Unraveling the Impact of Gut and Oral Microbiome on Gut Health in Inflammatory Bowel Diseases
Abstract
:1. Introduction
2. Overview of the Significance of Normal Microbiome
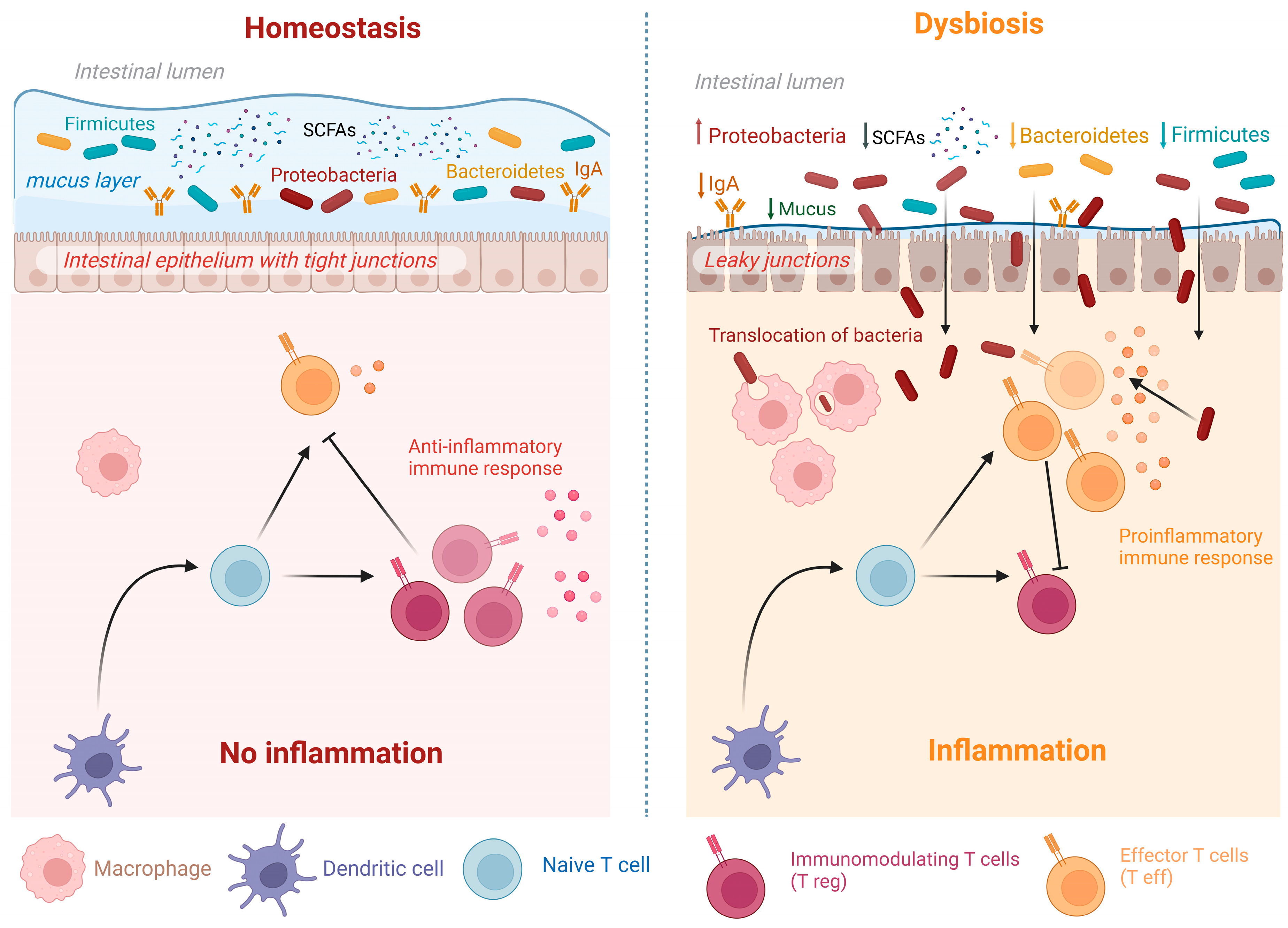
3. Dysbiosis and Relation to Diseases
4. Gut Microbiota Alterations in IBD and Relation to IBD Pathogenesis
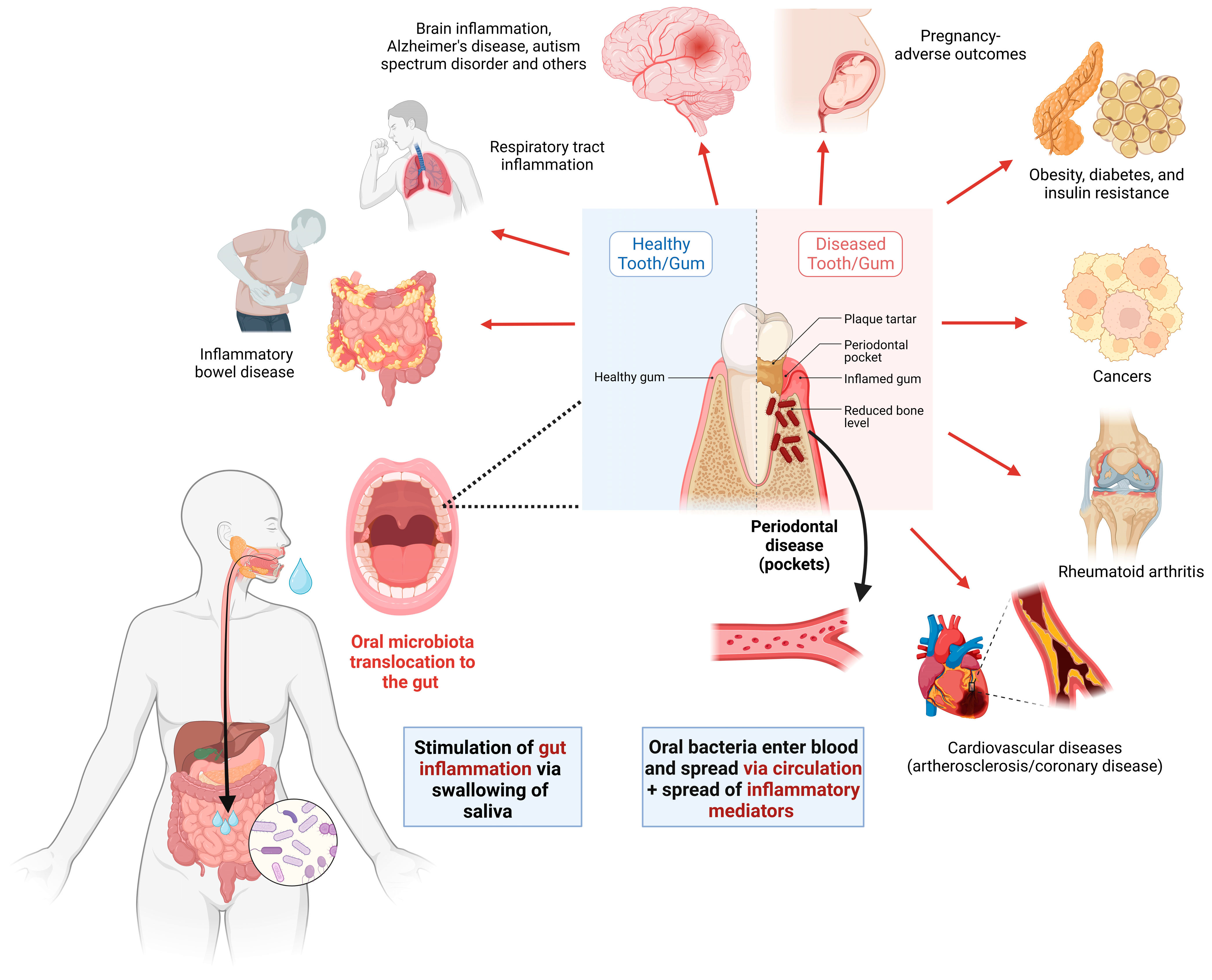
5. Oral–Gut Axis and the Role of the Oral Microbiome in IBD Pathogenesis
6. Oral Dysbiosis Link to Oral Diseases and Contribution to IBD
6.1. Oral Microbiome Alterations in CD
6.2. Oral Microbiome Alterations in UC
7. Factors Influencing Microbiome Composition and Their Impact on IBD Pathogenesis
7.1. Early Life Factors
7.2. Effect of Diet and Supplements
7.3. Effect of Exercise
7.4. IBD Drugs and Microbiota
8. Microbiome Modulation to Treat IBD
9. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Cheon, J.H. Incidence and Prevalence of Inflammatory Bowel Disease across Asia. Yonsei Med. J. 2021, 62, 99. [Google Scholar] [CrossRef]
- Seyedian, S.S.; Nokhostin, F.; Malamir, M.D. A review of the diagnosis, prevention, and treatment methods of inflammatory bowel disease. J. Med. Life 2019, 12, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, K.A.; Breuer, C.K. Pediatric inflammatory bowel disease. World J. Gastroenterol. 2006, 12, 3204–3212. [Google Scholar] [CrossRef]
- Xun, Z.; Zhang, Q.; Xu, T.; Chen, N.; Chen, F. Dysbiosis and Ecotypes of the Salivary Microbiome Associated With Inflammatory Bowel Diseases and the Assistance in Diagnosis of Diseases Using Oral Bacterial Profiles. Front. Microbiol. 2018, 9, 1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cococcioni, L.; Panelli, S.; Varotto-Boccazzi, I.; Carlo, D.D.; Pistone, D.; Leccese, G.; Zuccotti, G.V.; Comandatore, F. IBDs and the pediatric age: Their peculiarities and the involvement of the microbiota. Dig. Liver. Dis. 2021, 53, 17–25. [Google Scholar] [CrossRef]
- Carroll, M.W.; Kuenzig, M.E.; Mack, D.R.; Otley, A.R.; Griffiths, A.M.; Kaplan, G.G.; Bernstein, C.N.; Bitton, A.; Murthy, S.K.; Nguyen, G.C.; et al. The Impact of Inflammatory Bowel Disease in Canada 2018: Children and Adolescents with IBD. J. Can. Assoc. Gastroenterol. 2019, 2, S49–S67. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Kittana, M.; Ahmadani, A.; Al Marzooq, F.; Attlee, A. Dietary Fat Effect on the Gut Microbiome, and Its Role in the Modulation of Gastrointestinal Disorders in Children with Autism Spectrum Disorder. Nutrients 2021, 13, 3818. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.C.; Russell, R.K. Overview of paediatric IBD. Semin. Pediatr. Surg. 2017, 26, 344–348. [Google Scholar] [CrossRef]
- Rahman, B.; Al-Marzooq, F.; Saad, H.; Benzina, D.; Al Kawas, S. Dysbiosis of the Subgingival Microbiome and Relation to Periodontal Disease in Association with Obesity and Overweight. Nutrients 2023, 15, 826. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setubal, J.C.; Dias-Neto, E. Microbiomes. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2022; ISBN 978-0-12-809633-8. [Google Scholar]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.-H.; Zhu, C.-X.; Quan, Y.-S.; Yang, Z.-Y.; Wu, S.; Luo, W.-W.; Tan, B.; Wang, X.-Y. Relationship between intestinal microbiota and ulcerative colitis: Mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018, 24, 5. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Ghosh, S.; Whitley, C.S.; Haribabu, B.; Jala, V.R. Regulation of Intestinal Barrier Function by Microbial Metabolites. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1463–1482. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef] [Green Version]
