1. Introduction
Ulcerative colitis (UC) is one subtype of inflammatory bowel disease (IBD) [
1,
2]; it is a growing concern worldwide due to its high prevalence in developed nations and the significant rise in occurrences in developing countries [
1,
3]. It is characterized by inflammation confined to the mucosa, beginning in the rectum and spreading proximally in the colon [
1,
2,
4]. Although the precise cause of UC remains unclear, various factors, such as an imbalanced immune response, modified gut microbiota, hereditary predisposition, and external factors, have been suggested as possible contributors [
1,
5].
UC is an intestinal barrier disease caused initially by intestinal epithelial dysfunction [
1,
6]. Many structural components maintain a fragile balance to sustain the homeostasis of the intestinal barrier [
1]. The intestinal microbiota plays a significant role in improving barrier function by providing nutritional and dietary factors on the luminal surface [
1,
7]. The first layer of protection is formed by the underlying epithelium and the mucus layer that follows the intestinal lumen [
8,
9,
10]. Lamina propria is the innermost layer, which maintains the non-inflamed condition of the tissue [
1,
11]. Intestinal barrier deficiencies have been associated with a diverse range of diseases, thus indicating a novel therapeutic objective [
7,
12].
The intricate nature of the pathogenesis of UC suggests that the interaction between the host and intestinal microbiota is widely acknowledged as a pivotal factor [
5]. The disturbance of the microbiota leads to a rapid proliferation of pathogenic bacteria within the intestinal tract directly invading and damaging the epithelial cells and intestinal mucosal barrier [
5,
13]; intestinal mucosal barrier function declines, the shield function of the intestinal wall abates, and the translocation of the intestinal microbiota exacerbates the damage to the intestinal mucosal barrier, leading to a detrimental cycle and intensifying the inflammatory response within the intestines [
5,
14]. In addition, gut microbiota-derived metabolites secreted into the lumen directly interact with epithelium and immune cells and significantly influence the preservation of gut barrier integrity and the maintenance of intestinal homeostasis [
15,
16,
17].
Currently available treatments for UC, including aminosalicylic acid, corticosteroids, immunomodulators, monoclonal antibodies, and even surgeries, often have side effects, drug resistance, and poor clinical efficacy [
2,
18]. Hence, it is imperative to undertake an immediate investigation into a novel, secure, and efficacious strategy for patients with UC. Natural bioactive polysaccharides have attracted increasing attention for their exceptional advantages of safety, accessibility, and good biocompatibility [
19,
20,
21]. It is well established that most of them can be metabolized by the gut microbiota and can exert profound protection against oxidative stress, toxins, inflammatory responses, and tumors [
22]. Although the protection of natural polysaccharides against colitis has been recently extensively studied, the role of gut microbiota and gut microbiota-derived metabolite remains incompletely understood and confirmed.
Nostoc commune Vaucher. (
N. commune) is classified as a scarce macroscopic cyanobacterium and has long been appreciated worldwide as a healthy food and medicine [
23]. Owing to the noticeable phytophysiological stress property, the protective physiological and pharmacological activity of it and its extracts are of particular interest and have promising characteristics. The polysaccharides from it have been shown to have some favorable characteristics, including being identified as heteropolysaccharides, with an ultra-high molecular weight of 210 kDa, as well as accounting for up to 60% of the dry weight of
N. commune [
24], suggesting fairly compelling bioactivity. We, therefore, aimed to elucidate whether the polysaccharides were protective against the occurrence of UC and to understand and explain the role of gut microbiota. An integrated analysis of multi-omics data, including the microbiome, non-targeted metabolomics, and transcriptomics, with the macroscopic and histopathological examination, fully elaborated that the polysaccharides markedly attenuated intestinal mucosal barrier damage, which mainly compromises UC. Together, our results revealed that the polysaccharides from
N. commune predominantly strengthened the gut microbiota-mediated intestinal mucosal barrier to confer protection against UC in mice and exhibited dramatical prebiotic-like functions, offering an alternative or supplementary therapeutic approach for individuals diagnosed with IBD, especially UC.
2. Materials and Methods
2.1. Mice
All research conducted on mice was in accordance with and approved by the institutional Animal Ethics Committee at Shanxi Medical University. Male C57BL/6J mice were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) as mixed littermates and housed under SPF and pathogen-free conditions in the animal facility at the Yingze Campus Laboratory Animal Research Center of Shanxi Medical University. The mice were co-housed for a duration of one week prior to being randomly assigned to experimental groups in order to synchronize the gut microbiome across the individual mice and to minimize heterogeneity. The investigators did not employ blinding techniques for allocation during experiments and outcome assessment, unless specified sections explicitly incorporated blind assessment.
