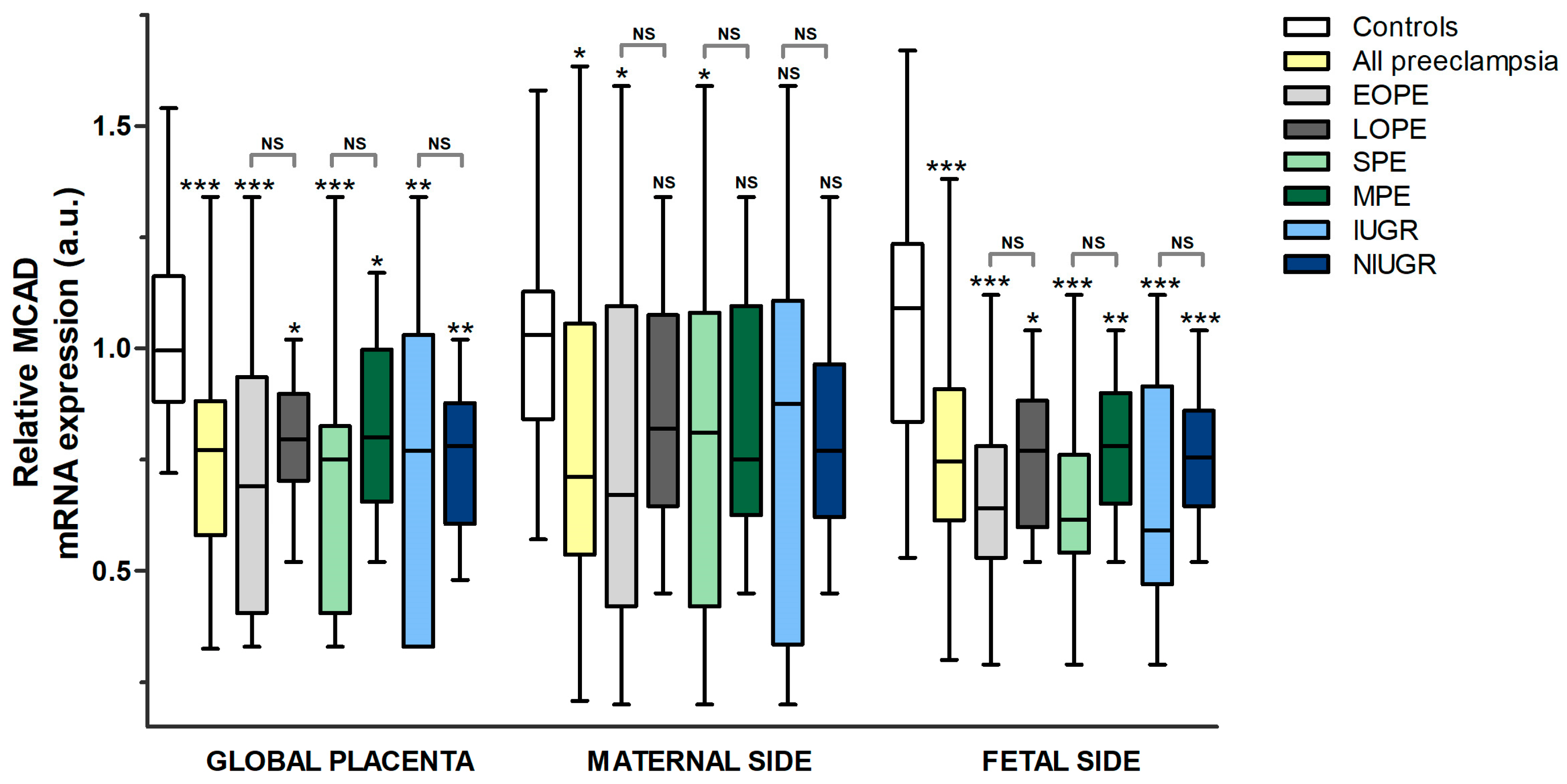
3.2.1. Comparison between Pre-Eclampsia and Controls
- LCHAD
- MCAD
- CPT1A
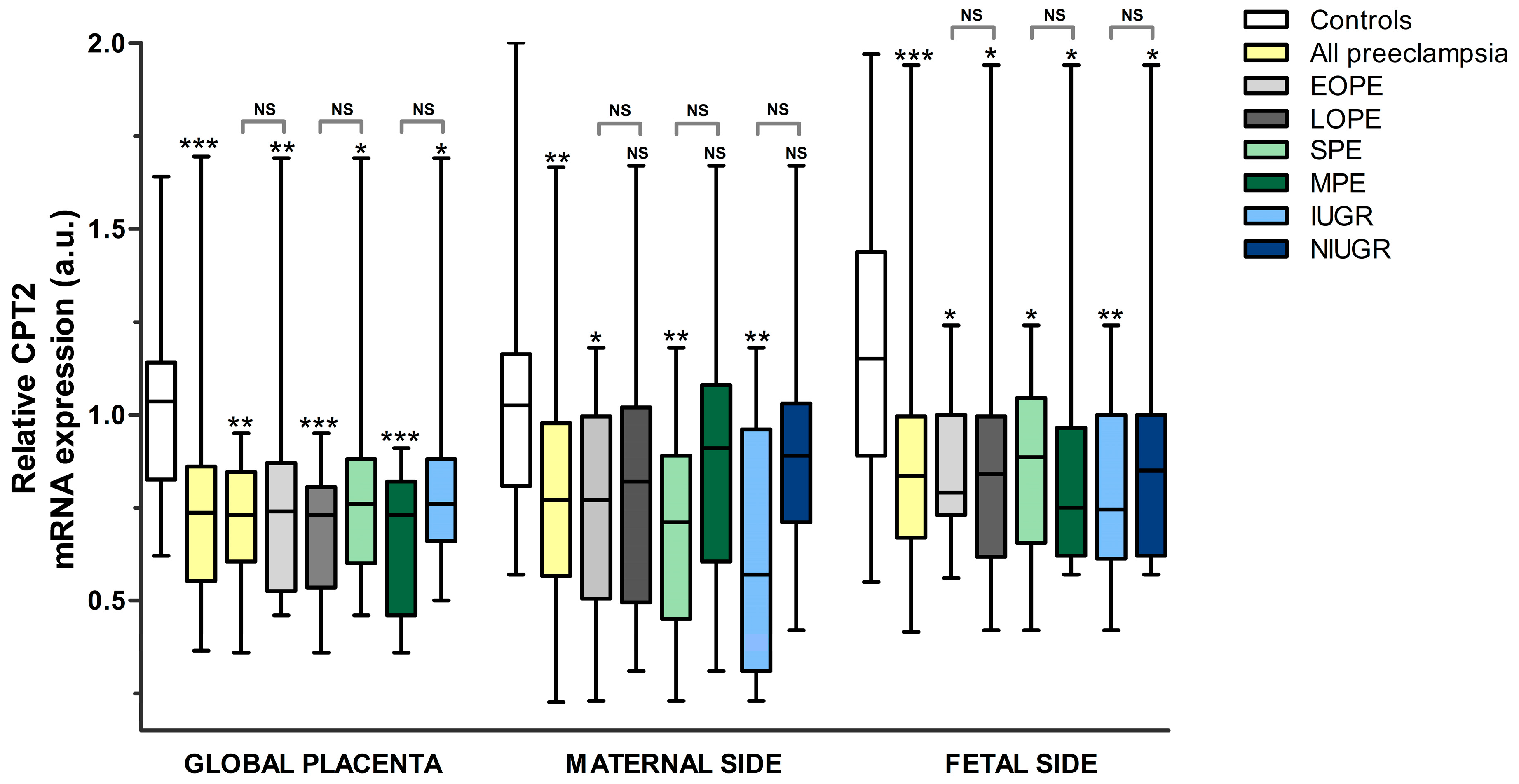
- CPT2
3.2.2. Comparison between the Different Subgroups of Pre-Eclampsia
3.2.3. Comparison between Sides of the Placenta in Each Subgroup
4. Discussion
4.1. Main Findings
4.2. Interpretation of Results
4.3. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFLP | Acute fatty liver of pregnancy |
| a.u. | Arbitrary units |
| cDNA | Complementary DNA |
| COX-2 | Cyclooxygenase-2 |
| CPT | Carnitine palmitoyltransferase |
| Cq | Quantification cycle |
| EOPE | Early-onset pre-eclampsia |
| FA | Fatty acid |
| FABPs | Fatty acid-binding proteins |
| FAO | Fatty acid oxidation |
| FAT/CD36 | Fatty acid translocase |
| FATPs | Fatty acid transport proteins |
| FFA | Free fatty acids |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| HDL | High-density lipoprotein |
| HELLP | Hemolysis, elevated liver function, and low platelets |
| IUGR | Intrauterine growth restriction |
| LC | Long chain |
| LCHAD | Long-chain 3-hydroxyacyl-CoA dehydrogenase |
| LDL | Low-density lipoprotein |
| L-NAME | Nω-nitro-L-arginine-methyl ester |
| LOPE | Late-onset pre-eclampsia |
| LPL | Lipoprotein lipase |
| MAPK | Mitogen-Activated Protein Kinases |
| MC | Medium chain |
| MCAD | Medium-chain acyl-CoA dehydrogenase |
| MPE | Mild pre-eclampsia |
| mRNA | Messenger RNA |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NF-κB | Nuclear factor-κB |
| NIUGR | Not intrauterine growth restriction |
| PlGF | Placental growth factor |
| PPARs | Peroxisome proliferator-activated receptors |
| PUFA | Polyunsaturated fatty acids |
| RPL30 | Ribosomal protein L30 |
| RT-qPCR | Real-time quantitative PCR |
| SC | Short-chain |