- Walker, W.A. Chapter 25—Dysbiosis. In The Microbiota in Gastrointestinal Pathophysiology; Floch, M.H., Ringel, Y., Allan Walker, W., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 227–232. ISBN 978-0-12-804024-9. [Google Scholar]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Huang, J.; Liu, W.; Kang, W.; He, Y.; Yang, R.; Mou, X.; Zhao, W. Effects of microbiota on anticancer drugs: Current knowledge and potential applications. eBioMedicine 2022, 83, 104197. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.Y.; Leung, W.K.; Cheung, K.S. Association between Gut Microbiota and SARS-CoV-2 Infection and Vaccine Immunogenicity. Microorganisms 2023, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef]
- Zhang, T.; Kayani, M.U.R.; Hong, L.; Zhang, C.; Zhong, J.; Wang, Z.; Chen, L. Dynamics of the Salivary Microbiome During Different Phases of Crohn’s Disease. Front. Cell. Infect. Microbiol. 2020, 10, 544704. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Wright, E.K.; Kamm, M.A.; Teo, S.M.; Inouye, M.; Wagner, J.; Kirkwood, C.D. Recent Advances in Characterizing the Gastrointestinal Microbiome in Crohn’s Disease: A Systematic Review. Inflamm. Bowel Dis. 2015, 21, 1219–1228. [Google Scholar] [CrossRef] [Green Version]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Said, H.S.; Suda, W.; Nakagome, S.; Chinen, H.; Oshima, K.; Kim, S.; Kimura, R.; Iraha, A.; Ishida, H.; Fujita, J.; et al. Dysbiosis of Salivary Microbiota in Inflammatory Bowel Disease and Its Association With Oral Immunological Biomarkers. DNA Res. 2014, 21, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in Inflammatory Bowel Disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Zhao, W.; Lan, P.; Mou, X. The microbiome in inflammatory bowel diseases: From pathogenesis to therapy. Protein Cell 2021, 12, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Gao, R.; Wu, X.; Sun, J.; Wan, J.; Wu, T.; Fichna, J.; Yin, L.; Chen, C. Characterization of Specific Signatures of the Oral Cavity, Sputum, and Ileum Microbiota in Patients With Crohn’s Disease. Front. Cell. Infect. Microbiol. 2022, 12, 864944. [Google Scholar] [CrossRef] [PubMed]
- de Alencar Junior, H.; Paiotti, A.P.R.; de Araújo Filho, H.B.; Oshima, C.T.F.; Miszputen, S.J.; Ambrogini-Júnior, O. The relationship between the commensal microbiota levels and Crohn’s disease activity. JGH Open 2020, 4, 784–789. [Google Scholar] [CrossRef]
- Fang, J.; Wang, H.; Zhou, Y.; Zhang, H.; Zhou, H.; Zhang, X. Slimy partners: The mucus barrier and gut microbiome in ulcerative colitis. Exp. Mol. Med. 2021, 53, 772–787. [Google Scholar] [CrossRef]
- Parsaei, M.; Sarafraz, N.; Moaddab, S.Y.; Ebrahimzadeh Leylabadlo, H. The importance of Faecalibacterium prausnitzii in human health and diseases. New Microbes New Infect. 2021, 43, 100928. [Google Scholar] [CrossRef] [PubMed]
- Nikitakis, N.G.; Papaioannou, W.; Sakkas, L.I.; Kousvelari, E. The autoimmunity-oral microbiome connection. Oral Dis. 2017, 23, 828–839. [Google Scholar] [CrossRef]
- Ma, X.; Lu, X.; Zhang, W.; Yang, L.; Wang, D.; Xu, J.; Jia, Y.; Wang, X.; Xie, H.; Li, S.; et al. Gut microbiota in the early stage of Crohn’s disease has unique characteristics. Gut Pathog. 2022, 14, 46. [Google Scholar] [CrossRef]
- Duranti, S.; Gaiani, F.; Mancabelli, L.; Milani, C.; Grandi, A.; Bolchi, A.; Santoni, A.; Lugli, G.A.; Ferrario, C.; Mangifesta, M.; et al. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. FEMS Microbiol. Ecol. 2016, 92, fiw191. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.H.; Dulai, P.S.; Vázquez-Baeza, Y.; Sauceda, C.; Daniel, N.; Gerner, R.R.; Batachari, L.E.; Malfavon, M.; Zhu, Q.; Weldon, K.; et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat. Microbiol. 2022, 7, 262–276. [Google Scholar] [CrossRef]
- Sankarasubramanian, J.; Ahmad, R.; Avuthu, N.; Singh, A.B.; Guda, C. Gut Microbiota and Metabolic Specificity in Ulcerative Colitis and Crohn’s Disease. Front. Med. 2020, 7, 606298. [Google Scholar] [CrossRef]