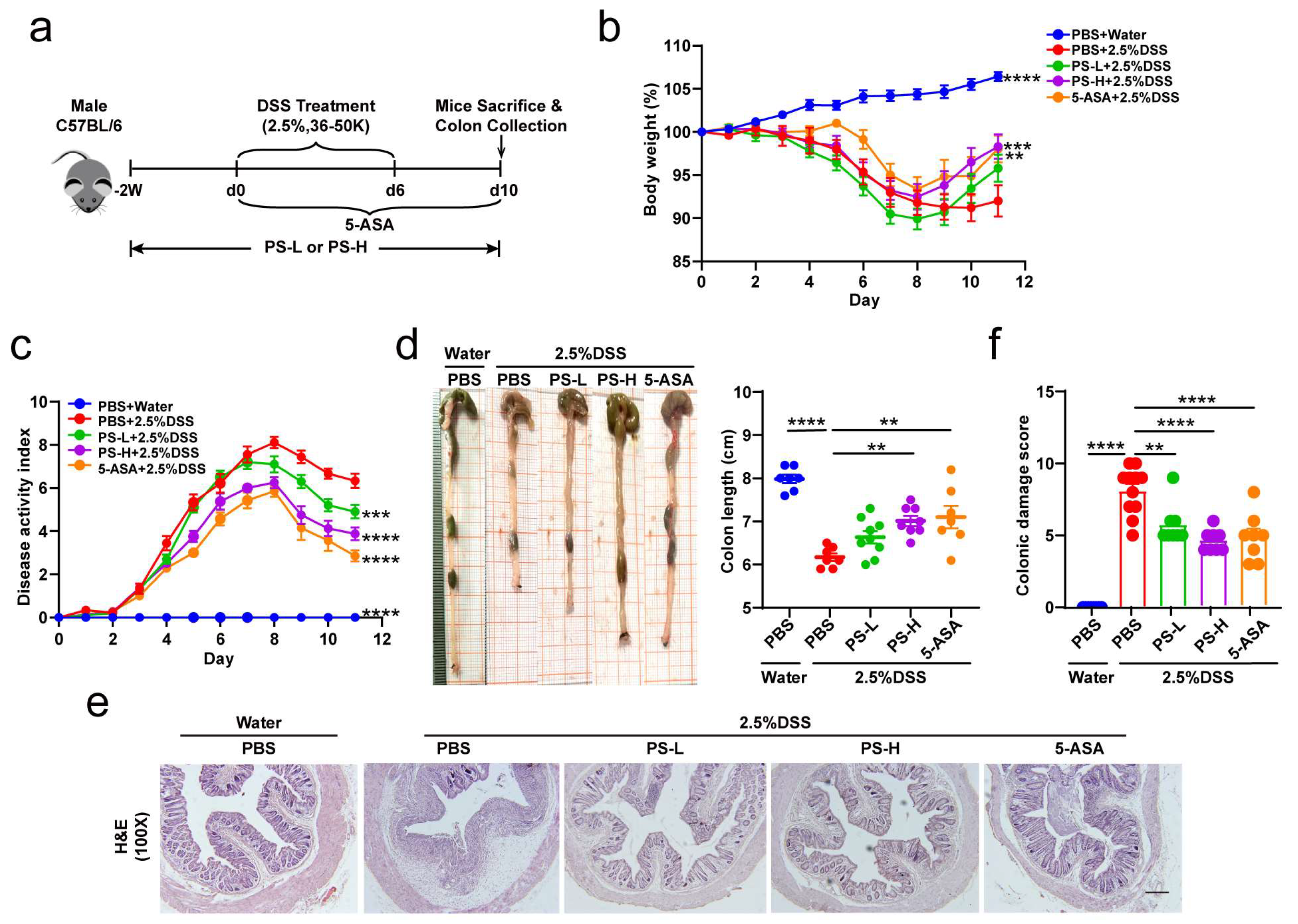
2.2. Induction of Acute DSS Colitis and Intervention
The model was conducted following a reported protocol with some minor modifications [
25,
26] (
Figure 1a). Briefly, mice received 2.5% dextran sodium sulfate (DSS, 36–50 kDa; MP Bio, Santa Ana, CA, USA) supplemented in the drinking water for 6 days, followed by normal water. PS was pre-administered at 200 mg/kg (PS-L) or 400 mg/kg (PS-H) by daily intragastric gavage that was started 2 weeks before being subjected to DSS-induced colitis and was sustained until the end of the experiment. The preparation and characteristics of polysaccharides from
N. commune (PS) were analyzed and described in our previous article [
24]. 5-aminosalicylic acid (5-ASA) was used as a positive control. Control healthy mice were exclusively given regular water. In some experiments, we pretreated mice by administering a cocktail of antibiotics (vancomycin 0.5 g/L, ampicillin 1.0 g/L, metronidazole 1.0 g/L, and neomycin 1.0 g/L) in drinking water [
27] for 4 weeks and replaced with a freshly prepared cocktail once every second day before DSS induction. Changes in body weight were determined daily over the 10 ds experimental period. Evaluation of disease activity index (DAI) was conducted by combining the parameters of weight loss, stool consistency, and rectal bleeding, as described previously [
28,
29]. The mean of the total score of the three parameters was calculated as the DAI. The sample of colonic content was collected for gut microbiome and metabolomics analysis. The colon was carefully isolated and the length was measured. Then, the proximal colon was kept for MPO (frozen immediately), the middle portion for RNA and protein isolation, and the rectal region for histological assessment and immunofluorescence staining [
26].
