Defining NAD(P)(H) Catabolism
Abstract
1. Introduction
2. NAD(P)(H) Metabolism
3. Non-Ribosylated Catabolites of NAD(P)(H)
4. Ribosylated Catabolites of NAD(P)(H)
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Arthur, H. The alcoholic ferment of yeast-juice. Proc. R. Soc. Lond. B 1906, 77, 405–420. [Google Scholar] [CrossRef]
- Harden, A. The alcoholic ferment of yeast-juice. Part II.-The coferment of yeast-juice. Proc. R. Soc. B Biol. Sci. 1906, 78, 369–375. [Google Scholar]
- Berger, F.; Ramirez-Hernandez, M.H.; Ziegler, M. The new life of a centenarian: Signalling functions of NAD(P). Trends Biochem. Sci. 2004, 29, 111–118. [Google Scholar] [CrossRef]
- Canto, C.; Auwerx, J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 291–298. [Google Scholar] [CrossRef]
- Amjad, S.; Nisar, S.; Bhat, A.A.; Shah, A.R.; Frenneaux, M.P.; Fakhro, K.; Haris, M.; Reddy, R.; Patay, Z.; Baur, J.; et al. Role of NAD+ in regulating cellular and metabolic signaling pathways. Mol. Metab. 2021, 49, 101195. [Google Scholar] [CrossRef] [PubMed]
- McReynolds, M.R.; Chellappa, K.; Chiles, E.; Jankowski, C.; Shen, Y.; Chen, L.; Descamps, H.C.; Mukherjee, S.; Bhat, Y.R.; Lingala, S.R.; et al. NAD+ flux is maintained in aged mice despite lower tissue concentrations. Cell Syst. 2021, 12, 1160–1172. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Raju, R.P. Regulation of NAD+ metabolism in aging and disease. Metabolism 2022, 126, 154923. [Google Scholar] [CrossRef]
- Chini, C.C.S.; Zeidler, J.D.; Kashyap, S.; Warner, G.; Chini, E.N. Evolving concepts in NAD+ metabolism. Cell Metab. 2021, 33, 1076–1087. [Google Scholar] [CrossRef]
- Alonso-Lavin, A.J.; Bajic, D.; Poyatos, J.F. Tolerance to NADH/NAD+ imbalance anticipates aging and anti-aging interventions. iScience 2021, 24, 102697. [Google Scholar] [CrossRef]
- Shabalin, K.; Nerinovski, K.; Yakimov, A.; Kulikova, V.; Svetlova, M.; Solovjeva, L.; Khodorkovskiy, M.; Gambaryan, S.; Cunningham, R.; Migaud, M.E.; et al. NAD Metabolome Analysis in Human Cells Using 1H NMR Spectroscopy. Int. J. Mol. Sci. 2018, 19, 3906. [Google Scholar] [CrossRef]
- Xiao, W.; Loscalzo, J. Metabolic Responses to Reductive Stress. Antioxid. Redox Signal. 2020, 32, 1330–1347. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Markhard, A.L.; Shah, H.; Sharma, R.; Skinner, O.S.; Clish, C.B.; Deik, A.; Patgiri, A.; Hsu, Y.-H.H.; Masia, R.; et al. Hepatic NADH reductive stress underlies common variation in metabolic traits. Nature 2020, 583, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Chini, C.C.S.; Peclat, T.R.; Gomez, L.S.; Zeidler, J.D.; Warner, G.M.; Kashyap, S.; Mazdeh, D.Z.; Hayat, F.; Migaud, M.E.; Paulus, A.; et al. Dihydronicotinamide Riboside Is a Potent NAD+ Precursor Promoting a Pro-Inflammatory Phenotype in Macrophages. Front. Immunol. 2022, 13, 840246. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef] [PubMed]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD+ Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Yang, F.; Deng, X.; Yu, Y.; Luo, L.; Chen, X.; Zheng, J.; Qiu, Y.; Xiao, F.; Xie, X.; Zhao, Y.; et al. Association of Human Whole Blood NAD+ Contents With Aging. Front. Endocrinol. 2022, 13, 829658. [Google Scholar] [CrossRef]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 2020, 5, 227. [Google Scholar] [CrossRef]
- Hikosaka, K.; Yaku, K.; Okabe, K.; Nakagawa, T. Implications of NAD metabolism in pathophysiology and therapeutics for neurodegenerative diseases. Nutr. Neurosci. 2021, 24, 371–383. [Google Scholar] [CrossRef]
- Cabrera-Rode, E.; Molina, G.; Arranz, C.; Vera, M.; Gonzalez, P.; Suarez, R.; Prieto, M.; Padron, S.; Leon, R.; Tillan, J.; et al. Effect of standard nicotinamide in the prevention of type 1 diabetes in first degree relatives of persons with type 1 diabetes. Autoimmunity 2006, 39, 333–340. [Google Scholar] [CrossRef]
- Feuz, M.B.; Meyer-Ficca, M.L.; Meyer, R.G. Beyond Pellagra-Research Models and Strategies Addressing the Enduring Clinical Relevance of NAD Deficiency in Aging and Disease. Cells 2023, 12, 500. [Google Scholar] [CrossRef]
- Poyan Mehr, A.; Tran, M.T.; Ralto, K.M.; Leaf, D.E.; Washco, V.; Messmer, J.; Lerner, A.; Kher, A.; Kim, S.H.; Khoury, C.C.; et al. De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat. Med. 2018, 24, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, L.; Zhang, L. NAD+ Metabolism as an Emerging Therapeutic Target for Cardiovascular Diseases Associated with Sudden Cardiac Death. Front. Physiol. 2020, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Poljsak, B.; Kovac, V.; Spalj, S.; Milisav, I. The Central Role of the NAD+ Molecule in the Development of Aging and the Prevention of Chronic Age-Related Diseases: Strategies for NAD+ Modulation. Int. J. Mol. Sci. 2023, 24, 2959. [Google Scholar] [CrossRef] [PubMed]