| SCHAD | Short-chain 3-hydroxyacyl-CoA dehydrogenase |
| sEng | Soluble endoglin |
| sFlt-1 | Soluble fms-like tyrosine kinase-1 |
| SPE | Severe pre-eclampsia |
| VLCAD | Very-long-chain acyl-coA dehydrogenase |
References
- Dimitriadis, E.; Rolnik, D.L.; Zhou, W.; Estrada-Gutierrez, G.; Koga, K.; Francisco, R.P.V.; Whitehead, C.; Hyett, J.; da Silva Costa, F.; Nicolaides, K.; et al. Pre-eclampsia. Nat. Rev. Dis. Primers 2023, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Lees, C.; Stampalija, T.; Baschat, A.A.; da Silva Costa, F.; Ferrazzi, E.; Figueras, F.; Hecher, K.; Kingdom, J.; Poon, L.C.; Salomon, L.J.; et al. ISUOG Practice Guidelines: Diagnosis and management of small-for-gestational-age fetus and fetal growth restriction. Ultrasound Obstet. Gynecol. 2020, 56, 298–312. [Google Scholar] [CrossRef]
- Magee, L.A.; Brown, M.A.; Hall, D.R.; Gupte, S.; Hennessy, A.; Karumanchi, S.A.; Kenny, L.C.; McCarthy, F.; Myers, J.; Poon, L.C.; et al. The 2021 International Society for the Study of Hypertension in Pregnancy classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 2022, 27, 148–169. [Google Scholar] [PubMed]
- Aye, I.; Aiken, C.E.; Charnock-Jones, D.S.; Smith, G.C.S. Placental energy metabolism in health and disease-significance of development and implications for preeclampsia. Am. J. Obstet. Gynecol. 2022, 226, S928–S944. [Google Scholar] [CrossRef] [PubMed]
- Sõber, S.; Reiman, M.; Kikas, T.; Rull, K.; Inno, R.; Vaas, P.; Teesalu, P.; Marti, J.M.L.; Mattila, P.; Laan, M. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci. Rep. 2015, 5, 13336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bi, S.; Liang, Y.; Huang, L.; Li, Y.; Huang, M.; Huang, B.; Deng, W.; Liang, J.; Gu, S.; et al. Integrated Metabolomic and Lipidomic Analysis in the Placenta of Preeclampsia. Front. Physiol. 2022, 13, 807583. [Google Scholar] [CrossRef]