2.3. Colonic Histopathology
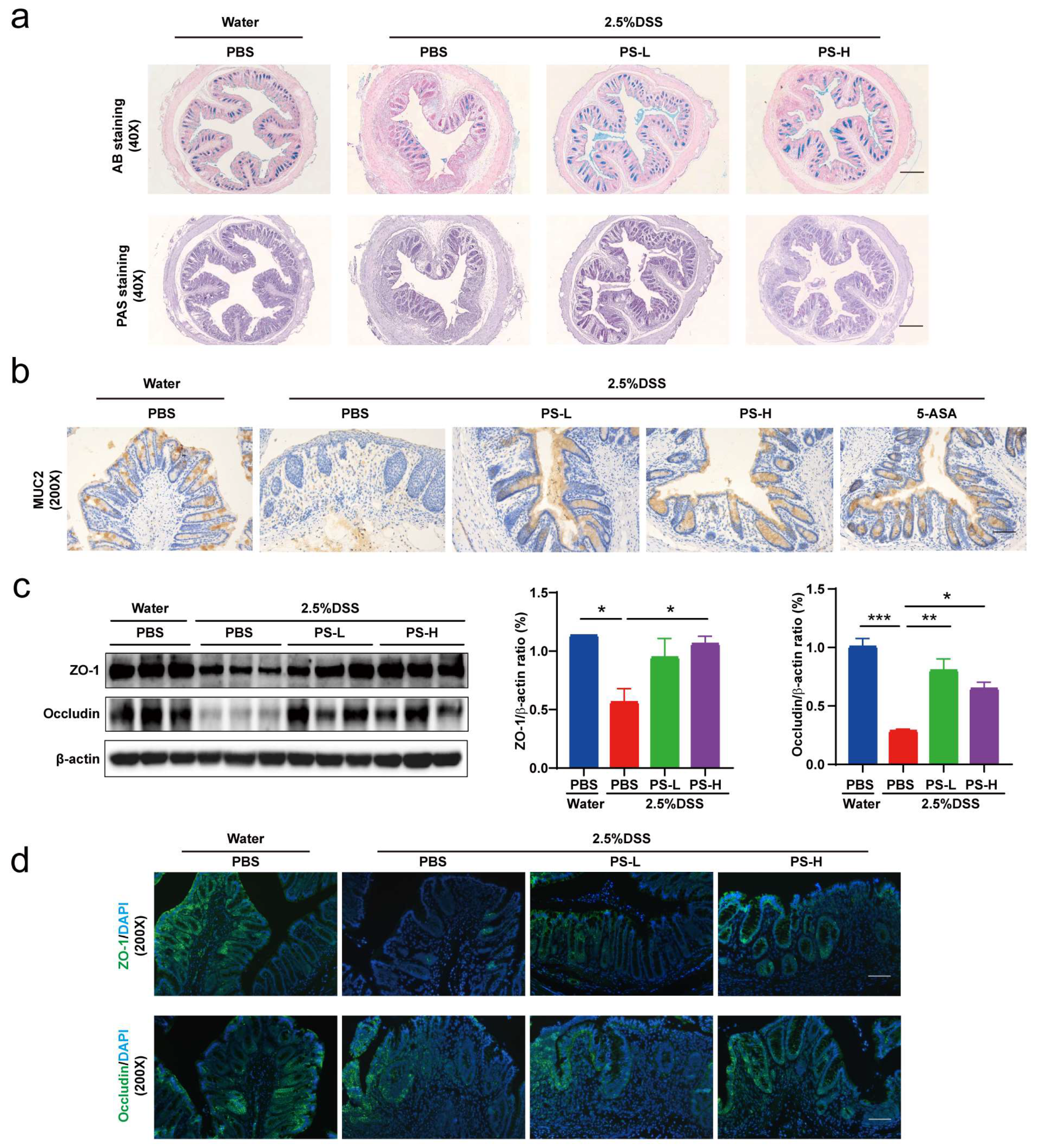
Colon segments were fixed in 4% paraformaldehyde for 24 h and embedded in paraffin. H&E staining was performed to evaluate colon pathological injury. The same rectal region of the colon from different groups was compared to analyze colitis severity. Histological damage was scored for epithelial damage and inflammatory cell infiltration in a blinded fashion. AB staining was performed to visualize the mucins. Data were analyzed using ImagePro Plus software (Version 6.0, Media Cybernetics, Rockville, MD, USA).
2.4. MPO Activity Measurement
Colonic MPO activity was measured by Myeloperoxidase Assay Kit (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions.