- Rajman, L.; Chwalek, K.; Sinclair, D.A. Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence. Cell Metab. 2018, 27, 529–547. [Google Scholar] [CrossRef]
- Camacho-Pereira, J.; Tarrago, M.G.; Chini, C.C.S.; Nin, V.; Escande, C.; Warner, G.M.; Puranik, A.S.; Schoon, R.A.; Reid, J.M.; Galina, A.; et al. CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. Cell Metab. 2016, 23, 1127–1139. [Google Scholar] [CrossRef]
- Tarrago, M.G.; Chini, C.C.S.; Kanamori, K.S.; Warner, G.M.; Caride, A.; de Oliveira, G.C.; Rud, M.; Samani, A.; Hein, K.Z.; Huang, R.; et al. A Potent and Specific CD38 Inhibitor Ameliorates Age-Related Metabolic Dysfunction by Reversing Tissue NAD+ Decline. Cell Metab. 2018, 27, 1081–1095. [Google Scholar] [CrossRef]
- Kang, B.E.; Choi, J.Y.; Stein, S.; Ryu, D. Implications of NAD+ boosters in translational medicine. Eur. J. Clin. Investig. 2020, 50, e13334. [Google Scholar] [CrossRef] [PubMed]
- Katsyuba, E.; Mottis, A.; Zietak, M.; De Franco, F.; van der Velpen, V.; Gariani, K.; Ryu, D.; Cialabrini, L.; Matilainen, O.; Liscio, P.; et al. De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature 2018, 563, 354–359. [Google Scholar] [CrossRef]
- Khaidizar, F.D.; Bessho, Y.; Nakahata, Y. Nicotinamide Phosphoribosyltransferase as a Key Molecule of the Aging/Senescence Process. Int. J. Mol. Sci. 2021, 22, 3709. [Google Scholar] [CrossRef]
- Gardell, S.J.; Hopf, M.; Khan, A.; Dispagna, M.; Hampton Sessions, E.; Falter, R.; Kapoor, N.; Brooks, J.; Culver, J.; Petucci, C.; et al. Boosting NAD+ with a small molecule that activates NAMPT. Nat. Commun. 2019, 10, 3241. [Google Scholar] [CrossRef]
- Dutta, T.; Kapoor, N.; Mathew, M.; Chakraborty, S.S.; Ward, N.P.; Prieto-Farigua, N.; Falzone, A.; DeLany, J.P.; Smith, S.R.; Coen, P.M.; et al. Source of nicotinamide governs its metabolic fate in cultured cells, mice, and humans. Cell Rep. 2023, 42, 112218. [Google Scholar] [CrossRef]
- Janssens, G.; Grevendonk, L.; Perez, R.; Schomakers, B.; van den Bosch, J.; Geurts, J.; van Weeghek, M.; Schrauwen, P.; Houtkooper, R.; Hoeks, J. Healthy aging and muscle function are positively associated with NAD+ abundance in humans. Nature Aging 2022, 2, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Connell, N.J.; Grevendonk, L.; Fealy, C.E.; Moonen-Kornips, E.; Bruls, Y.M.H.; Schrauwen-Hinderling, V.B.; de Vogel, J.; Hageman, R.; Geurts, J.; Zapata-Perez, R.; et al. NAD+-Precursor Supplementation With L-Tryptophan, Nicotinic Acid, and Nicotinamide Does Not Affect Mitochondrial Function or Skeletal Muscle Function in Physically Compromised Older Adults. J. Nutr. 2021, 151, 2917–2931. [Google Scholar] [CrossRef] [PubMed]
- Pirinen, E.; Auranen, M.; Khan, N.A.; Brilhante, V.; Urho, N.; Pessia, A.; Hakkarainen, A.; Ulla Heinonen, J.K.; Schmidt, M.S.; Haimilahti, K.; et al. Niacin Cures Systemic NAD+ Deficiency and Improves Muscle Performance in Adult-Onset Mitochondrial Myopathy. Cell Metab. 2020, 32, 144. [Google Scholar] [CrossRef] [PubMed]
- Bogan, K.L.; Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: A molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008, 28, 115–130. [Google Scholar] [CrossRef]
- Jacobson, M.K.; Jacobson, E.L. Vitamin B3 in Health and Disease: Toward the Second Century of Discovery. Methods Mol. Biol. 2018, 1813, 3–8. [Google Scholar] [CrossRef]
- Elhassan, Y.S.; Kluckova, K.; Fletcher, R.S.; Schmidt, M.S.; Garten, A.; Doig, C.L.; Cartwright, D.M.; Oakey, L.; Burley, C.V.; Jenkinson, N.; et al. Nicotinamide Riboside Augments the Aged Human Skeletal Muscle NAD+ Metabolome and Induces Transcriptomic and Anti-inflammatory Signatures. Cell Rep. 2019, 28, 1717–1728.e6. [Google Scholar] [CrossRef]
- Martens, C.R.; Denman, B.A.; Mazzo, M.R.; Armstrong, M.L.; Reisdorph, N.; McQueen, M.B.; Chonchol, M.; Seals, D.R. Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD+ in healthy middle-aged and older adults. Nat. Commun. 2018, 9, 1286. [Google Scholar] [CrossRef]
- Airhart, S.E.; Shireman, L.M.; Risler, L.J.; Anderson, G.D.; Nagana Gowda, G.A.; Raftery, D.; Tian, R.; Shen, D.D.; O’Brien, K.D. An open-label, non-randomized study of the pharmacokinetics of the nutritional supplement nicotinamide riboside (NR) and its effects on blood NAD+ levels in healthy volunteers. PLoS ONE 2017, 12, e0186459. [Google Scholar] [CrossRef]
- Yamamoto, T.; Byun, J.; Zhai, P.; Ikeda, Y.; Oka, S.; Sadoshima, J. Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PLoS ONE 2014, 9, e98972. [Google Scholar] [CrossRef]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef]