- Gu, W.; Zhang, L.; Han, T.; Huang, H.; Chen, J. Dynamic Changes in Gut Microbiome of Ulcerative Colitis: Initial Study from Animal Model. JIR 2022, 15, 2631–2647. [Google Scholar] [CrossRef]
- Mar, J.S.; LaMere, B.J.; Lin, D.L.; Levan, S.; Nazareth, M.; Mahadevan, U.; Lynch, S.V. Disease Severity and Immune Activity Relate to Distinct Interkingdom Gut Microbiome States in Ethnically Distinct Ulcerative Colitis Patients. mBio 2016, 7, e01072-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Han, M.; Liu, S.; Fan, L.; Shi, H.; Li, P. Composition and diverse differences of intestinal microbiota in ulcerative colitis patients. Front. Cell. Infect. Microbiol. 2022, 12, 953962. [Google Scholar] [CrossRef]
- do Nascimento, R.D.P.; da Fonseca Machado, A.P.; Galvez, J.; Cazarin, C.B.B.; Maróstica Junior, M.R. Ulcerative colitis: Gut microbiota, immunopathogenesis and application of natural products in animal models. Life Sci. 2020, 258, 118129. [Google Scholar] [CrossRef]
- Qi, Y.; Zang, S.; Wei, J.; Yu, H.; Yang, Z.; Wu, H.; Kang, Y.; Tao, H.; Yang, M.; Jin, L.; et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics 2021, 113, 664–676. [Google Scholar] [CrossRef]
- Abdullah, N.; Al-Marzooq, F.; Mohamad, S.; Abd Rahman, N.; Chi Ngo, H.; Perera Samaranayake, L. Intraoral appliances for in situ oral biofilm growth: A systematic review. J. Oral Microbiol. 2019, 11, 1647757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef] [Green Version]
- Elmaghrawy, K.; Hussey, S.; Moran, G.P. The Oral Microbiome in Pediatric IBD: A Source of Pathobionts or Biomarkers? Front. Pediatr. 2021, 8, 620254. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I.; Yamazaki, K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 2019, 11, 1586422. [Google Scholar] [CrossRef] [Green Version]
- Read, E.; Curtis, M.A.; Neves, J.F. The role of oral bacteria in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 731–742. [Google Scholar] [CrossRef]
- Cecoro, G.; Annunziata, M.; Iuorio, M.T.; Nastri, L.; Guida, L. Periodontitis, Low-Grade Inflammation and Systemic Health: A Scoping Review. Medicina 2020, 56, 272. [Google Scholar] [CrossRef] [PubMed]
- Ray, K. The oral–gut axis in IBD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 532. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Jiao, Y.; Gillilland, M.G.; Hayashi, A.; Imai, J.; Sugihara, K.; Miyoshi, M.; Brazil, J.C.; Kuffa, P.; et al. The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell 2020, 182, 447–462.e14. [Google Scholar] [CrossRef] [PubMed]
- Al-Rawi, N.; Al-Marzooq, F. The Relation between Periodontopathogenic Bacterial Levels and Resistin in the Saliva of Obese Type 2 Diabetic Patients. J. Diabetes Res. 2017, 2017, 2643079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Rawi, N.H.; Al-Marzooq, F.; Al-Nuaimi, A.S.; Hachim, M.Y.; Hamoudi, R. Salivary microRNA 155, 146a/b and 203: A pilot study for potentially non-invasive diagnostic biomarkers of periodontitis and diabetes mellitus. PLoS ONE 2020, 15, e0237004. [Google Scholar] [CrossRef]
- Bartlett, A.; Gullickson, R.G.; Singh, R.; Ro, S.; Omaye, S.T. The Link between Oral and Gut Microbiota in Inflammatory Bowel Disease and a Synopsis of Potential Salivary Biomarkers. Appl. Sci. 2020, 10, 6421. [Google Scholar] [CrossRef]
- Docktor, M.J.; Paster, B.J.; Abramowicz, S.; Ingram, J.; Wang, Y.E.; Correll, M.; Jiang, H.; Cotton, S.L.; Kokaras, A.S.; Bousvaros, A. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, B.; Gao, H.; He, C.; Hua, R.; Liang, C.; Xin, S.; Wang, Y.; Xu, J. Insight into the Relationship between Oral Microbiota and the Inflammatory Bowel Disease. Microorganisms 2022, 10, 1868. [Google Scholar] [CrossRef]
- Kodukula, K.; Faller, D.V.; Harpp, D.N.; Kanara, I.; Pernokas, J.; Pernokas, M.; Powers, W.R.; Soukos, N.S.; Steliou, K.; Moos, W.H. Gut Microbiota and Salivary Diagnostics: The Mouth Is Salivating to Tell Us Something. BioResearch Open Access 2017, 6, 123–132. [Google Scholar] [CrossRef]