- Magee, L.A.; Nicolaides, K.H.; von Dadelszen, P. Preeclampsia. N. Engl. J. Med. 2022, 386, 1817–1832. [Google Scholar] [CrossRef]
- Erez, O.; Romero, R.; Jung, E.; Chaemsaithong, P.; Bosco, M.; Suksai, M.; Gallo, D.M.; Gotsch, F. Preeclampsia and eclampsia: The conceptual evolution of a syndrome. Am. J. Obstet. Gynecol. 2022, 226, S786–S803. [Google Scholar] [CrossRef]
- Abascal-Saiz, A.; Duque-Alcorta, M.; Fioravantti, V.; Antolín, E.; Fuente-Luelmo, E.; Haro, M.; Ramos-Álvarez, M.P.; Perdomo, G.; Bartha, J.L. The Relationship between Angiogenic Factors and Energy Metabolism in Preeclampsia. Nutrients 2022, 14, 2172. [Google Scholar] [CrossRef]
- Rakheja, D.; Bennett, M.J.; Foster, B.M.; Domiati-Saad, R.; Rogers, B.B. Evidence for fatty acid oxidation in human placenta, and the relationship of fatty acid oxidation enzyme activities with gestational age. Placenta 2002, 23, 447–450. [Google Scholar] [CrossRef]
- Herrera, E.; Amusquivar, E.; Lopez-Soldado, I.; Ortega, H. Maternal lipid metabolism and placental lipid transfer. Horm. Res. 2006, 65, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.E.J.; Watkins, O.C.; Mah, T.K.L.; Cracknell-Hazra, V.K.B.; Pillai, R.A.; Selvam, P.; Islam, M.O.; Sharma, N.; Cazenave-Gassiot, A.; Bendt, A.K.; et al. Increasing maternal age associates with lower placental CPT1B mRNA expression and acylcarnitines, particulary in overweight women. Front. Physiol. 2023, 14, 1166827. [Google Scholar] [CrossRef] [PubMed]
- Bentebibel, A. Study of the mechanisms of inhibition of carnitine palmitoyltransferase I activity. Ph.D. Thesis, University of Barcelona, Barcelona, Spain, 2009. Available online: http://hdl.handle.net/2445/36293 (accessed on 7 July 2023). (In Spanish).