- Yi, L.; Maier, A.B.; Tao, R.; Lin, Z.; Vaidya, A.; Pendse, S.; Thasma, S.; Andhalkar, N.; Avhad, G.; Kumbhar, V. The efficacy and safety of beta-nicotinamide mononucleotide (NMN) supplementation in healthy middle-aged adults: A randomized, multicenter, double-blind, placebo-controlled, parallel-group, dose-dependent clinical trial. Geroscience 2023, 45, 29–43. [Google Scholar] [CrossRef]
- Nadeeshani, H.; Li, J.; Ying, T.; Zhang, B.; Lu, J. Nicotinamide mononucleotide (NMN) as an anti-aging health product—Promises and safety concerns. J. Adv. Res. 2022, 37, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Zapata-Perez, R.; Tammaro, A.; Schomakers, B.V.; Scantlebery, A.M.L.; Denis, S.; Elfrink, H.L.; Giroud-Gerbetant, J.; Canto, C.; Lopez-Leonardo, C.; McIntyre, R.L.; et al. Reduced nicotinamide mononucleotide is a new and potent NAD+ precursor in mammalian cells and mice. FASEB J. 2021, 35, e21456. [Google Scholar] [CrossRef] [PubMed]
- Giroud-Gerbetant, J.; Joffraud, M.; Giner, M.P.; Cercillieux, A.; Bartova, S.; Makarov, M.V.; Zapata-Perez, R.; Sanchez-Garcia, J.L.; Houtkooper, R.H.; Migaud, M.E.; et al. A reduced form of nicotinamide riboside defines a new path for NAD+ biosynthesis and acts as an orally bioavailable NAD+ precursor. Mol. Metab. 2019, 30, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Mohammed, F.S.; Zhang, N.; Sauve, A.A. Dihydronicotinamide riboside is a potent NAD+ concentration enhancer in vitro and in vivo. J. Biol. Chem. 2019, 294, 9295–9307. [Google Scholar] [CrossRef] [PubMed]
- Reiten, O.K.; Wilvang, M.A.; Mitchell, S.J.; Hu, Z.; Fang, E.F. Preclinical and clinical evidence of NAD+ precursors in health, disease, and ageing. Mech. Ageing Dev. 2021, 199, 111567. [Google Scholar] [CrossRef]
- Bieganowski, P.; Brenner, C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004, 117, 495–502. [Google Scholar] [CrossRef]
- Ratajczak, J.; Joffraud, M.; Trammell, S.A.; Ras, R.; Canela, N.; Boutant, M.; Kulkarni, S.S.; Rodrigues, M.; Redpath, P.; Migaud, M.E.; et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 2016, 7, 13103. [Google Scholar] [CrossRef]
- Fletcher, R.S.; Ratajczak, J.; Doig, C.L.; Oakey, L.A.; Callingham, R.; Da Silva Xavier, G.; Garten, A.; Elhassan, Y.S.; Redpath, P.; Migaud, M.E.; et al. Nicotinamide riboside kinases display redundancy in mediating nicotinamide mononucleotide and nicotinamide riboside metabolism in skeletal muscle cells. Mol. Metab. 2017, 6, 819–832. [Google Scholar] [CrossRef]
- Ciarlo, E.; Joffraud, M.; Hayat, F.; Giner, M.P.; Giroud-Gerbetant, J.; Sanchez-Garcia, J.L.; Rumpler, M.; Moco, S.; Migaud, M.E.; Canto, C. Nicotinamide Riboside and Dihydronicotinic Acid Riboside Synergistically Increase Intracellular NAD+ by Generating Dihydronicotinamide Riboside. Nutrients 2022, 14, 2752. [Google Scholar] [CrossRef]
- Li, J.; Koczor, C.A.; Saville, K.M.; Hayat, F.; Beiser, A.; McClellan, S.; Migaud, M.E.; Sobol, R.W. Overcoming Temozolomide Resistance in Glioblastoma via Enhanced NAD+ Bioavailability and Inhibition of Poly-ADP-Ribose Glycohydrolase. Cancers 2022, 14, 3572. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, N.; Zhang, G.; Sauve, A.A. NRH salvage and conversion to NAD+ requires NRH kinase activity by adenosine kinase. Nat. Metab. 2020, 2, 364–379. [Google Scholar] [CrossRef] [PubMed]
- Grozio, A.; Mills, K.F.; Yoshino, J.; Bruzzone, S.; Sociali, G.; Tokizane, K.; Lei, H.C.; Cunningham, R.; Sasaki, Y.; Migaud, M.E.; et al. Slc12a8 is a nicotinamide mononucleotide transporter. Nat. Metab. 2019, 1, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, J.; Baur, J.A.; Imai, S.I. NAD+ Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018, 27, 513–528. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.F.; Yoshida, S.; Stein, L.R.; Grozio, A.; Kubota, S.; Sasaki, Y.; Redpath, P.; Migaud, M.E.; Apte, R.S.; Uchida, K.; et al. Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metab. 2016, 24, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Yegutkin, G.G. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: Functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. [Google Scholar] [CrossRef]
- Kropotov, A.; Kulikova, V.; Nerinovski, K.; Yakimov, A.; Svetlova, M.; Solovjeva, L.; Sudnitsyna, J.; Migaud, M.E.; Khodorkovskiy, M.; Ziegler, M.; et al. Equilibrative Nucleoside Transporters Mediate the Import of Nicotinamide Riboside and Nicotinic Acid Riboside into Human Cells. Int. J. Mol. Sci. 2021, 22, 1391. [Google Scholar] [CrossRef]
- Kropotov, A.; Kulikova, V.; Solovjeva, L.; Yakimov, A.; Nerinovski, K.; Svetlova, M.; Sudnitsyna, J.; Plusnina, A.; Antipova, M.; Khodorkovskiy, M.; et al. Purine nucleoside phosphorylase controls nicotinamide riboside metabolism in mammalian cells. J. Biol. Chem. 2022, 298, 102615. [Google Scholar] [CrossRef]