2.5. Immunohistochemistry
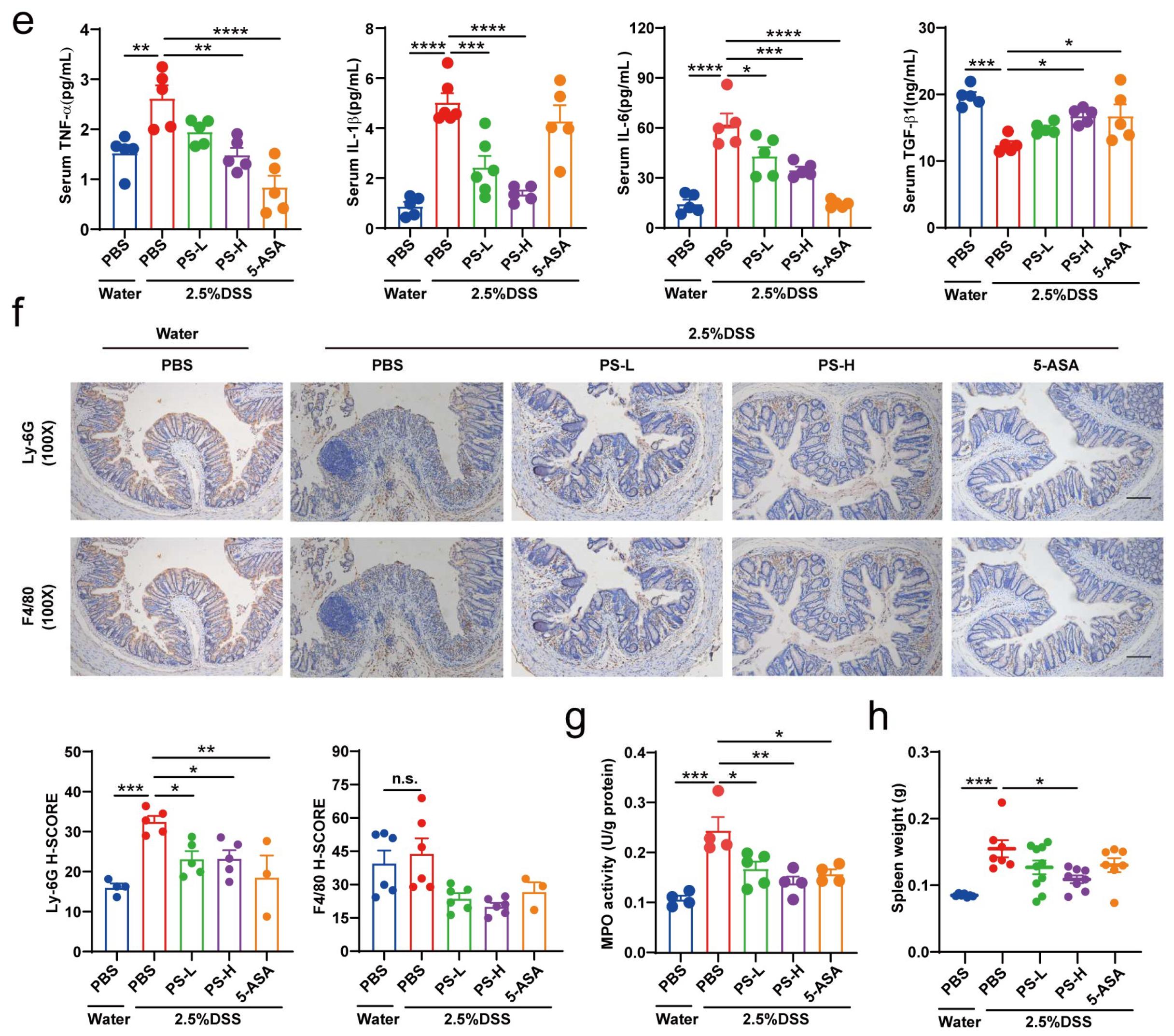
Antigens were recovered by treatment of samples at 98 °C for 10 min in 10 mM citrate buffer (pH 6.0). For immunohistochemical analysis, endogenous peroxidase was blocked with 10% H2O2 for 20 min and nonspecific antigens were blocked with serum for 30 min at room temperature. The slides were then incubated with specific primary antibodies at 4 °C for 12 h. Antibodies used were: MUC2 (1:2000, Proteintech, Wuhan, China), Ly-6 g (1:80, Proteintech), and F4/80 (1:2000, Proteintech). Slides were then treated with HRP-conjugated secondary antibodies. Expression of the stained markers was calculated using a histochemistry score (H-SCORE = 3x% of strongly staining cells + 2x% of moderately staining cells + 1x% of weakly staining cells).