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Bartha, J.L.; Visiedo, F.; Fernandez-Deudero, A.; Bugatto, F.; Perdomo, G. Decreased mitochondrial fatty acid oxidation in placentas from women with preeclampsia. Placenta 2012, 33, 132–134. [Google Scholar] [CrossRef] [PubMed]
- Abascal-Saiz, A.; Fuente-Luelmo, E.; Haro, M.; de la Calle, M.; Ramos-Álvarez, M.P.; Perdomo, G.; Bartha, J.L. Placental Compartmentalization of Lipid Metabolism: Implications for Singleton and Twin Pregnancies. Reprod. Sci. 2021, 28, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Sibai, B.M. Diagnosis, controversies, and management of the syndrome of hemolysis, elevated liver enzymes, and low platelet count. Obs. Gynecol. 2004, 103, 981–991. [Google Scholar] [CrossRef] [PubMed]
- NCBI (National Center for Biotechnology Information). Primer-BLAST: A Tool for Finding Specific Primers. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast (accessed on 1 November 2021).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Murthi, P.; Fitzpatrick, E.; Borg, A.J.; Donath, S.; Brennecke, S.P.; Kalionis, B. GAPDH, 18S rRNA and YWHAZ are suitable endogenous reference genes for relative gene expression studies in placental tissues from human idiopathic fetal growth restriction. Placenta 2008, 29, 798–801. [Google Scholar] [CrossRef]
- Panagodimou, E.; Koika, V.; Markatos, F.; Kaponis, A.; Adonakis, G.; Georgopoulos, N.A.; Markantes, G.K. Expression stability of ACTB, 18S, and GAPDH in human placental tissues from subjects with PCOS and controls: GAPDH expression is increased in PCOS. Hormones 2022, 21, 329–333. [Google Scholar] [CrossRef]
- Wajner, M.; Amaral, A.U. Mitochondrial dysfunction in fatty acid oxidation disorders: Insights from human and animal studies. Biosci. Rep. 2015, 36, e00281. [Google Scholar] [CrossRef] [PubMed]
- Ribas, G.S.; Vargas, C.R. Evidence that Oxidative Disbalance and Mitochondrial Dysfunction are Involved in the Pathophysiology of Fatty Acid Oxidation Disorders. Cell. Mol. Neurobiol. 2022, 42, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Brassier, A.; Ottolenghi, C.; Boddaert, N.; Sonigo, P.; Attié-Bitach, T.; Millischer-Bellaiche, A.E.; Baujat, G.; Cormier-Daire, V.; Valayannopoulos, V.; Seta, N.; et al. Prenatal symptoms and diagnosis of inherited metabolic diseases. Arch. Pediatr. 2012, 19, 959–969. (In French) [Google Scholar] [CrossRef] [PubMed]
- Oey, N.A.; Ruiter, J.P.; Attie-Bitach, T.; Ijlst, L.; Wanders, R.J.; Wijburg, F.A. Fatty acid oxidation in the human fetus: Implications for fetal and adult disease. J. Inherit. Metab. Dis. 2006, 29, 71–75. [Google Scholar] [CrossRef]
- Shekhawat, P.; Bennett, M.J.; Sadovsky, Y.; Nelson, D.M.; Rakheja, D.; Strauss, A.W. Human placenta metabolizes fatty acids: Implications for fetal fatty acid oxidation disorders and maternal liver diseases. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E1098–E1105. [Google Scholar] [CrossRef]
- Tyni, T.; Ekholm, E.; Pihko, H. Pregnancy complications are frequent in long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency. Am. J. Obstet. Gynecol. 1998, 178, 603–608. [Google Scholar] [CrossRef]