- Yaku, K.; Palikhe, S.; Izumi, H.; Yoshida, T.; Hikosaka, K.; Hayat, F.; Karim, M.; Iqbal, T.; Nitta, Y.; Sato, A.; et al. BST1 regulates nicotinamide riboside metabolism via its glycohydrolase and base-exchange activities. Nat. Commun. 2021, 12, 6767. [Google Scholar] [CrossRef]
- Kulikova, V.; Shabalin, K.; Nerinovski, K.; Yakimov, A.; Svetlova, M.; Solovjeva, L.; Kropotov, A.; Khodorkovskiy, M.; Migaud, M.E.; Ziegler, M.; et al. Degradation of Extracellular NAD+ Intermediates in Cultures of Human HEK293 Cells. Metabolites 2019, 9, 293. [Google Scholar] [CrossRef]
- Makarov, M.V.; Hayat, F.; Graves, B.; Sonavane, M.; Salter, E.A.; Wierzbicki, A.; Gassman, N.R.; Migaud, M.E. Chemical and Biochemical Reactivity of the Reduced Forms of Nicotinamide Riboside. ACS Chem. Biol. 2021, 16, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Segal, R.; Stein, G. 1010. The effect of acids on dihydronicotinamide derivatives. J. Chem. Soc. 1960, 5254–5257. [Google Scholar] [CrossRef]
- Berthiaume, J.M.; Kurdys, J.G.; Muntean, D.M.; Rosca, M.G. Mitochondrial NAD+/NADH Redox State and Diabetic Cardiomyopathy. Antioxid. Redox Signal. 2019, 30, 375–398. [Google Scholar] [CrossRef] [PubMed]
- Makarov, M.V.; Trammell, S.A.J.; Migaud, M.E. The chemistry of the vitamin B3 metabolome. Biochem. Soc. Trans. 2019, 47, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Savvidou, S. Pellagra: A non-eradicated old disease. Clin. Pract. 2014, 4, 637. [Google Scholar] [CrossRef]
- Maeta, A.; Sano, M.; Fukuwatari, T.; Shibata, K. Simultaneous measurement of nicotinamide and its catabolites, nicotinamide N-oxide, N1-methyl-2-pyridone-5-carboxamide, and N1-methyl-4-pyridone-3-carboxamide, in mice urine. Biosci. Biotechnol. Biochem. 2014, 78, 1306–1309. [Google Scholar] [CrossRef] [PubMed]
- Slominska, E.M.; Adamski, P.; Lipinski, M.; Swierczynski, J.; Smolenski, R.T. Liquid chromatographic/mass spectrometric procedure for measurement of NAD catabolites in human and rat plasma and urine. Nucleosides Nucleotides Nucleic Acids 2006, 25, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Morley, N.H.; Storvick, C.A.; Duryee, F.; Edwards, M.; Irgens-Møller, I. Oxidized Pyridine Nucleotides in Various Fractions of the Blood and Niacin and Tryptophan Metabolites in the Urine of Women on a Controlled Adequate Dietary. J. Nutr. 1957, 63, 539–554. [Google Scholar] [CrossRef]
- Hayat, F.; Sonavane, M.; Makarov, M.V.; Trammell, S.A.J.; McPherson, P.; Gassman, N.R.; Migaud, M.E. The biochemical pathways of nicotinamide-derived pyridones. Int. J. Mol. Sci. 2021, 22, 1145. [Google Scholar] [CrossRef]
- Rutkowski, P.; Slominska, E.M.; Wolyniec, W.; Smolenski, R.T.; Szolkiewicz, M.; Swierczynski, J.; Rutkowski, B. Nicotinamide metabolites accumulate in the tissues of uremic rats. J. Ren. Nutr. 2008, 18, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Shibata, K.; Morita, N.; Shibata, Y.; Fukuwatari, T. Enzymes that control the conversion of L-tryptophan-nicotinamide and the urinary excretion ratio (N1-methyl-2-pyridone-5-carboxamide + N1-methyl-4-pyridone-3-carboxamide)/N1-methylnicotinamide in mice. Biosci. Biotechnol. Biochem. 2013, 77, 2105–2111. [Google Scholar] [CrossRef] [PubMed]
- Shibata, K.; Fukuwatari, T. Pyridone compounds, catabolites of NAD are new uremic toxins. Bitamin 2007, 81, 571–574. [Google Scholar]
- Li, D.; Tian, Y.J.; Guo, J.; Sun, W.P.; Lun, Y.Z.; Guo, M.; Luo, N.; Cao, Y.; Cao, J.M.; Gong, X.J.; et al. Nicotinamide supplementation induces detrimental metabolic and epigenetic changes in developing rats. Br. J. Nutr. 2013, 110, 2156–2164. [Google Scholar] [CrossRef]
- Sonavane, M.; Hayat, F.; Makarov, M.; Migaud, M.E.; Gassman, N.R. Dihydronicotinamide riboside promotes cell-specific cytotoxicity by tipping the balance between metabolic regulation and oxidative stress. PLoS ONE 2020, 15, e0242174. [Google Scholar] [CrossRef]
- Shi, W.; Hegeman, M.A.; Doncheva, A.; Bekkenkamp-Grovenstein, M.; de Boer, V.C.J.; Keijer, J. High Dose of Dietary Nicotinamide Riboside Induces Glucose Intolerance and White Adipose Tissue Dysfunction in Mice Fed a Mildly Obesogenic Diet. Nutrients 2019, 11, 2439. [Google Scholar] [CrossRef]
- Sun, W.P.; Li, D.; Lun, Y.Z.; Gong, X.J.; Sun, S.X.; Guo, M.; Jing, L.X.; Zhang, L.B.; Xiao, F.C.; Zhou, S.S. Excess nicotinamide inhibits methylation-mediated degradation of catecholamines in normotensives and hypertensives. Hypertens. Res. 2012, 35, 180–185. [Google Scholar] [CrossRef]
- Shats, I.; Williams, J.G.; Liu, J.; Makarov, M.V.; Wu, X.; Lih, F.B.; Deterding, L.J.; Lim, C.; Xu, X.; Randall, T.A.; et al. Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metab. 2020, 31, 564–579.e7. [Google Scholar] [CrossRef]
- Chellappa, K.; McReynolds, M.R.; Lu, W.; Zeng, X.; Makarov, M.; Hayat, F.; Mukherjee, S.; Bhat, Y.R.; Lingala, S.R.; Shima, R.T.; et al. NAD precursors cycle between host tissues and the gut microbiome. Cell Metab. 2022, 34, 1947–1959.e5. [Google Scholar] [CrossRef]