- Yin, C.; Chen, J.; Wu, X.; Liu, Y.; He, Q.; Cao, Y.; Huang, Y.-E.; Liu, S. Preterm Birth Is Correlated With Increased Oral Originated Microbiome in the Gut. Front. Cell. Infect. Microbiol. 2021, 11, 537. [Google Scholar] [CrossRef]
- Tan, C.X.W.; Brand, H.S.; Kalender, B.; De Boer, N.K.H.; Forouzanfar, T.; de Visscher, J.G.A.M. Dental and periodontal disease in patients with inflammatory bowel disease. Clin. Oral Investig. 2021, 25, 5273–5280. [Google Scholar] [CrossRef]
- Nazir, M.A. Prevalence of periodontal disease, its association with systemic diseases and prevention. Int. J. Health Sci. 2017, 11, 72–80. [Google Scholar]
- Bao, J.; Li, L.; Zhang, Y.; Wang, M.; Chen, F.; Ge, S.; Chen, B.; Yan, F. Periodontitis may induce gut microbiota dysbiosis via salivary microbiota. Int. J. Oral Sci. 2022, 14, 32. [Google Scholar] [CrossRef]
- Harikishan, G.; Reddy, N.R.; Prasad, H.; Anitha, S. Oral Crohn’s disease without intestinal manifestations. J. Pharm. Bioallied Sci. 2012, 4, S431–S434. [Google Scholar] [CrossRef]
- Somineni, H.K.; Weitzner, J.H.; Venkateswaran, S.; Dodd, A.; Prince, J.; Karikaran, A.; Sauer, C.G.; Abramowicz, S.; Zwick, M.E.; Cutler, D.J.; et al. Site- and Taxa-Specific Disease-Associated Oral Microbial Structures Distinguish Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2021, 27, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Lucas López, R.; Grande Burgos, M.J.; Gálvez, A.; Pérez Pulido, R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: A state of the science review. APMIS 2017, 125, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Molinero, N.; Taladrid, D.; Zorraquín-Peña, I.; de Celis, M.; Belda, I.; Mira, A.; Bartolomé, B.; Moreno-Arribas, M.V. Ulcerative Colitis Seems to Imply Oral Microbiome Dysbiosis. Curr. Issues Mol. Biol. 2022, 44, 1513–1527. [Google Scholar] [CrossRef]
- Kojima, A.; Nakano, K.; Wada, K.; Takahashi, H.; Katayama, K.; Yoneda, M.; Higurashi, T.; Nomura, R.; Hokamura, K.; Muranaka, Y.; et al. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci. Rep. 2012, 2, 332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brito, F.; Zaltman, C.; Carvalho, A.T.P.; Fischer, R.G.; Persson, R.; Gustafsson, A.; Figueredo, C.M.S. Subgingival microflora in inflammatory bowel disease patients with untreated periodontitis. Eur. J. Gastroenterol. Hepatol. 2013, 25, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Mok, J.; Gowans, M.; Ong, D.E.H.; Hartono, J.L.; Lee, J.W.J. Oral Microbiome of Crohn’s Disease Patients With and Without Oral Manifestations. J. Crohn’s Colitis 2022, 16, 1628–1636. [Google Scholar] [CrossRef]
- Abdelbary, M.M.H.; Hatting, M.; Bott, A.; Dahlhausen, A.; Keller, D.; Trautwein, C.; Conrads, G. The oral-gut axis: Salivary and fecal microbiome dysbiosis in patients with inflammatory bowel disease. Front. Cell. Infect. Microbiol. 2022, 12, 1010853. [Google Scholar] [CrossRef] [PubMed]
- Kelsen, J.; Bittinger, K.; Pauly-Hubbard, H.; Posivak, L.; Grunberg, S.; Baldassano, R.; Lewis, J.D.; Wu, G.D.; Bushman, F.D. Alterations of the Subgingival Microbiota in Pediatric Crohn’s Disease Studied Longitudinally in Discovery and Validation Cohorts. Inflamm. Bowel Dis. 2015, 21, 2797–2805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmaghrawy, K.; Fleming, P.; Fitzgerald, K.; Cooper, S.; Dominik, A.; Hussey, S.; Moran, G.P. The Oral Microbiome in Treatment-Naïve Paediatric IBD Patients Exhibits Dysbiosis Related to Disease Severity that Resolves Following Therapy. J. Crohn’s Colitis 2022, 17, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.; Xu, L.; Qian, Y.; Sun, Z.; Yu, D.; Huang, J.; Zhou, X.; Wang, Y.; Zhang, T.; Ren, R.; et al. Evolution of the Gut Microbiome in Early Childhood: A Cross-Sectional Study of Chinese Children. Front. Microbiol. 2020, 11, 439. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Yoo, J.Y.; Valeria Ozorio Dutra, S.; Morgan, K.H.; Groer, M. The Association between Early-Life Gut Microbiota and Long-Term Health and Diseases. J. Clin. Med. 2021, 10, 459. [Google Scholar] [CrossRef]