- Natarajan, S.K.; Ibdah, J.A. Role of 3-Hydroxy Fatty Acid-Induced Hepatic Lipotoxicity in Acute Fatty Liver of Pregnancy. Int. J. Mol. Sci. 2018, 19, 322. [Google Scholar] [CrossRef]
- Oey, N.A.; Boer, M.E.J.D.; Ruiter, J.P.N.; Wanders, R.J.A.; Duran, M.; Waterham, H.R.; Boer, K.; van der Post, J.A.M.; Wijburg, F.A. High activity of fatty acid oxidation enzymes in human placenta: Implications for fetal-maternal disease. J. Inherit. Metab. Dis. 2003, 26, 385–392. [Google Scholar] [CrossRef]
- Bartha, J.L.; Bugatto, F.; Fernandez-Deudero, A.; Fernandez-Macias, R.; Perdomo, G. Tissue specific expression of human fatty acid oxidation enzyme genes in late pregnancy. Lipids Health Dis. 2016, 15, 200. [Google Scholar] [CrossRef]
- Oey, N.A.; Boer, M.E.J.D.; Wijburg, F.A.; Vekemans, M.; Augé, J.; Steiner, C.; Wanders, R.J.A.; Waterham, H.R.; Ruiter, J.P.N.; Attié-Bitach, T. Long-chain fatty acid oxidation during early human development. Pediatr. Res. 2005, 57, 755–759. [Google Scholar] [CrossRef]
- Ma, R.Q.; Sun, M.N.; Yang, Z. Inhibition of nitric oxide synthase lowers fatty acid oxidation in preeclampsia-like mice at early gestational stage. Chin. Med. J. 2011, 124, 3141–3147. [Google Scholar] [PubMed]
- Sun, M.N.; Yang, Z.; Ma, R.Q. Interaction of fatty acid oxidation with oxidative stress in preeclampsia-like mouse model at multiple stages of gestation. Zhonghua Yi Xue Za Zhi 2011, 91, 2343–2347. (In Chinese) [Google Scholar] [PubMed]
- Sun, X.L.; Yang, Z.; Wang, W.; Wang, X.Y.; Wang, J.L.; Wu, S.Y. Interaction mechanism and influence between fatty acid oxidation in trophoblast cells and p38MAPK signal transduction pathway of severe preeclampsia. Zhonghua Fu Chan Ke Za Zhi 2013, 48, 853–857. (In Chinese) [Google Scholar] [PubMed]
- Sun, X.L.; Yang, Z.; Wang, J.L.; Sun, M.N.; Wu, S.Y.; Wang, X.Y. Correlation between severe preeclampsia and abnormal expression of long-chain fatty acid oxidative enzyme. Zhonghua Yi Xue Za Zhi 2011, 91, 2026–2029. (In Chinese) [Google Scholar] [PubMed]
- Li, F.; Yang, Z.; Zhang, A.; Sun, X.; Wang, J.; Meng, R. The changes of LCHAD in preeclampsia with different clinical features and the correlation with NADPH P47-phox, p38MAPK-α, COX-2 and serum FFA and TG. Zhonghua Fu Chan Ke Za Zhi 2015, 50, 92–100. (In Chinese) [Google Scholar]
- Wang, J.L.; Yang, Z.; Wang, R.; Zhu, J.M. Interaction among abnormal fatty acid oxidation, endothelial function disorder, and oxidative stress in the onset of severe preeclampsia. Zhonghua Yi Xue Za Zhi 2008, 88, 1471–1475. (In Chinese) [Google Scholar]
- Ding, X.; Yang, Z.; Han, Y.; Yu, H. Correlation of long-chain fatty acid oxidation with oxidative stress and inflammation in pre-eclampsia-like mouse models. Placenta 2015, 36, 1442–1449. [Google Scholar] [CrossRef]
- Zhou, X.; Han, T.L.; Chen, H.; Baker, P.N.; Qi, H.; Zhang, H. Impaired mitochondrial fusion, autophagy, biogenesis and dysregulated lipid metabolism is associated with preeclampsia. Exp. Cell Res. 2017, 359, 195–204. [Google Scholar] [CrossRef]