2.6. Full-Length 16S rRNA Gene Sequencing
The DNA was extracted from the colonic content samples using a QIAamp DNA Isolation Kit (Qiagen, Hilden, Germany). Nanodrop 2000 was used to check the quantity and quality of the extracted DNA. The full-length bacterial 16S rRNA gene was amplified with primer pairs: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) by an ABI GeneAmp 9700 PCR thermocycler (ABI, Los Angeles, CA, USA). Purified amplicons were pooled in an equimolar and a DNA library was constructed using the SMRTbell Express Template Prep Kit 2.0 (PacBio, Menlo Park, CA, USA). Purified SMRTbell libraries were sequenced on a Pacbio Sequel II System (PacBio, Menlo Park, CA, USA) according to the standard protocols by Majorbio Bio-Pharm Technology (Shanghai, China). The raw 16S rRNA gene sequencing reads were demultiplexed to circular consensus sequence (CCS) reads by SMRTLink version 8.0 and length-filtered (<1000 or >1800 bp). Operational taxonomic units (OTUs) with 97% similarity cutoff were clustered using UPARSE version 7.1; chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.11 against the 16S rRNA database (Silva v138) with a confidence threshold of 0.7. All obtained raw sequence datasets have been uploaded to the NCBI Sequence Read Archive (SRA) with the accession number PRJNA988420.
2.7. LC-MS Metabolomic Analysis Technology
Each 50 mg colon tissue sample was added to 400 μL of pre-cooled methanol–water solution (1:1, v/v), precipitated, disrupted, and centrifuged. The supernatant was collected for mass spectrometry analysis. Samples were separated by a Thermo Fisher Scientific Vanquish Horizon system. To mitigate the influence of fluctuations in instrument detection signals, a sequential analysis of samples was conducted. Within the sample queue, a quality control (QC) sample was interspersed at regular intervals among every 10 experimental samples; it was applied to assess and appraise the system’s stability and the dependability of the experimental data. Electrospray ionization (ESI) was employed for the identification and quantification of both positive and negative ions. Samples were separated by UHPLC and analyzed by Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The original data were processed by Progenesis QI software (Version 2.3, Waters Corporation, Milford, CT, USA) and exported as a three-dimensional data matrix in CSV format. Internal standard peaks, as well as any known false positive peaks and the redundancy, were removed. Peaks were merged and HMDB (
http://www.hmdb.ca/ (accessed on 1 January 2019)), Metlin (
https://metlin.scripps.edu/ (accessed on 1 January 2017)), and Majorbio Database were retrieved. The extracted data from databases were uploaded to the Majorbio cloud platform (
https://cloud.majorbio.com (accessed on 1 September 2016)) and preprocessed for missing value deletion and recoding, normalization, QC samples RSD assessment, and log10 logarithmization. The data matrix was by performed variance analysis; then, R package ropls (Version 1.6.2) was used to carry out multidimensional statistical analysis, including unsupervised principal component analysis (PCA) and orthogonal least partial squares discriminant analysis (OPLS-DA). Metabolites based on variable importance in the projection (VIP) and the p-value of the Student’s
t-test were identified as differential metabolites (VIP > 1,
p < 0.05). KEGG (
http://www.genome.jp/kegg/ (accessed on 18 September 2021)) database was retrieved for metabolic pathway enrichment of differential metabolites between two groups. Scipy (Python packages, Version1.0.0) was employed to detect statistically significantly enriched pathways utilizing Fisher’s exact test.
2.8. Transcriptome Data Processing and Analysis
Total RNA underwent processing using Agilent 2100 Bioanalyzer in accordance with the manufacturer’s instructions (Agilent, Santa Clara, CA, USA). cDNA quantity and labeling efficiency were assessed using Agilent 2100 Bioanalyzer and ABI StepOne Plus Real-Time PCR and sequenced using the Illumina NovaSeq 6000 platform. The raw reads were filtered using the filter software fastq (Version 0.19.5); the comparison of gene expression in the RNA seq reads was performed using the HiSat2 software (Version 2.1.0). The reconstruction of transcripts for each sample was accomplished using StringTie (Version 2.1.2). Transcripts per million reads (TPM) were used to calculate gene expression levels. RSEM (Version 1.3.3) was used to quantify gene abundances. The DESeq2 software (Version 1.24.0) was utilized for differential expression analysis.
2.9. RNA and Protein Preparation
Colonic RNA and protein were extracted by E.Z.N.A. DNA/RNA/Protein Kit (Omega Bio-Tek, Guangzhou, China), providing a rapid and easy method for the isolation of total RNA, genomic DNA, and protein from animal tissues and allowing single processing of multiple samples in less than 40 min.
2.10. Western Blotting
Proteins were extracted from colon tissues and then quantitated using a Super-Rapid Protein Quantification Kit (Abbkine Scientific, Wuhan, China). Equal amounts of protein (25 μg) from different samples were separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked for 1 h at room temperature in blocking buffer (5% skimmed milk or 5% BSA in TBST), then probed with ZO-1 rabbit monoclonal Ab (1:2000, Affinity Biosciences, Changzhou, China), occludin rabbit monoclonal Ab (1:2000, Proteintech). Mouse monoclonal anti-β-actin Ab (1:10,000, Bio World, Nanjing, China) was used as the loading control. Images were captured using the ChemiDoc MP imaging system (Bio-Rad, Hercules, CA, USA). Blotted bands were quantified with Image J software (Version, 1.8.0.112, NIH).