- Agrawal, M.; Sabino, J.; Frias-Gomes, C.; Hillenbrand, C.M.; Soudant, C.; Axelrad, J.E.; Shah, S.C.; Ribeiro-Mourão, F.; Lambin, T.; Peter, I.; et al. Early life exposures and the risk of inflammatory bowel disease: Systematic review and meta-analyses. EClinicalMedicine 2021, 36, 100884. [Google Scholar] [CrossRef] [PubMed]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzello, F.; Spisni, E.; Giovanardi, E.; Imbesi, V.; Salice, M.; Alvisi, P.; Valerii, M.C.; Gionchetti, P. Implications of the Westernized Diet in the Onset and Progression of IBD. Nutrients 2019, 11, 1033. [Google Scholar] [CrossRef] [Green Version]
- Chiba, M.; Nakane, K.; Komatsu, M. Westernized Diet is the Most Ubiquitous Environmental Factor in Inflammatory Bowel Disease. Perm. J. 2019, 23, 18–107. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-E.; Kim, K.S.; Koh, H.; Lee, D.-W.; Kang, N.J. Diet-Induced Host–Microbe Interactions: Personalized Diet Strategies for Improving Inflammatory Bowel Disease. Curr. Dev. Nutr. 2022, 6, nzac110. [Google Scholar] [CrossRef]
- Constante, M.; Fragoso, G.; Calvé, A.; Samba-Mondonga, M.; Santos, M.M. Dietary Heme Induces Gut Dysbiosis, Aggravates Colitis, and Potentiates the Development of Adenomas in Mice. Front. Microbiol. 2017, 8, 1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahalhal, A.; Williams, J.M.; Johnson, S.; Ellaby, N.; Duckworth, C.A.; Burkitt, M.D.; Liu, X.; Hold, G.L.; Campbell, B.J.; Pritchard, D.M.; et al. Oral iron exacerbates colitis and influences the intestinal microbiome. PLoS ONE 2018, 13, e0202460. [Google Scholar] [CrossRef] [Green Version]
- Reifen, R.; Matas, Z.; Zeidel, L.; Berkovitch, Z.; Bujanover, Y. Iron Supplementation May Aggravate Inflammatory Status of Colitis in a Rat Model. Dig. Dis. Sci. 2000, 45, 394–397. [Google Scholar] [CrossRef]
- Carrier, J.C.; Aghdassi, E.; Jeejeebhoy, K.; Allard, J.P. Exacerbation of dextran sulfate sodium-induced colitis by dietary iron supplementation: Role of NF-κB. Int. J. Color. Dis. 2006, 21, 381–387. [Google Scholar] [CrossRef]
- Malesza, I.J.; Bartkowiak-Wieczorek, J.; Winkler-Galicki, J.; Nowicka, A.; Dzięciołowska, D.; Błaszczyk, M.; Gajniak, P.; Słowińska, K.; Niepolski, L.; Walkowiak, J.; et al. The Dark Side of Iron: The Relationship between Iron, Inflammation and Gut Microbiota in Selected Diseases Associated with Iron Deficiency Anaemia—A Narrative Review. Nutrients 2022, 14, 3478. [Google Scholar] [CrossRef]
- Lee, T.; Clavel, T.; Smirnov, K.; Schmidt, A.; Lagkouvardos, I.; Walker, A.; Lucio, M.; Michalke, B.; Schmitt-Kopplin, P.; Fedorak, R.; et al. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut 2017, 66, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Adamji, M.; Day, A.S. An overview of the role of exclusive enteral nutrition for complicated Crohn’s disease. Intest. Res. 2019, 17, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Gubatan, J.; Boye, T.L.; Temby, M.; Sojwal, R.S.; Holman, D.R.; Sinha, S.R.; Rogalla, S.R.; Nielsen, O.H. Gut Microbiome in Inflammatory Bowel Disease: Role in Pathogenesis, Dietary Modulation, and Colitis-Associated Colon Cancer. Microorganisms 2022, 10, 1371. [Google Scholar] [CrossRef]
- Kong, C.; Yan, X.; Liu, Y.; Huang, L.; Zhu, Y.; He, J.; Gao, R.; Kalady, M.F.; Goel, A.; Qin, H.; et al. Ketogenic diet alleviates colitis by reduction of colonic group 3 innate lymphoid cells through altering gut microbiome. Signal Transduct. Target. Ther. 2021, 6, 154. [Google Scholar] [CrossRef]