- Ding, X.; Yang, Z.; Han, Y.; Yu, H. Fatty acid oxidation changes and the correlation with oxidative stress in different preeclampsia-like mouse models. PLoS ONE 2014, 9, e109554. [Google Scholar] [CrossRef]
- Chavan-Gautam, P.; Rani, A.; Freeman, D.J. Distribution of Fatty Acids and Lipids During Pregnancy. Adv. Clin. Chem. 2018, 84, 209–239. [Google Scholar]
- Yang, X.; Xu, P.; Zhang, F.; Zhang, L.; Zheng, Y.; Hu, M.; Wang, L.; Han, T.-L.; Peng, C.; Wang, L.; et al. AMPK Hyper-Activation Alters Fatty Acids Metabolism and Impairs Invasiveness of Trophoblasts in Preeclampsia. Cell. Physiol. Biochem. 2018, 49, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Laivuori, H.; Gallaher, M.; Collura, L.; Crombleholme, W.; Markovic, N.; Rajakumar, A.; Hubel, C.; Roberts, J.; Powers, R. Relationships between maternal plasma leptin, placental leptin mRNA and protein in normal pregnancy, pre-eclampsia and intrauterine growth restriction without pre-eclampsia. Mol. Hum. Reprod. 2006, 12, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chao, B.; Xu, J.; Liu, Z.; Tao, Y.; He, J.; Wang, J.; Yang, H.; Luo, X.; Qi, H. CPT1A modulates PI3K/Akt/mTOR pathway to promote preeclampsia. Placenta 2023, 133, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.G.; Niezen-Koning, K.E.; van Gennip, A.H.; Aarnoudse, J.G. Increased plasma carnitine concentrations in preeclampsia. Obs. Gynecol. 2004, 103, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.; Hui, X.; Song, Y. Metabolomics Applied to Cord Serum in Preeclampsia Newborns: Implications for Neonatal Outcomes. Front. Pediatr. 2022, 10, 869381. [Google Scholar] [CrossRef] [PubMed]
- Reiss, J.D.; Chang, A.L.; Mayo, J.A.; Bianco, K.; Lee, H.C.; Stevenson, D.K.; Shaw, G.M.; Aghaeepour, N.; Sylvester, K.G. Newborn screen metabolic panels reflect the impact of common disorders of pregnancy. Pediatr. Res. 2022, 92, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Deng, W.; Cui, W.; Xie, Q.; Zhao, G.; Wu, X.; Dai, L.; Chen, D.; Yu, B. Analysis of amino acid and acyl carnitine profiles in maternal and fetal serum from preeclampsia patients. J. Matern. Fetal Neonatal Med. 2020, 33, 2743–2750. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Wu, H.; Gao, Y.; Zheng, J.; Zhang, J. Maternal High-Fat Diet Impairs Placental Fatty Acid β-Oxidation and Metabolic Homeostasis in the Offspring. Front. Nutr. 2022, 9, 849684. [Google Scholar] [CrossRef]
- Sun, X.L.; Yang, Z.; Wang, X.Y.; Wang, J.L.; Wu, S.Y. Effects of expression of mitochondria long-chain fatty acid oxidative enzyme with different chain lengths of free fatty acids in trophoblast cells. Zhonghua Yi Xue Za Zhi 2012, 92, 2034–2037. (In Chinese) [Google Scholar]
- Yu, H.; Yang, Z.; Ding, X.; Wang, Y.; Han, Y. Correlation between the different chain lengths of free fatty acid oxidation and ability of trophoblastic invasion. Chin. Med. J. 2014, 127, 3378–3382. [Google Scholar]