- Mrochek, J.E.; Jolley, R.L.; Young, D.S.; Turner, W.J. Metabolic response of humans to ingestion of nicotinic acid and nicotinamide. Clin. Chem. 1976, 22, 1821–1827. [Google Scholar] [CrossRef]
- Shibata, K.; Fukuwatari, T.; Suzuki, C. Pharmacological doses of nicotinic acid and nicotinamide are independently metabolized in rats. J. Nutr. Sci. Vitaminol. 2014, 60, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Inamadugu, J.K.; Damaramadugu, R.; Mullangi, R.; Ponneri, V. Simultaneous determination of niacin and its metabolites--nicotinamide, nicotinuric acid and N-methyl-2-pyridone-5-carboxamide--in human plasma by LC-MS/MS and its application to a human pharmacokinetic study. Biomed. Chromatogr. 2010, 24, 1059–1074. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Cheng, M.L.; Fan, C.M.; Hong, C.Y.; Shiao, M.S. Nicotinuric acid: A potential marker of metabolic syndrome through a metabolomics-based approach. Diabetes Care 2013, 36, 1729–1731. [Google Scholar] [CrossRef]
- Real, A.M.; Hong, S.; Pissios, P. Nicotinamide N-oxidation by CYP2E1 in human liver microsomes. Drug Metab. Dispos. 2013, 41, 550–553. [Google Scholar] [CrossRef]
- Fukuwatari, T.; Wada, H.; Sasaki, R.; Shibata, K. Effects of excess nicotinamide administration on the urinary excretion of nicotinamide N-oxide and nicotinuric acid by rats. Biosci. Biotechnol. Biochem. 2004, 68, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Pissios, P. Nicotinamide N-Methyltransferase: More Than a Vitamin B3 Clearance Enzyme. Trends Endocrinol. Metab. 2017, 28, 340–353. [Google Scholar] [CrossRef]
- Nejabati, H.R.; Mihanfar, A.; Pezeshkian, M.; Fattahi, A.; latifi, Z.; Safaie, N.; Valiloo, M.; Jodati, A.R.; Nouri, M. N1-methylnicotinamide (MNAM) as a guardian of cardiovascular system. J. Cell. Physiol. 2018, 233, 6386–6394. [Google Scholar] [CrossRef]
- Blazejczyk, A.; Switalska, M.; Chlopicki, S.; Marcinek, A.; Gebicki, J.; Nowak, M.; Nasulewicz-Goldeman, A.; Wietrzyk, J. 1-methylnicotinamide and its structural analog 1,4-dimethylpyridine for the prevention of cancer metastasis. J. Exp. Clin. Cancer Res. 2016, 35, 110. [Google Scholar] [CrossRef]
- Campagna, R.; Mateuszuk, L.; Wojnar-Lason, K.; Kaczara, P.; Tworzydlo, A.; Kij, A.; Bujok, R.; Mlynarski, J.; Wang, Y.; Sartini, D.; et al. Nicotinamide N-methyltransferase in endothelium protects against oxidant stress-induced endothelial injury. Biochim Biophys Acta Mol Cell Res 2021, 1868, 119082. [Google Scholar] [CrossRef]
- Takahashi, R.; Kanda, T.; Komatsu, M.; Itoh, T.; Minakuchi, H.; Urai, H.; Kuroita, T.; Shigaki, S.; Tsukamoto, T.; Higuchi, N.; et al. The significance of NAD + metabolites and nicotinamide N-methyltransferase in chronic kidney disease. Sci. Rep. 2022, 12, 6398. [Google Scholar] [CrossRef]
- Komatsu, M.; Kanda, T.; Urai, H.; Kurokochi, A.; Kitahama, R.; Shigaki, S.; Ono, T.; Yukioka, H.; Hasegawa, K.; Tokuyama, H.; et al. NNMT activation can contribute to the development of fatty liver disease by modulating the NAD+ metabolism. Sci. Rep. 2018, 8, 8637. [Google Scholar] [CrossRef]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Gaare, J.J.; Dolle, C.; Brakedal, B.; Brugger, K.; Haugarvoll, K.; Nido, G.S.; Tzoulis, C. Nicotinamide riboside supplementation is not associated with altered methylation homeostasis in Parkinson’s disease. iScience 2023, 26, 106278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chan, L.; Zhou, S. Trigonelline: A plant alkaloid with therapeutic potential for diabetes and central nervous system disease. Curr. Med. Chem. 2012, 19, 3523–3531. [Google Scholar] [CrossRef]
- Mohamadi, N.; Sharififar, F.; Pournamdari, M.; Ansari, M. A Review on Biosynthesis, Analytical Techniques, and Pharmacological Activities of Trigonelline as a Plant Alkaloid. J. Diet. Suppl. 2018, 15, 207–222. [Google Scholar] [CrossRef]
- Konstantinidis, N.; Franke, H.; Schwarz, S.; Lachenmeier, D.W. Risk Assessment of Trigonelline in Coffee and Coffee By-Products. Molecules 2023, 28, 3460. [Google Scholar] [CrossRef]
- Rutkowski, P.; Malgorzewicz, S.; Slominska, E.; Renke, M.; Lysiak-Szydlowska, W.; Swierczynski, J.; Rutkowski, B. Interrelationship between uremic toxicity and oxidative stress. J. Ren. Nutr. 2006, 16, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Terao, M.; Garattini, E.; Romao, M.J.; Leimkuhler, S. Evolution, expression, and substrate specificities of aldehyde oxidase enzymes in eukaryotes. J. Biol. Chem. 2020, 295, 5377–5389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.S.; Li, D.; Sun, W.P.; Guo, M.; Lun, Y.Z.; Zhou, Y.M.; Xiao, F.C.; Jing, L.X.; Sun, S.X.; Zhang, L.B.; et al. Nicotinamide overload may play a role in the development of type 2 diabetes. World J. Gastroenterol. 2009, 15, 5674–5684. [Google Scholar] [CrossRef]