- Fritsch, J.; Garces, L.; Quintero, M.A.; Pignac-Kobinger, J.; Santander, A.M.; Fernández, I.; Ban, Y.J.; Kwon, D.; Phillips, M.C.; Knight, K.; et al. Low-Fat, High-Fiber Diet Reduces Markers of Inflammation and Dysbiosis and Improves Quality of Life in Patients With Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 2021, 19, 1189–1199.e30. [Google Scholar] [CrossRef]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxidative Med. Cell. Longev. 2017, 2017, 3831972. [Google Scholar] [CrossRef]
- Clauss, M.; Gérard, P.; Mosca, A.; Leclerc, M. Interplay Between Exercise and Gut Microbiome in the Context of Human Health and Performance. Front. Nutr. 2021, 8, 637010. [Google Scholar] [CrossRef] [PubMed]
- Dziewiecka, H.; Buttar, H.S.; Kasperska, A.; Ostapiuk–Karolczuk, J.; Domagalska, M.; Cichoń, J.; Skarpańska-Stejnborn, A. Physical activity induced alterations of gut microbiota in humans: A systematic review. BMC Sports Sci. Med. Rehabil. 2022, 14, 122. [Google Scholar] [CrossRef] [PubMed]
- Koutouratsas, T.; Philippou, A.; Kolios, G.; Koutsilieris, M.; Gazouli, M. Role of exercise in preventing and restoring gut dysbiosis in patients with inflammatory bowel diseases: A review. World J. Gastroenterol. 2021, 27, 5037–5046. [Google Scholar] [CrossRef] [PubMed]
- Franzin, M.; Stefančič, K.; Lucafò, M.; Decorti, G.; Stocco, G. Microbiota and Drug Response in Inflammatory Bowel Disease. Pathogens 2021, 10, 211. [Google Scholar] [CrossRef]
- Alkhatry, M.; Al-Rifai, A.; Annese, V.; Georgopoulos, F.; Jazzar, A.N.; Khassouan, A.M.; Koutoubi, Z.; Nathwani, R.; Taha, M.S.; Limdi, J.K. First United Arab Emirates consensus on diagnosis and management of inflammatory bowel diseases: A 2020 Delphi consensus. World J. Gastroenterol. 2020, 26, 6710–6769. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- Lahad, A.; Weiss, B. Current therapy of pediatric Crohn’s disease. World J. Gastrointest. Pathophysiol. 2015, 6, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Stournaras, E.; Qian, W.; Pappas, A.; Hong, Y.Y.; Shawky, R.; Investigators, U.I.B.; Raine, T.; Parkes, M. Thiopurine monotherapy is effective in ulcerative colitis but significantly less so in Crohn’s disease: Long-term outcomes for 11928 patients in the UK inflammatory bowel disease bioresource. Gut 2021, 70, 677–686. [Google Scholar] [CrossRef]
- Cuffari, C. Diagnostic Considerations in Pediatric Inflammatory Bowel Disease Management. Gastroenterol. Hepatol. 2009, 5, 775–783. [Google Scholar]
- Kolho, K.-L.; Korpela, K.; Jaakkola, T.; Pichai, M.V.A.; Zoetendal, E.G.; Salonen, A.; de Vos, W.M. Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am. J. Gastroenterol. 2015, 110, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Fijan, S. Microorganisms with Claimed Probiotic Properties: An Overview of Recent Literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Stefano, M.; Santonocito, S.; Polizzi, A.; Mauceri, R.; Troiano, G.; Lo Giudice, A.; Romano, A.; Mascitti, M.; Isola, G. A Reciprocal Link between Oral, Gut Microbiota during Periodontitis: The Potential Role of Probiotics in Reducing Dysbiosis-Induced Inflammation. Int. J. Mol. Sci. 2023, 24, 1084. [Google Scholar] [CrossRef]
- Selvamani, S.; Mehta, V.; Ali El Enshasy, H.; Thevarajoo, S.; El Adawi, H.; Zeini, I.; Pham, K.; Varzakas, T.; Abomoelak, B. Efficacy of Probiotics-Based Interventions as Therapy for Inflammatory Bowel Disease: A Recent Update. Saudi J. Biol. Sci. 2022, 29, 3546–3567. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, M.; Pu, J.; Zhu, Z.; Gao, Z.; Zhou, Q.; Gu, Q.; Li, P. Probiotics and Their Metabolites Ameliorate Inflammatory Bowel Disease: A Critical Review. Infect. Microbes Dis. 2021, 3, 4. [Google Scholar] [CrossRef]
- Marteau, P.; Lémann, M.; Seksik, P.; Laharie, D.; Colombel, J.F.; Bouhnik, Y.; Cadiot, G.; Soulé, J.C.; Bourreille, A.; Metman, E.; et al. Ineffectiveness of Lactobacillus johnsonii LA1 for prophylaxis of postoperative recurrence in Crohn’s disease: A randomised, double blind, placebo controlled GETAID trial. Gut 2006, 55, 842–847. [Google Scholar] [CrossRef]