- Yu, H.; Yang, Z.; Ding, X.; Wang, Y.; Han, Y. Effects of serum from patients with early-onset pre-eclampsia, HELLP syndrome, and antiphospholipid syndrome on fatty acid oxidation in trophoblast cells. Arch. Gynecol. Obstet. 2015, 292, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Livergood, M.C.; Nakagawa, P.; Wu, J.; Sigmund, C.D. Role of the Peroxisome Proliferator Activated Receptors in Hypertension. Circ. Res. 2021, 128, 1021–1039. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.; Siersbaek, M.; Mandrup, S. PPARs: Fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 2012, 23, 631–639. [Google Scholar] [CrossRef]
- Kadam, L.; Kohan-Ghadr, H.R.; Drewlo, S. The balancing act—PPAR-gamma’s roles at the maternal-fetal interface. Syst. Biol. Reprod. Med. 2015, 161, 65–71. [Google Scholar] [CrossRef]
- Li, Y.; Chen, W.L.; Liu, L.; Gu, H. Expression of peroxisome proliferator-activated receptors, fatty acid binding protein-4 in placenta and their correlations with the prognosis of pre-eclampsia. Zhonghua Fu Chan Ke Za Zhi 2017, 52, 443–448. (In Chinese) [Google Scholar]
- Grimaldi, B.; Kohan-Ghadr, H.R.; Drewlo, S. The Potential for Placental Activation of PPARgamma to Improve the Angiogenic Profile in Preeclampsia. Cells 2022, 11, 3514. [Google Scholar] [CrossRef]
- Psilopatis, I.; Vrettou, K.; Fleckenstein, F.N.; Theocharis, S. The Role of Peroxisome Proliferator-Activated Receptors in Preeclampsia. Cells 2023, 12, 647. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Kodama, T.; Yamamoto, Y.; Ebrahimi, M.; Miyazaki, H.; Yasumoto, Y.; Kagawa, Y.; Sawada, T.; Owada, Y.; Tokuda, N. Omega-3 fatty acids transport through the placenta. Asian J. Med. Biol. Res. 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Khaire, A.A.; Thakar, S.R.; Wagh, G.N.; Joshi, S.R. Placental lipid metabolism in preeclampsia. J. Hypertens. 2021, 39, 127–134. [Google Scholar] [CrossRef]
- Ferrazzi, E.; Stampalija, T.; Aupont, J.E. The evidence for late-onset pre-eclampsia as a maternogenic disease of pregnancy. Fetal Matern. Med. Rev. 2013, 24, 18–31. [Google Scholar] [CrossRef]
- Rana, S.; Lemoine, E.; Granger, J.P.; Karumanchi, S.A. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ. Res. 2019, 124, 1094–1112. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Romero, R.; Yeo, L.; Gomez-Lopez, N.; Chaemsaithong, P.; Jaovisidha, A.; Gotsch, F.; Erez, O. The etiology of preeclampsia. Am. J. Obs. Gynecol. 2022, 226, S844–S866. [Google Scholar] [CrossRef] [PubMed]
| MATERNAL RESULTS | Control n = 24 | EOPE n = 12 | LOPE n = 11 | p-Value |
|---|---|---|---|---|
| Maternal age (years) | 37.0 (7.0) | 36.0 (16.0) | 35.0 (3.0) | 0.761 |
| Ethnic Group: | 0.352 | |||
| 18 [75.0%] | 7 [58.3%] | 7 [63.6%] | |
| 6 [25.0%] | 5 [41.7%] | 3 [27.3%] | |
| 0 | 0 | 1 [9.1%] | |
| Pregravid body mass index (kg/m2) | 23.14 ± 2.73 | 22.05 ± 3.07 | 25.23 ± 3.73 | 0.636 |
| Body mass index classification: | 0.243 | |||
| 0 | 1 [8.3%] | 0 | |
| 19 [79.2%] | 6 [50.0%] | 6 [54.5%] | |
| 3 [12.5%] | 2 [16.6%] | 4 [36.4%] | |
| 0 | 0 | 1 [9.1%] | |
| Gestational weight gain (kg) | 15.0 (9.0) | 10.0 (6.0) | 14.0 (8.0) | 0.269 |
| OBSTETRIC RESULTS | ||||
| Gestational age at study (weeks) | 39.17 ± 1.12 | 31.75 ± 3.44 | 36.91 ± 1.22 | a 1 × 10−6 * b 0.013 * c 1 × 10−6 * |
| Parity: | ||||