- Lenglet, A.; Liabeuf, S.; Bodeau, S.; Louvet, L.; Mary, A.; Boullier, A.; Lemaire-Hurtel, A.S.; Jonet, A.; Sonnet, P.; Kamel, S.; et al. N-methyl-2-pyridone-5-carboxamide (2PY)-Major Metabolite of Nicotinamide: An Update on an Old Uremic Toxin. Toxins 2016, 8, 339. [Google Scholar] [CrossRef]
- Shibata, K.; Kawada, T.; Iwai, K. Simultaneous micro-determination of nicotinamide and its major metabolites, N1-methyl-2-pyridone-5-carboxamide and N1-methyl-4-pyridone-3-carboxamide, by high-performance liquid chromatography. J. Chromatogr. 1988, 424, 23–28. [Google Scholar] [CrossRef]
- Slominska, E.M.; Rutkowski, P.; Smolenski, R.T.; Szutowicz, A.; Rutkowski, B.; Swierczynski, J. The age-related increase in N-methyl-2-pyridone-5-carboxamide (NAD catabolite) in human plasma. Mol. Cell. Biochem. 2004, 267, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Gholson, R.K.; Raica, N. Isolation and identification of two new nicotinamide metabolites. J. Biol. Chem. 1969, 244, 3277–3282. [Google Scholar] [CrossRef] [PubMed]
- Trammell, S.A.; Schmidt, M.S.; Weidemann, B.J.; Redpath, P.; Jaksch, F.; Dellinger, R.W.; Li, Z.; Abel, E.D.; Migaud, M.E.; Brenner, C. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 2016, 7, 12948. [Google Scholar] [CrossRef] [PubMed]
- Mierzejewska, P.; Gawlik-Jakubczak, T.; Jablonska, P.; Czajkowski, M.; Kutryb-Zajac, B.; Smolenski, R.T.; Matuszewski, M.; Slominska, E.M. Nicotinamide metabolism alterations in bladder cancer: Preliminary studies. Nucleosides Nucleotides Nucleic Acids 2018, 37, 687–695. [Google Scholar] [CrossRef]
- Slominska, E.M.; Smolenski, R.T.; Szolkiewicz, M.; Leaver, N.; Rutkowski, B.; Simmonds, H.A.; Swierczynski, J. Accumulation of plasma N-methyl-2-pyridone-5-carboxamide in patients with chronic renal failure. Mol. Cell. Biochem. 2002, 231, 83–88. [Google Scholar] [CrossRef]
- Deen, C.P.J.; Veen, A.V.; Gomes-Neto, A.W.; Geleijnse, J.M.; Berg, K.; Heiner-Fokkema, M.R.; Kema, I.P.; Bakker, S.J.L. Urinary Excretion of N1-Methylnicotinamide and N1-Methyl-2-Pyridone-5-Carboxamide and Mortality in Kidney Transplant Recipients. Nutrients 2020, 12, 2059. [Google Scholar] [CrossRef]
- Deen, C.P.J.; van der Veen, A.; Gomes-Neto, A.W.; Geleijnse, J.M.; Borgonjen-van den Berg, K.J.; Heiner-Fokkema, M.R.; Kema, I.P.; Bakker, S.J.L. Urinary Excretion of N1-methyl-2-pyridone-5-carboxamide and N1-methylnicotinamide in Renal Transplant Recipients and Donors. J. Clin. Med. 2020, 9, 437. [Google Scholar] [CrossRef]
- Rutkowski, B.; Slominska, E.; Szolkiewicz, M.; Smolenski, R.T.; Striley, C.; Rutkowski, P.; Swierczynski, J. N-methyl-2-pyridone-5-carboxamide: A novel uremic toxin? Kidney Int. Suppl. 2003, 63, S19–S21. [Google Scholar] [CrossRef]
- Giner, M.P.; Christen, S.; Bartova, S.; Makarov, M.V.; Migaud, M.E.; Canto, C.; Moco, S. A Method to Monitor the NAD+ Metabolome-From Mechanistic to Clinical Applications. Int. J. Mol. Sci. 2021, 22, 10598. [Google Scholar] [CrossRef]
- Mills, G.C.; Davis, N.J.; Lertratanangkoon, K. Isolation and Identification of 1-Ribosyl Pyridone Nucleosides from Human Urine. Nucleosides Nucleotides 1989, 8, 415–430. [Google Scholar] [CrossRef]
- Dutta, S.P.; Crain, P.F.; McCloskey, J.A.; Chheda, G.B. Isolation and characterization of 1-β-D-ribofuranosylpyridin-4-one-3-carboxamide from human urine. Life Sci. 1979, 24, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Romaszko, P.; Slominska, E.M.; Orlewska, C.; Lipinski, M.; Smolenski, R.T. Metabolism of 4-pyridone-3-carboxamide-1-beta-D-ribonucleoside (4PYR) in rodent tissues and in vivo. Mol. Cell. Biochem. 2011, 351, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Carrey, E.A.; Smolenski, R.T.; Edbury, S.M.; Laurence, A.; Marinaki, A.M.; Duley, J.A.; Zhu, L.; Goldsmith, D.J.; Simmonds, H.A. Origin and characteristics of an unusual pyridine nucleotide accumulating in erythrocytes: Positive correlation with degree of renal failure. Clin. Chim. Acta 2003, 335, 117–129. [Google Scholar] [CrossRef]
- Slominska, E.M.; Carrey, E.A.; Foks, H.; Orlewska, C.; Wieczerzak, E.; Sowinski, P.; Yacoub, M.H.; Marinaki, A.M.; Simmonds, H.A.; Smolenski, R.T. A novel nucleotide found in human erythrocytes, 4-pyridone-3-carboxamide-1-beta-D-ribonucleoside triphosphate. J. Biol. Chem. 2006, 281, 32057–32064. [Google Scholar] [CrossRef]
- Laurence, A.; Edbury, S.M.; Marinaki, A.M.; Smolenski, R.T.; Goldsmith, D.J.; Simmonds, H.A.; Carrey, E.A. 4-pyridone-3-carboxamide ribonucleoside triphosphate accumulating in erythrocytes in end stage renal failure originates from tryptophan metabolism. Clin. Exp. Med. 2007, 7, 135–141. [Google Scholar] [CrossRef]