- Vinderola, G.; Sanders, M.E.; Salminen, S. The Concept of Postbiotics. Foods 2022, 11, 1077. [Google Scholar] [CrossRef]
- Martyniak, A.; Medyńska-Przęczek, A.; Wędrychowicz, A.; Skoczeń, S.; Tomasik, P.J. Prebiotics, Probiotics, Synbiotics, Paraprobiotics and Postbiotic Compounds in IBD. Biomolecules 2021, 11, 1903. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, W.; Feng, C.; Kwok, L.-Y.; He, Q.; Sun, Z. Stronger gut microbiome modulatory effects by postbiotics than probiotics in a mouse colitis model. Npj Sci. Food 2022, 6, 53. [Google Scholar] [CrossRef]
- Baunwall, S.M.D.; Lee, M.M.; Eriksen, M.K.; Mullish, B.H.; Marchesi, J.R.; Dahlerup, J.F.; Hvas, C.L. Faecal microbiota transplantation for recurrent Clostridioides difficile infection: An updated systematic review and meta-analysis. EClinicalMedicine 2020, 29–30, 100642. [Google Scholar] [CrossRef] [PubMed]
- de Fátima Caldeira, L.; Borba, H.H.; Tonin, F.S.; Wiens, A.; Fernandez-Llimos, F.; Pontarolo, R. Fecal microbiota transplantation in inflammatory bowel disease patients: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0238910. [Google Scholar] [CrossRef]
- Kedia, S.; Virmani, S.; Vuyyuru, S.K.; Kumar, P.; Kante, B.; Sahu, P.; Kaushal, K.; Farooqui, M.; Singh, M.; Verma, M.; et al. Faecal microbiota transplantation with anti-inflammatory diet (FMT-AID) followed by anti-inflammatory diet alone is effective in inducing and maintaining remission over 1 year in mild to moderate ulcerative colitis: A randomised controlled trial. Gut 2022, 71, 2401–2413. [Google Scholar] [CrossRef] [PubMed]
- Haifer, C.; Paramsothy, S.; Kaakoush, N.O.; Saikal, A.; Ghaly, S.; Yang, T.; Luu, L.D.W.; Borody, T.J.; Leong, R.W. Lyophilised oral faecal microbiota transplantation for ulcerative colitis (LOTUS): A randomised, double-blind, placebo-controlled trial. Lancet Gastroenterol. Hepatol. 2022, 7, 141–151. [Google Scholar] [CrossRef] [PubMed]
| Sample Type | IBD Type | Age Group | Sex | Sample Size | Geographical Region | Bacteria | Finding: Increased/Decreased | Reference |
|---|---|---|---|---|---|---|---|---|
| Saliva | CD | Adults | Sex was not considered—not mentioned | 2 | Japan | Klebsiella, Fusobacterium, and Veillonella | Increased | [12] |
| Saliva | CD | Adults | 34 males and 31 females | 65 | Singapore | Actinobacteria and Proteobacteria | Increased | [72] |
| Firmicutes, Bacteriodetes | Decreased | |||||||
| Saliva | CD | Adults | 392 males and 276 females | 668 | North America | Fusobacterium nucleatum, Heamophilus parainfluenzae, Veillonella parvula, Eikenella corrodens, and Gemella moribillum | Increased | [28] |
| Bacteroides vulgatus and Bacteroides caccae | Decreased | |||||||
| Saliva | CD and UC | Adults | 38 males and 21 females | 59 | Japan | Neisseria (phy. Proteobacteria.), Gemella (phy. Firmicutes), Proteobacteria, Neisseria, and Haemophilus | Decreased | [31] |
| Bacteroidetes and Prevotella | Increased | |||||||
| Saliva | CD | Adults | 57 males and 34 females | 91 | China | Phyla: Firmicutes, Bacteroidetes, and Proteobacteria Genera: Streptococcus, Neisseria, Prevotella, Haemophilus, and Veillonella | Increased | [27] |
| Saliva | CD and UC | Adults | 14 males and 12 females | 26 | Germany | Phyla: Fusobacteria, Proteobacteria, and Patescibacteria Genera: Neisseria, Streptococcus, Haemophilus, Porphyromonas, and Fusobacterium | Decreased | [73] |
| Phyla: Firmicutes, Bacteroidetes, and Actinobacteria Genera: Veillonella and Prevotella | Increased | |||||||
| Saliva | UC | Adults | Sex was not considered—not mentioned | 21 | Spain | Staphylococcus and Neisseria | Increased | [69] |
| Peptostreptococcaceae, Atopobiaceae, Lachnospiraceae, and Ruminococcaceae | Decreased | |||||||