2.11. ELISA
Serum pro- and anti-inflammatory cytokines (TNF-α/IL-1β/IL-6/TGF-1β/IL-10) were measured by a Mouse High Sensitivity ELISA Kit (MULTISCIENCES, Hangzhou, China) according to the manufacturer’s instructions.
2.12. Quantification and Statistical Analysis
Mice were assigned at random to each group for all mouse studies. Immunostaining and histological scoring were performed in a blinded fashion. Group sizes of 3 mice or above were sufficient to reach a statistical power of at least 80%. The data were first analyzed for normal distribution. Statistical significance was determined by using unpaired Student’s t-tests (comparison of two groups), Mann–Whitney tests (comparison of two groups, non-parametric data), or one-way ANOVA (comparison of three or more groups), followed by Bonferroni’s post hoc test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data conforming to normal distribution were presented as mean ± SEM and statistical tests were carried out using GraphPad Prism software version 8.0.
4. Discussion
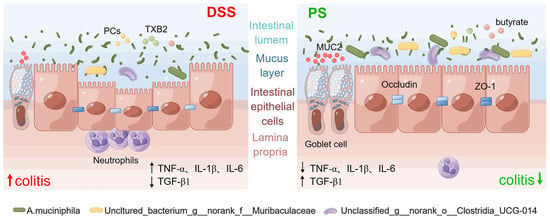
In this study, we determined the protective effect of natural bioactive polysaccharides on UC in mice and described that gut microbiota was the crucial event in mediating the prebiotic-like function. Compared with the untreated group, supplementation of the polysaccharides from N. commune exhibited a superior capacity to improve intestinal mucosal barrier function, including improving intestinal dysregulation, alleviating intestinal inflammation and reducing inflammatory infiltration, and repairing intestinal epithelial tight junction integrity. We then conducted a cocktail of antibiotics to validate the mediation of gut microbiota. Furthermore, we found that inflammation and immune function-associated signaling pathways were involved in the protection.
UC used to be more common in Western high-income countries, but the epidemiological paradigm has changed over the past two decades, with rates stabilizing in Western countries and increasing substantially in emerging economies [
1,
2,
3]. The pathogenesis of UC is multifaceted and still not entirely understood [
1]; but, it has been recognized that UC is an intestinal barrier disease [
1] characterized by inflammation confined to the mucosa [
1,
2,
3] and initially caused by either an epithelial cell or structural intestinal epithelial dysfunction [
1,
6]. As a physical, chemical, immune, and biological barrier, the intestinal barrier prevents pathogens, toxins, and allergens from spreading to other tissues, organs, and blood [
22], while allowing nutrients to be absorbed and immune responses to be triggered and limiting the spread of potentially harmful microorganisms and antigens [
12]. Few luminal antigens enter the lamina propria when there is a functional intestinal barrier, which is made up of epithelial cells and a mucus layer on top. The immune cells in the lamina propria are prevented from mounting a pro-inflammatory immunological response by the current tolerance mechanisms [
1]. These tolerance mechanisms, however, breakdown as the barrier breach widens and more luminal antigens pass through it, leading to the stimulation of local immune cells, the production of chemokines, and the subsequent infiltration of immune cells that worsen the inflammatory process [
1,
14]. The intestinal barrier assumes a crucial role in the modulation of the immune system and, hence, in health [
12,
14]. Intestinal barrier dysfunctions have been linked to a diverse array of diseases. The fundamental treatment goal ought to be the preservation of barrier function [
1,
12], which designates a new therapeutic target [
12].
The pathogenesis of UC is complicated; however, there is a close relationship between it and intestinal microbiota [
5,
32]. Studies have shown that the disruption of resident microbiota may be the major factor in inducing IBD [
5]. Probiotics can effectively induce and maintain remission in UC patients, indicating that optimizing intestinal microbiota is the key to treat UC [
5,
33].
A. muciniphila, populating the mucus layer of the gastrointestinal tract, maintains intestinal immunity and regulates gut barrier functions [
34]. In this study, we found that the relative abundance of
A. muciniphila exhibited absolute predominance in the colonic microbiome of mice pretreated with PS, acting as the central differential species (
Figure 2e,f). It was therefore reasonable to speculate that
A. muciniphila improved mucus thickness [
34] by regulating ZO-1 and occludin expression (
Figure 4a,c). Additionally,
A. muciniphila reinstated the amount of goblet cells [
34] and enhanced both the expression and secretion of MUC2 (
Figure 4a,b), accompanied by the reduced level of pro-inflammatory cytokine TNF-α (
Figure 4e). Our results also showed that
uncultured_bacterium_g__norank_f__Muribaculaceae was enriched after the treatment with PS (
Figure 2e,f), which was identified as one possible target microorganism in the development of colitis [
35]. Previous studies reported that some members of
norank_f_Muribaculaceae could hinder the growth and colonization of pathogenic bacteria in the intestine by occupying ecological space [
36]. We also observed a notable increase in
unclassified_g__norank_f__norank_o__Clostridia_UCG-014 in the gut microbiota of PS-treated mice (
Figure 2e,f).