| 6 [25.0%] | 6 [50.0%] | 6 [45.5%] | 0.120 |
| 4 [16.6%] | 3 [25.0%] | 2 [18.2%] | 0.380 |
| 11 [45.8%] | 3 [25.0%] | 2 [18.2%] | 0.419 |
| 9 [37.5%] | 2 [16.7%] | 4 [36.6%] | 0.333 |
| Twin pregnancy | 4 [16.7%] | 2 [16.7%] | 3 [27.4%] | 0.612 |
| Mode of pregnancy: | 0.553 | |||
| 19 [79.2%] | 9 [75.0%] | 9 [81.8%] | |
| 0 | 1 [8.3%] | 0 | |
| 5 [20.8%] | 2 [16.7%] | 2 [18.2%] | |
| Obstetric reason for cesarean section: | ||||
| A.—Fetal: | ||||
| 7 [29.3%] | 1 [8.3%] | 1 [9.1%] | |
| 1 [4.1%] | 0 | 0 | |
| 0 | 1 [8.3%] | 1 [9.1%] | |
| 0 | 2 [16.7%] | 1 [9.1%] | |
| B.—Maternal: | ||||
| 0 | 8 [66.7%] | 5 [45.4%] | |
| 0 | 4 [33.3%] | 1 [9.1%] | |
| 10 [41.7%] | 0 | 0 | |
| 4 [16.7%] | 0 | 1 [9.1%] | |
| 0 | 0 | 1 [9.1%] | |
| 1 [4.1%] | 0 | 0 | |
| 1 [4.1%] | 0 | 0 | |
| 0 | 0 | 1 [9.1%] | |
| PERINATAL RESULTS | n = 28 | n = 14 | n = 14 | |
| EFW (g) | 3081.4 ± 578.9 | 1284.9 ± 473.8 | 2345.83 ± 471.9 | a 1 × 10−6 * b 1.2 × 10−4 * c 4 × 10−5 * |
| Centile EFW | 65.0 (63.0) | 4.0 (7.0) | 70.0 (70.0) | a 5 × 10−5 * b 0.719 c 0.001 * |
| Fetal growth restriction: | ||||
| 1 [43.6%] | 3 [21.4%] | 0 | a 0.022 * b 0.252 c 0.024 * |
| 0 | 8 [57.1%] | 3 [21.4%] | a 2 × 10−4 * b 0.001 * c 5 × 10−4 * |
| Neonatal birth weight (g) | 3320.0 (600.0) | 1600.0 (540.0) | 2440.0 (1045.0) | a 2 × 10−6 * b 0.019 * c 0.029 * |
| Neonatal birth weight centile | 66.0 (32.0) | 3.0 (5.0) | 51.0 (92.0) | a 1 × 10−6 * b 0.719 c 0.001 * |
| Neonatal sex: | 0.358 | |||
| 12 [42.9%] | 3 [21.4%] | 6 [42.9%] | |
| 16 [57.1%] | 11 [78.6%] | 8 [57.1%] | |
| Umbilical artery pH at birth | 7.30 ± 0.06 | 7.28 ± 0.07 | 7.28 ± 0.07 | 0.500 |
| Placental weight | 581.68 ± 130.85 | 319.49 ± 116.64 | 487.26 ± 136.21 | a 3 × 10−6 * b 0.122 c 0.101 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abascal-Saiz, A.; Fuente-Luelmo, E.; Haro, M.; Fioravantti, V.; Antolín, E.; Ramos-Álvarez, M.P.; Bartha, J.L. Decreased Fatty Acid Oxidation Gene Expression in Pre-Eclampsia According to the Onset and Presence of Intrauterine Growth Restriction. Nutrients 2023, 15, 3877. https://doi.org/10.3390/nu15183877
Abascal-Saiz A, Fuente-Luelmo E, Haro M, Fioravantti V, Antolín E, Ramos-Álvarez MP, Bartha JL. Decreased Fatty Acid Oxidation Gene Expression in Pre-Eclampsia According to the Onset and Presence of Intrauterine Growth Restriction. Nutrients. 2023; 15(18):3877. https://doi.org/10.3390/nu15183877
Chicago/Turabian StyleAbascal-Saiz, Alejandra, Eva Fuente-Luelmo, María Haro, Victoria Fioravantti, Eugenia Antolín, María P. Ramos-Álvarez, and José L. Bartha. 2023. "Decreased Fatty Acid Oxidation Gene Expression in Pre-Eclampsia According to the Onset and Presence of Intrauterine Growth Restriction" Nutrients 15, no. 18: 3877. https://doi.org/10.3390/nu15183877
APA StyleAbascal-Saiz, A., Fuente-Luelmo, E., Haro, M., Fioravantti, V., Antolín, E., Ramos-Álvarez, M. P., & Bartha, J. L. (2023). Decreased Fatty Acid Oxidation Gene Expression in Pre-Eclampsia According to the Onset and Presence of Intrauterine Growth Restriction. Nutrients, 15(18), 3877. https://doi.org/10.3390/nu15183877