- Slominska, E.M.; Orlewska, C.; Yuen, A.; Osman, L.; Romaszko, P.; Sokolowska, E.; Foks, H.; Simmonds, H.A.; Yacoub, M.H.; Smolenski, R.T. Metabolism of 4-pyridone-3-carboxamide-1-beta-D-ribonucleoside triphosphate and its nucleoside precursor in the erythrocytes. Nucleosides Nucleotides Nucleic Acids 2008, 27, 830–834. [Google Scholar] [CrossRef]
- Synesiou, E.; Fairbanks, L.D.; Simmonds, H.A.; Slominska, E.M.; Smolenski, R.T.; Carrey, E.A. 4-Pyridone-3-carboxamide-1-beta-D-ribonucleoside triphosphate (4PyTP), a novel NAD metabolite accumulating in erythrocytes of uremic children: A biomarker for a toxic NAD analogue in other tissues? Toxins 2011, 3, 520–537. [Google Scholar] [CrossRef]
- de Rosa, M.; Pennati, A.; Pandini, V.; Monzani, E.; Zanetti, G.; Aliverti, A. Enzymatic oxidation of NADP+ to its 4-oxo derivative is a side-reaction displayed only by the adrenodoxin reductase type of ferredoxin-NADP+ reductases. FEBS J. 2007, 274, 3998–4007. [Google Scholar] [CrossRef]
- Bossi, R.T.; Aliverti, A.; Raimondi, D.; Fischer, F.; Zanetti, G.; Ferrari, D.; Tahallah, N.; Maier, C.S.; Heck, A.J.; Rizzi, M.; et al. A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase. Biochemistry 2002, 41, 8807–8818. [Google Scholar] [CrossRef]
- Aliverti, A.; Pandini, V.; Pennati, A.; de Rosa, M.; Zanetti, G. Structural and functional diversity of ferredoxin-NADP+ reductases. Arch. Biochem. Biophys. 2008, 474, 283–291. [Google Scholar] [CrossRef]
- den Braver-Sewradj, S.P.; den Braver, M.W.; Toorneman, R.M.; van Leeuwen, S.; Zhang, Y.; Dekker, S.J.; Vermeulen, N.P.E.; Commandeur, J.N.M.; Vos, J.C. Reduction and Scavenging of Chemically Reactive Drug Metabolites by NAD(P)H:Quinone Oxidoreductase 1 and NRH:Quinone Oxidoreductase 2 and Variability in Hepatic Concentrations. Chem. Res. Toxicol. 2018, 31, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, T.P.; Björklund, M. NQO2 Is a Reactive Oxygen Species Generating Off-Target for Acetaminophen. Mol. Pharm. 2014, 11, 4395–4404. [Google Scholar] [CrossRef] [PubMed]
- Hayat, F.; Makarov, M.V.; Belfleur, L.; Migaud, M.E. Synthesis of Mixed Dinucleotides by Mechanochemistry. Molecules 2022, 27, 3229. [Google Scholar] [CrossRef] [PubMed]
- Waszczuk-Jankowska, M.; Markuszewski, M.J.; Markuszewski, M.; Kaliszan, R. Comparison of RP-HPLC columns used for determination of nucleoside metabolic patterns in urine of cancer patients. Bioanalysis 2012, 4, 1185–1194. [Google Scholar] [CrossRef]
- Mierzejewska, P.; Kunc, M.; Zabielska-Kaczorowska, M.A.; Kutryb-Zajac, B.; Pelikant-Malecka, I.; Braczko, A.; Jablonska, P.; Romaszko, P.; Koszalka, P.; Szade, J.; et al. An unusual nicotinamide derivative, 4-pyridone-3-carboxamide ribonucleoside (4PYR), is a novel endothelial toxin and oncometabolite. Exp. Mol. Med. 2021, 53, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Intrieri, M.; Calcagno, G.; Oriani, G.; Pane, F.; Zarrilli, F.; Cataldo, P.T.; Foggia, M.; Piazza, M.; Salvatore, F.; Sacchetti, L. Pseudouridine and 1-ribosylpyridin-4-one-3-carboxamide (PCNR) serum concentrations in human immunodeficiency virus type 1-infected patients are independent predictors for AIDS progression. J. Infect. Dis. 1996, 174, 199–203. [Google Scholar] [CrossRef]
- Carrey, E.A.; Smolenski, R.T.; Edbury, S.M.; Laurence, A.; Marinaki, A.M.; Duley, J.A.; Zhu, L.M.; Goldsmith, D.J.; Simmonds, H.A. An unusual pyridine nucleotide accumulating in erythrocytes: Its identity and positive correlation with degree of renal failure. Nucleosides Nucleotides Nucleic Acids 2004, 23, 1135–1139. [Google Scholar] [CrossRef]
- Xu, G.; Schmid, H.R.; Lu, X.; Liebich, H.M.; Lu, P. Excretion pattern investigation of urinary normal and modified nucleosides of breast cancer patients by RP-HPLC and factor analysis method. Biomed. Chromatogr. 2000, 14, 459–463. [Google Scholar] [CrossRef]
- Schram, K.H. Urinary nucleosides. Mass Spectrom. Rev. 1998, 17, 131–251. [Google Scholar] [CrossRef]
- Slominska, E.M.; Borkowski, T.; Rybakowska, I.; Abramowicz-Glinka, M.; Orlewska, C.; Smolenski, R.T. In vitro and cellular effects of 4-pyridone-3-carboxamide riboside on enzymes of nucleotide metabolism. Nucleosides Nucleotides Nucleic Acids 2014, 33, 353–357. [Google Scholar] [CrossRef]
- Rutkowski, B.; Rutkowski, P.; Slominska, E.; Smolenski, R.T.; Swierczynski, J. Cellular toxicity of nicotinamide metabolites. J. Ren. Nutr. 2012, 22, 95–97. [Google Scholar] [CrossRef]
- Romaszko, P.; Slominska, E.M.; Smolenski, R.T. Effect of 4-pyridone-3-carboxamide ribonucleoside (4PYR)-potential cardiovascular toxin in perfused rat heart. Nucleosides Nucleotides Nucleic Acids 2014, 33, 333–337. [Google Scholar] [CrossRef]
- Pelikant-Malecka, I.; Sielicka, A.; Kaniewska, E.; Smolenski, R.T.; Slominska, E.M. 4-Pyridone-3-carboxamide-1beta-D-ribonucleoside metabolism in endothelial cells and its impact on cellular energetic balance. Nucleosides Nucleotides Nucleic Acids 2014, 33, 338–341. [Google Scholar] [CrossRef]