| Saliva | UC | Adults | Sex was not considered—not mentioned | 92 | China | Streptococcus and Enterobacteriaceae | Increased | [5] |
| Lachnospiraceae and Prevotella | Decreased | |||||||
| CD | Villanella | Increased | ||||||
| Neisseriaceae and Haemophilus | Decreased | |||||||
| Saliva | CD | Adults | 18 males and 13 females | 30 | China | Saccharibacteria (TM7), Absconditabacteria (SR1), Actinobacteria, Bulleidia, Parvimonas, and Prevotella | Increased | [47] |
| Rothia, Corynebacterium, and Mycoplasma | Decreased | |||||||
| UC | Saccharibacteria (TM7), Absconditabacteria (SR1), Actinobacteria, Leptotrichia, and Atopobium | Increased | ||||||
| Rothia, Corynebacterium, and Mycoplasma | Decreased | |||||||
| Subgingival plaque samples | CD | Adults | 22 males and 23 females | 45 | Brazil | Periodontitis sites: Bacteroides ureolyticus, Campylobacter gracilis, P. melaninogenica, S. aureus, S. anginosus, Streptococcus intermedius, S. mitis, and S. mutans Gingivitis sites: Parvimonas micra, Prevotella melaninogenica, Peptostreptococcus anaerobius, Staphylococcus aureus, Streptococcus anginosus, Streptococcus mitis, S. mutans, and Treponema denticola | Increased | [71] |
| UC | Periodontitis sites: Bacteroides ureolyticus, Campylobacter gracilis, P. melaninogenica, S. aureus, S. anginosus, Streptococcus intermedius, and S. mutans Gingivitis sites: P. anaerobius and S. aureus | Increased | ||||||
| Gingivitis sites: P. micra, S. anginosus, and S. mitis, | Decreased | |||||||
| Tongue and buccal mucosal brushings | CD | Pediatrics | 62 males and 52 females | 114 | USA | Fusobacteria and Firmicutes. | Decreased | [59] |
| UC | Fusobacteria | Decreased | ||||||
| Spirochaetes, Synergistetes, and Bacteroidetes | Increased | |||||||
| Subgingival plaque samples | CD | Pediatrics | Sex was not considered—not mentioned | 156 | USA | Alloprevotella, Campylobacter, Catonella, Fusobacterium, Porphyromonas, Prevotella, Selenomonas, and Veillonella | Decreased | [74] |
| Capnocytophaga, Rothia, and TM7. | Increased | |||||||
| Tongue and buccal mucosal swabs | CD | Pediatrics | Male:female ratio: 2.6:1 in IBD 2:1 in healthy control | 248 | Ireland | Prevotella, Fusobacterium, Leptotrichia, Rothia, Porphyromonas Veillonella, Oribacterium, Peptostreptococcaceae, and Lachnoanaerobaculum | Decreased | [75] |
| Lachnospiraceae, Oribacterium, Catonella, Stomatobaculum, and Ruminococcaceae | Decreased in association with severe IBD | |||||||
| ‘IBD-associated’ taxa Eikenella and Pseudopropionibacterium spp. | Decreased after therapy | |||||||
| Ottowia, Pseudopropionobacterium, Lautropia, Staphylococcus, Pseudomonas and Corynebacterium species, Eikenella, and Streptococcus species. | Increased | |||||||
| Lactobacillus, Streptococcus, Staphylococcus, and Klebsiella spp. | Increased in those with severe IBD | |||||||
| ‘Health-associated’ taxa: Veillonella spp. and Oribacterium spp. | Increased after therapy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elzayat, H.; Mesto, G.; Al-Marzooq, F. Unraveling the Impact of Gut and Oral Microbiome on Gut Health in Inflammatory Bowel Diseases. Nutrients 2023, 15, 3377. https://doi.org/10.3390/nu15153377
Elzayat H, Mesto G, Al-Marzooq F. Unraveling the Impact of Gut and Oral Microbiome on Gut Health in Inflammatory Bowel Diseases. Nutrients. 2023; 15(15):3377. https://doi.org/10.3390/nu15153377
Chicago/Turabian StyleElzayat, Hala, Ghaidaa Mesto, and Farah Al-Marzooq. 2023. "Unraveling the Impact of Gut and Oral Microbiome on Gut Health in Inflammatory Bowel Diseases" Nutrients 15, no. 15: 3377. https://doi.org/10.3390/nu15153377
APA StyleElzayat, H., Mesto, G., & Al-Marzooq, F. (2023). Unraveling the Impact of Gut and Oral Microbiome on Gut Health in Inflammatory Bowel Diseases. Nutrients, 15(15), 3377. https://doi.org/10.3390/nu15153377