Clostridia_UCG-014 has commonly been reported as a pro-inflammatory bacterium, but it has also been reported to reduce in UC [
37], indicating that the multiple members of
Clostridia_UCG-014 may play different roles in the gut. The relative abundance of
norank_f_norank_o_Clostridia_UCG-014 has been shown to increase during the first 7 days post-UC and then decrease [
38], consistent with our results harvested on the 10th day post-DSS induction.
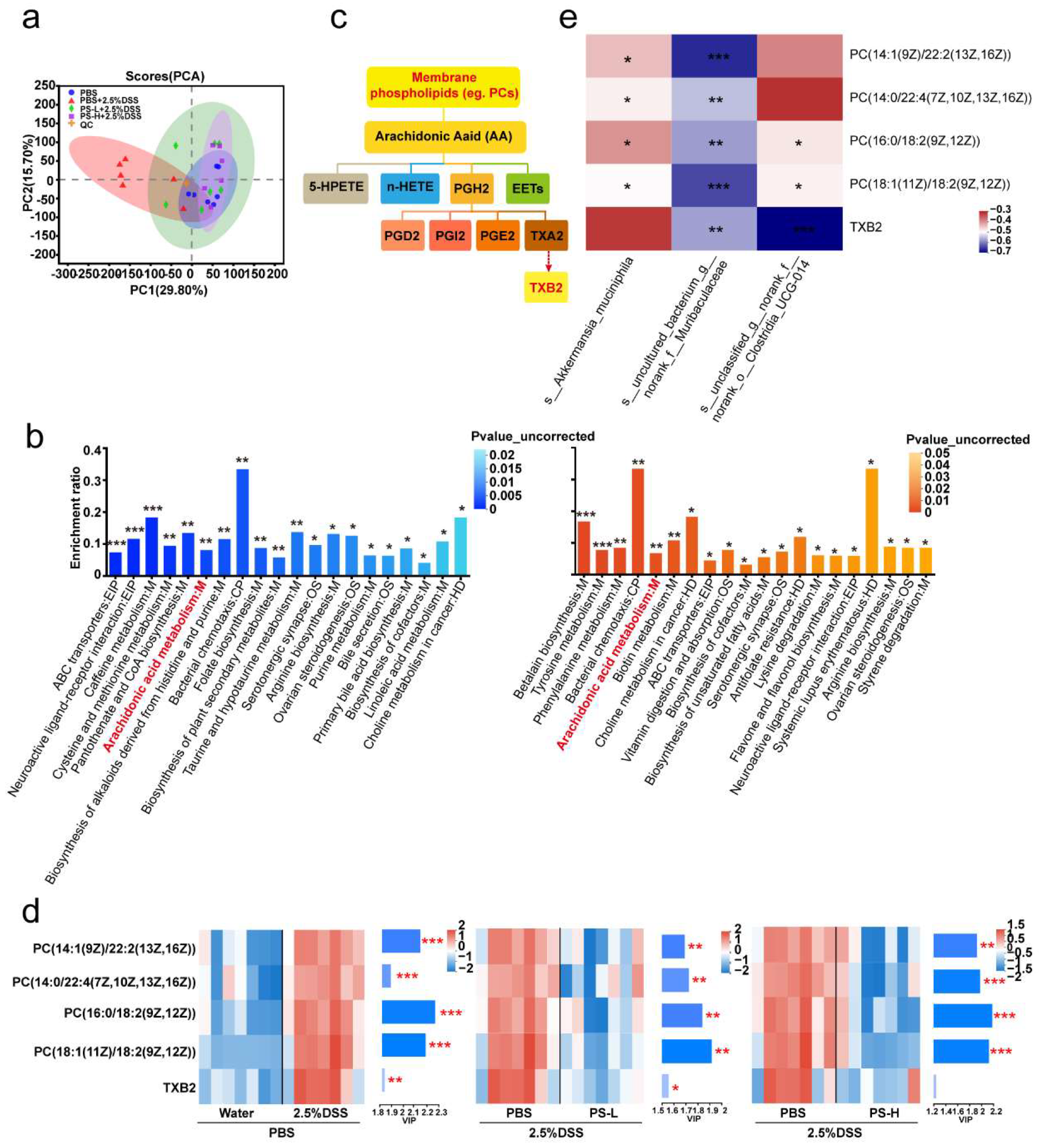
One of the major functions of the gut microbiota is the digestion of dietary components and the production of active metabolites to directly affect human physiology including gut barrier function [
15,
16,
17]. Here, we also examined the changes by metabolomics; the KEGG pathway enrichment analysis showed the differential metabolites (PCs and TXB 2) mapped to arachidonic acid (AA) metabolism (
Figure 3b,c). AA and its metabolites have attracted a lot of attention in cardiovascular and cancer biology [
39], particularly concerning inflammatory processes and disease (
Figure 3c). PC, a typical eukaryotic membrane phospholipid, is present in only about 15% of all bacterial species [
40]. However, relatively little has been reported about the relation of PC in colitis and gut barrier function. The most intriguing aspect of PC formation in bacteria lies in its critical involvement in symbiotic and pathogenic microbe–host interactions [
40]. In some organisms, the absence of PC also interferes with motility, bacterial growth, and stress response [
40], indicating that PC plays an important role in infection and inflammation (
Figure 3d). Spearman correlation analysis in
Figure 3e showed that
uncultured_bacterium_g__norank_f__Muribaculaceae exhibited a significantly negative correlation with the level of all PCs (
R < −0.54,
p < 0.01). We can speculate that
uncultured_bacterium_g__norank_f__Muribaculaceae could inhibit the growth and colonization of PC-producing bacteria and then occupy ecological space. TXB 2 is the stable metabolite of TXA 2, which possesses pro-inflammatory action in AA metabolism [
41]. The generation of TXA 2 can be estimated by the measurement of TXB 2. Accordingly, we observed a significant reduction in TXB 2 level after PS intervention obtained from the colonic content of mice (
Figure 3e) and was significantly and negatively correlated with
unclassified_g__norank_f__norank_o__Clostridia_UCG-014 (
Figure 3e).