- Pelikant-Malecka, I.; Sielicka, A.; Kaniewska, E.; Smolenski, R.T.; Slominska, E.M. Endothelial toxicity of unusual nucleotide metabolites. Pharmacol. Rep. 2015, 67, 818–822. [Google Scholar] [CrossRef]
- Zabielska, M.; Kutryb-Zajac, B.; Zukowska, P.; Slominska, E.; Smolenski, R. Effects of 4-Pyridone-3-carboxamide-1beta-D-ribonucleoside on adenine nucleotide catabolism in the aortic wall; Implications for atherosclerosis in ApoE-/-LDLR-/- mice. Nucleosides Nucleotides Nucleic Acids 2016, 35, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Pelikant-Malecka, I.; Kaniewska-Bednarczuk, E.; Szrok, S.; Sielicka, A.; Sledzinski, M.; Orlewska, C.; Smolenski, R.T.; Slominska, E.M. Metabolic pathway of 4-pyridone-3-carboxamide-1beta-d-ribonucleoside and its effects on cellular energetics. Int. J. Biochem. Cell Biol. 2017, 88, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, P.; Mierzejewska, P.; Kutryb-Zajac, B.; Rzyman, W.; Dziadziuszko, R.; Polanska, J.; Sitkiewicz, M.; Smolenski, R.T.; Slominska, E.M. Increased plasma concentration of 4-pyridone-3-carboxamide-1-ss-D-ribonucleoside (4PYR) in lung cancer. Preliminary studies. Nucleosides Nucleotides Nucleic Acids 2019, 38, 781–787. [Google Scholar] [CrossRef]
- Koszalka, P.; Kutryb-Zajac, B.; Mierzejewska, P.; Tomczyk, M.; Wietrzyk, J.; Serafin, P.K.; Smolenski, R.T.; Slominska, E.M. 4-Pyridone-3-carboxamide-1-beta-D-ribonucleoside (4PYR)-A Novel Oncometabolite Modulating Cancer-Endothelial Interactions in Breast Cancer Metastasis. Int. J. Mol. Sci. 2022, 23, 5774. [Google Scholar] [CrossRef] [PubMed]
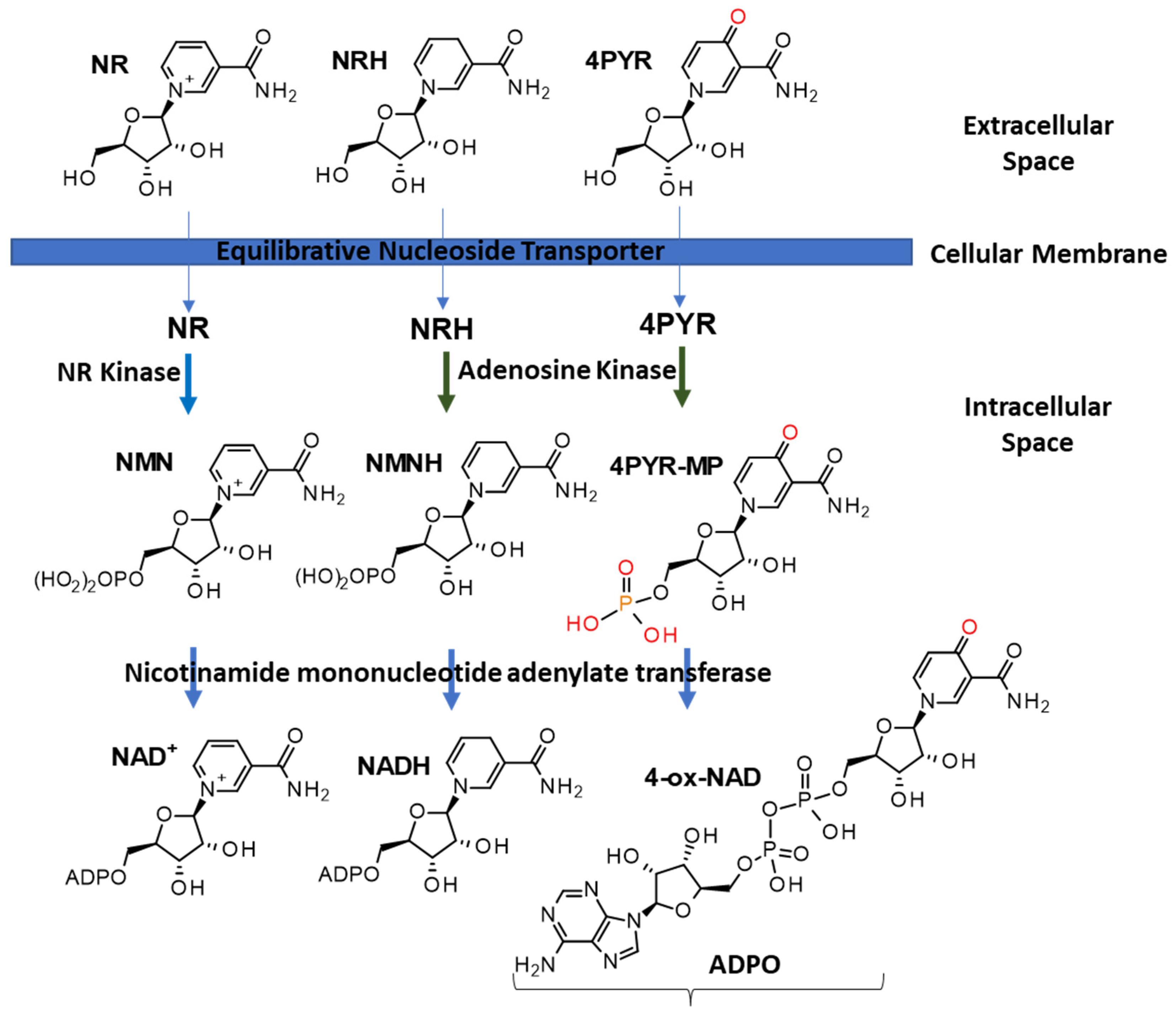
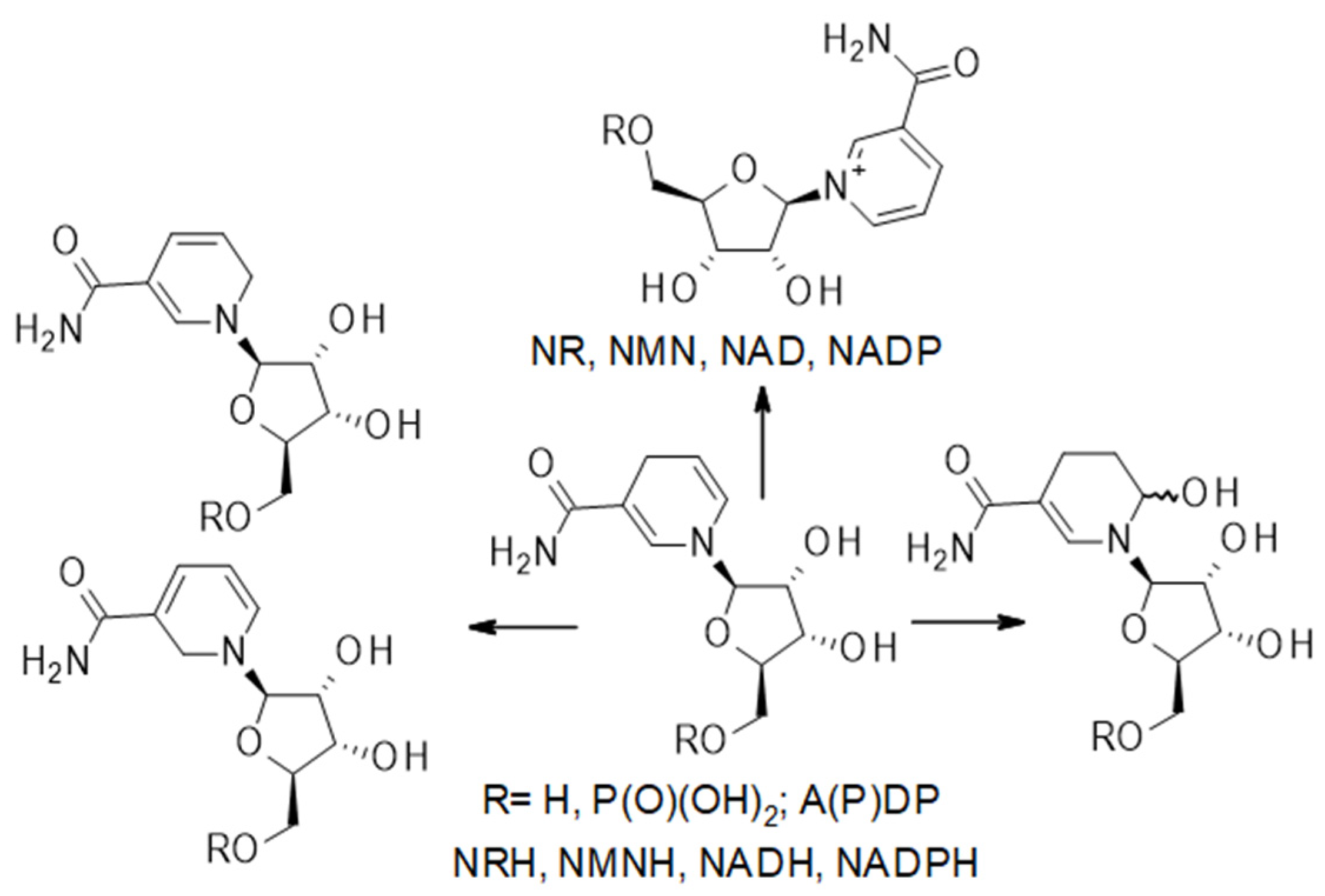
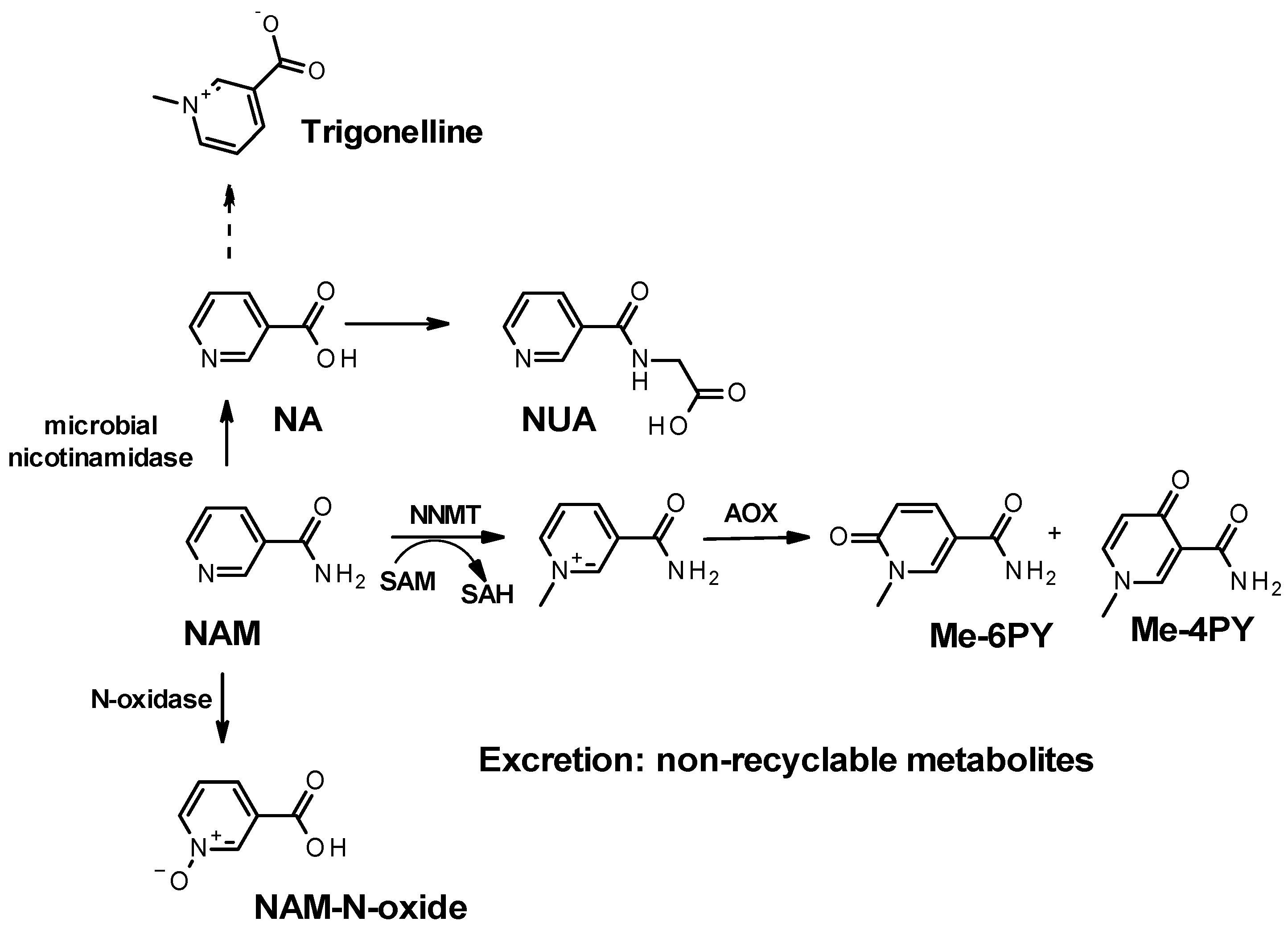
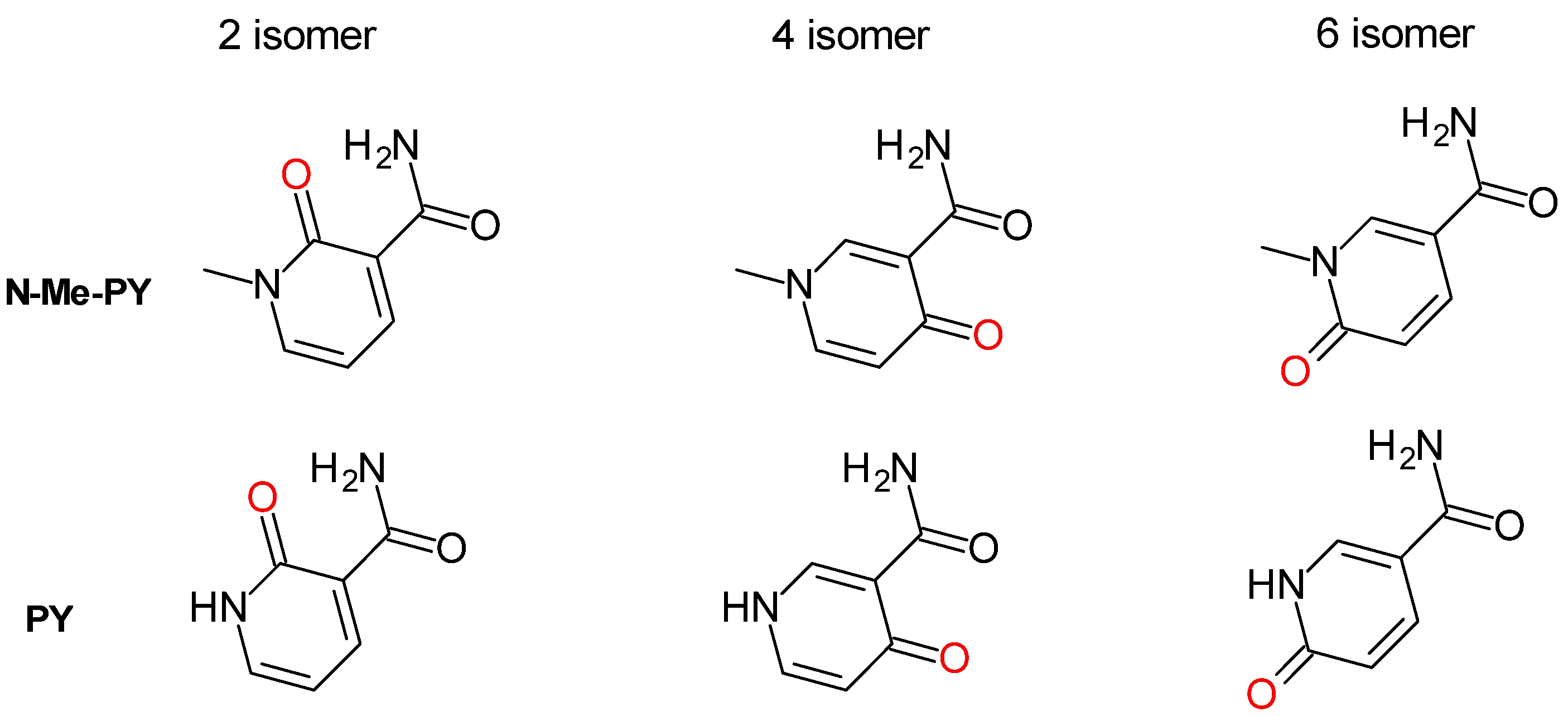
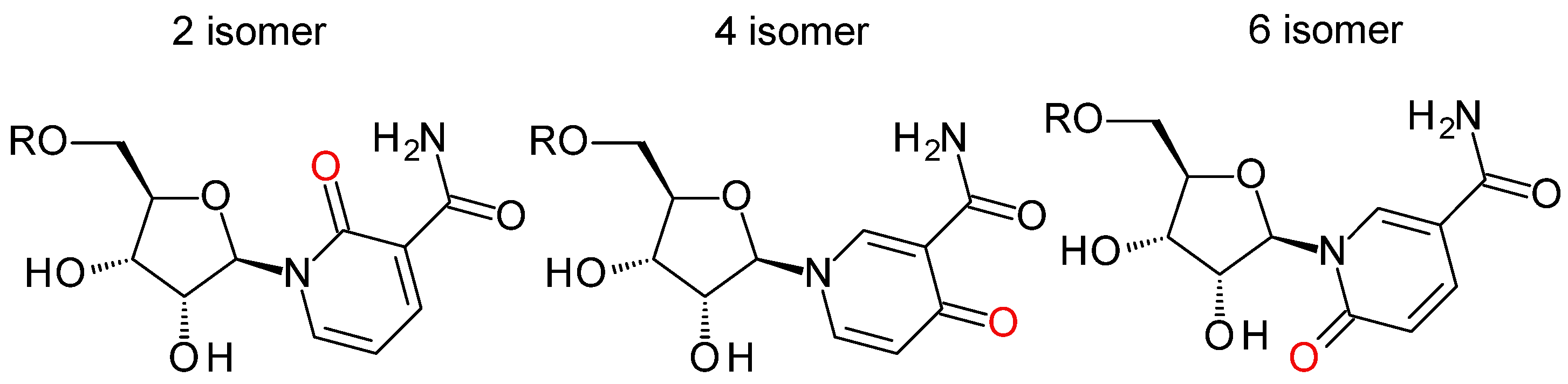
| Name | Abbreviation in Text | Formula | Measurable in | Reported in | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | S/P | T | U | B | S/P | T | U | |||||
| Niacinamide/nicotinamide | anabolites | nicotinamide | NAM | C6H6N2O | ||||||||
| nicotinamide riboside | NR | C11H15N2O5+ | ||||||||||
| nicotinamide riboside, reduced form | NRH | C11H16N2O5 | ||||||||||
| nicotinamide mononucleotide | NMN | C11H16N2O8P+ | ||||||||||
| nicotinamide mononucleotide, reduced form | NMNH | C11H17N2O8P | ||||||||||
| nicotinamide adenine dinucleotide | NAD | C21H28N7O14P2+ | ||||||||||
| nicotinamide adenine dinucleotide, reduced form | NADH | C21H29N7O14P2 | ||||||||||
| nicotinamide adenine dinucleotide phosphate | NADP | C21H29N7O17P3+ | ||||||||||
| nicotinamide adenine dinucleotide phosphate, reduced form | NADPH | C21H30N7O17P3 | ||||||||||
| catabolites | methyl-nicotinamide | N-Me-Nam | C7H9N2O+ | |||||||||
| methyl-2/4/6-pyridone | Me-2/4/6-PY | C7H8N2O2 | ||||||||||
| 2/4/6-hydroxy-nicotinamide | 2/4/6-PY | C6H6N2O2 | ||||||||||
| nicotinamide N-oxide | NAM-N-oxide | C6H6N2O2 | ||||||||||
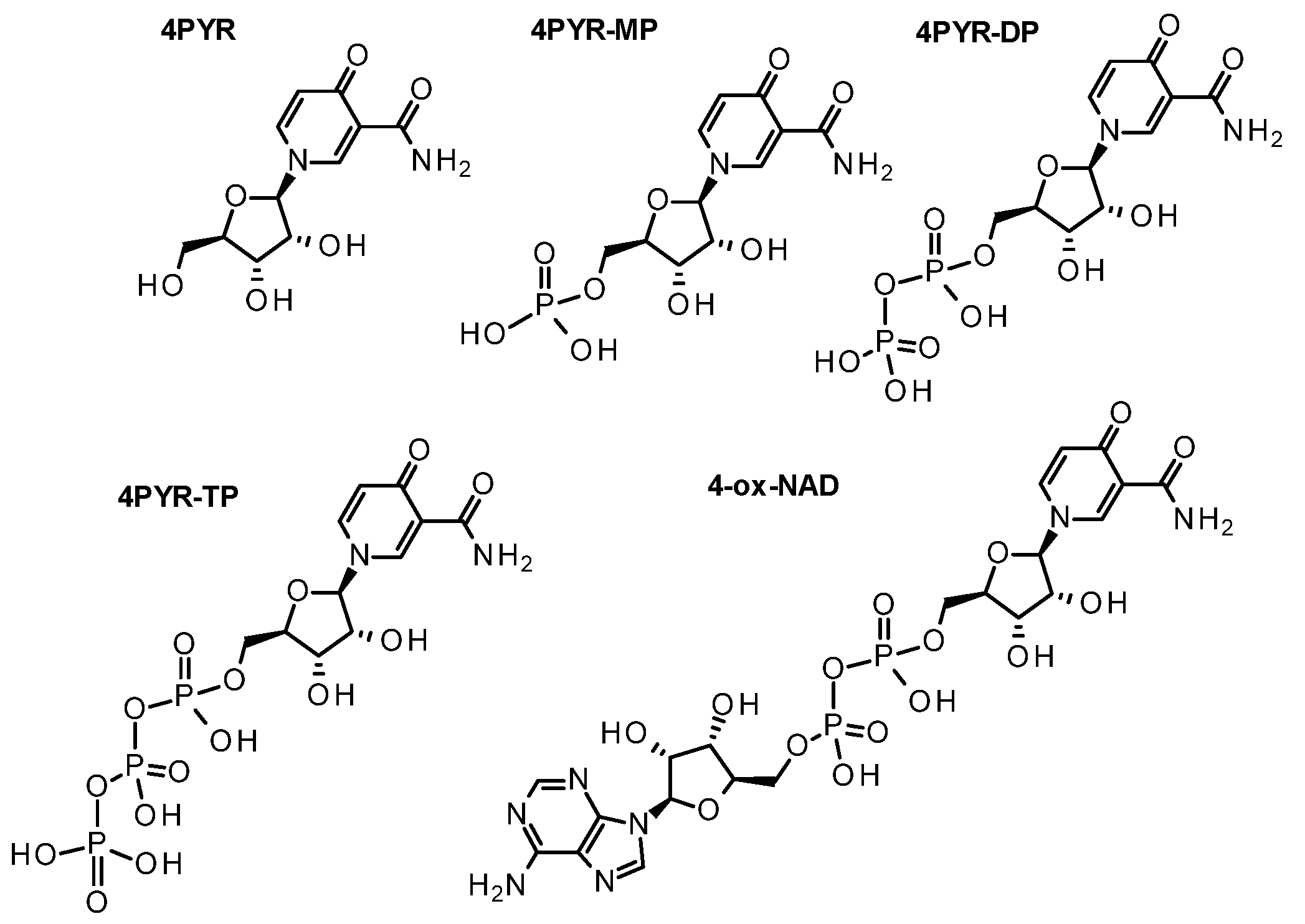
| 2/4/6-pyridone carboxamide riboside | 2/4/6-PYR | C11H14N2O6 | ||||||||||
| 2/4/6-pyridone carboxamide mononucleotide | 2/4/6-PYR-MP | C11H15N2O9P | ||||||||||
| 2/4/6-pyridone carboxamide riboside diphosphate | 2/4/6-PYR-DP | C11H16N2O12P2 | ||||||||||
| 2/4/6-pyridone carboxamide riboside triphosphate | 2/4/6-PYR-TP | C11H17N2O15P3 | ||||||||||
| 2/4/6-pyridone adenine dinucleotide | 2/4/6-ox-NAD | C21H27N7O15P2 | ||||||||||
| 2/4/6-pyridone adenine dinucleotide phosphate | 2/4/6-ox-NADP | C21H28N7O18P3 | ||||||||||
| Name | Abbreviation in text | Formula | Measurable in | Reported in | ||||||||
| B | S/P | T | U | B | S/P | T | U | |||||
| niacin | anabolites | Nicotinic acid | NA | C6H5NO2 | ||||||||
| nicotinic acid riboside | NAR | C11H14NO6+ | ||||||||||
| nicotinic acid mononucleotide | NAMN | C11H15NO9P+ | ||||||||||
| nicotinic acid adenine dinucleotide | NAAD | C21H27N6O15P2+ | ||||||||||
| catabolites | nicotinuric acid | NUA | C8H8N2O3 | |||||||||
| trigonelline | Trig | C7H8NO2+ | ||||||||||

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhuguru, J.; Dellinger, R.W.; Migaud, M.E. Defining NAD(P)(H) Catabolism. Nutrients 2023, 15, 3064. https://doi.org/10.3390/nu15133064
Dhuguru J, Dellinger RW, Migaud ME. Defining NAD(P)(H) Catabolism. Nutrients. 2023; 15(13):3064. https://doi.org/10.3390/nu15133064
Chicago/Turabian StyleDhuguru, Jyothi, Ryan W. Dellinger, and Marie E. Migaud. 2023. "Defining NAD(P)(H) Catabolism" Nutrients 15, no. 13: 3064. https://doi.org/10.3390/nu15133064
APA StyleDhuguru, J., Dellinger, R. W., & Migaud, M. E. (2023). Defining NAD(P)(H) Catabolism. Nutrients, 15(13), 3064. https://doi.org/10.3390/nu15133064