It should be recognized that currently available treatments for UC, including aminosalicylic acid, corticosteroids, immunomodulators, monoclonal antibodies, and even surgeries, often have side effects, drug resistance, and poor clinical efficacy [
18,
19]. Natural bioactive polysaccharides have attracted increasing attention for their exceptional advantages of safety, accessibility, and good biocompatibility [
19,
21]. It is well established that most of them can be metabolized by the gut microbiota and can exert profound protection against oxidative stress, toxins, inflammatory response, and tumors [
22]. Although the protection of natural polysaccharides against colitis has been recently extensively studied, the role of gut microbiota and gut microbiota-derived metabolite remains incompletely understood and confirmed. The polysaccharides from
N. commune have some favorable characteristics, including being identified as a heteropolysaccharide with an ultra-high molecular weight of 210 kDa, as well as accounting for up to 60% of the dry weight of
N. commune [
24], strongly implying fairly compelling bioactivity. In this study, we conducted a cocktail of antibiotics to deplete intestinal microbiota from mice for 4 consecutive weeks using a combined low dose of antibiotics for maintaining continuously, thus demonstrating intestinal microbiota-mediated protective effects of polysaccharides from
N. commune on colitis (
Figure 5).
The damage to the structure and function of the intestinal epithelial cell barrier can disrupt the balance in the mucosal immune system and trigger an inflammatory response, exacerbating the damage to the intestinal epithelial barrier [
6,
22]. Cytokines are crucial targets for inflammation management and play a key role in T cell differentiation and regulation [
22]. TNF-α is a potent pro-inflammatory mediator implicated in intestinal barrier defects [
12], which frequently collaborates with IL-1β and IL-6 to enhance the inflammatory response by promoting the recruitment and activation of other inflammatory elements, resulting in increased production and release of inflammatory mediators [
22] (
Figure 4e). The gut is the primary metabolic site for polysaccharides, while the ultimate fate of it regulated by polysaccharides is always fascinating. Transcriptomics analysis showed that differential genes of colon tissue from mice in each group enriched in inflammation and immune system-associated signaling pathway and signaling molecule and interaction, including FoxO, mTOR, cAMP, PI3K-Akt, MAPK signaling pathway, antigen processing and presentation, chemokine signaling pathway, and cytokine–cytokine receptor interaction (
Figure 6d). The development of IBD is attributed to polygenic and multifactorial pathology via activation of certain signaling pathways [
4]. In IBD pathophysiology, aberrant signaling pathways cause deregulation of the inflammatory response. The IBD-related signaling pathways mainly include NF-κB, MAPK, JAK/STAT, and PI3K/TLR4 signaling pathways [
4]. Of these, the PI3K pathway is a cell-surface-receptor-controlled signal transduction pathway that is involved in the regulation of important leukocyte processes, such as activation, proliferation, and recruitment [
4]. PI3K can regulate the anti-apoptotic signals and pro-inflammatory signals triggered by LPS and TNF-α [
4]. Disruptions of signaling pathways act on the intestinal barrier, resulting in the unrestricted release of pro-inflammatory cytokines [
4]. Therefore, we are prone to speculate that polysaccharides of
N. commune could inhibit the expression of pro-inflammatory cytokines and protect the damaged intestinal barrier by regulating inflammation and immune function-associated signaling pathways (
Figure 6d).
In summary, our work integrated multi-omics profiling to reveal that the polysaccharides from N. commune predominantly enhanced the intestinal microbiota-mediated intestinal mucosal barrier to confer protection against UC in mice and exhibited dramatical prebiotic-like functions, providing an alternative or complementary treatment for IBD, especially UC.