Serotonin Transporter (SERT) Expression Modulates the Composition of the Western-Diet-Induced Microbiota in Aged Female Mice
Abstract
1. Introduction
2. Methods
2.1. Animals
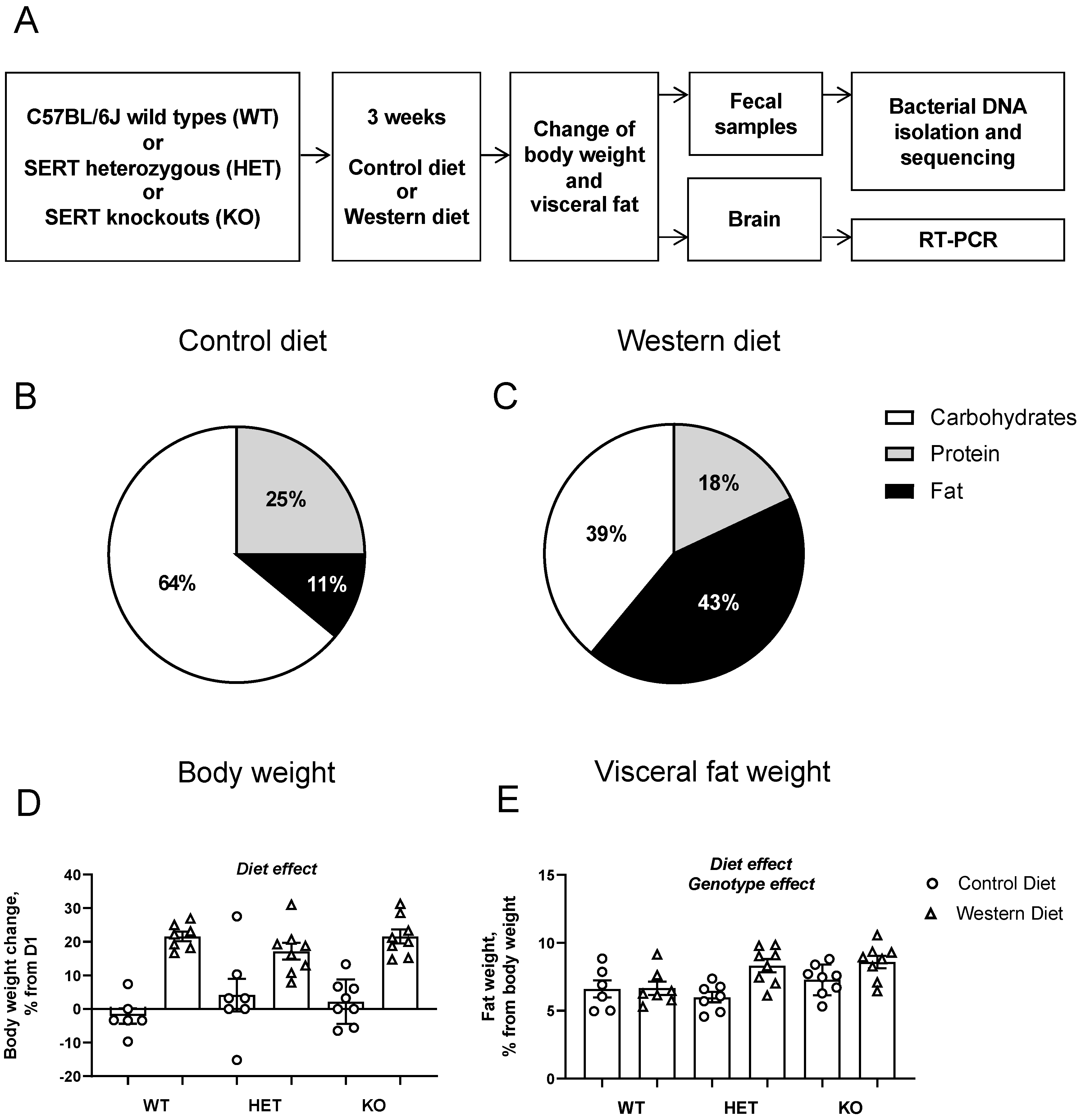
2.2. Study Design and Dietary Intervention
2.3. Bacterial DNA Isolation and Sequencing
2.4. Bioinformatics
2.5. Statistical Analysis
2.6. Community Analyses of Microbiota
2.7. Compositional Analyses on the Taxonomic Data
2.8. Exploratory Association Analyses between Gut Microbiota and Behavioral, Metabolic and Brain Measures
3. Results
3.1. Physiological Changes in Host Metabolism and Brain Gene Expression
3.2. Community Analyses of Gut Microbiota
3.3. Compositional Analyses of Gut Microbiota
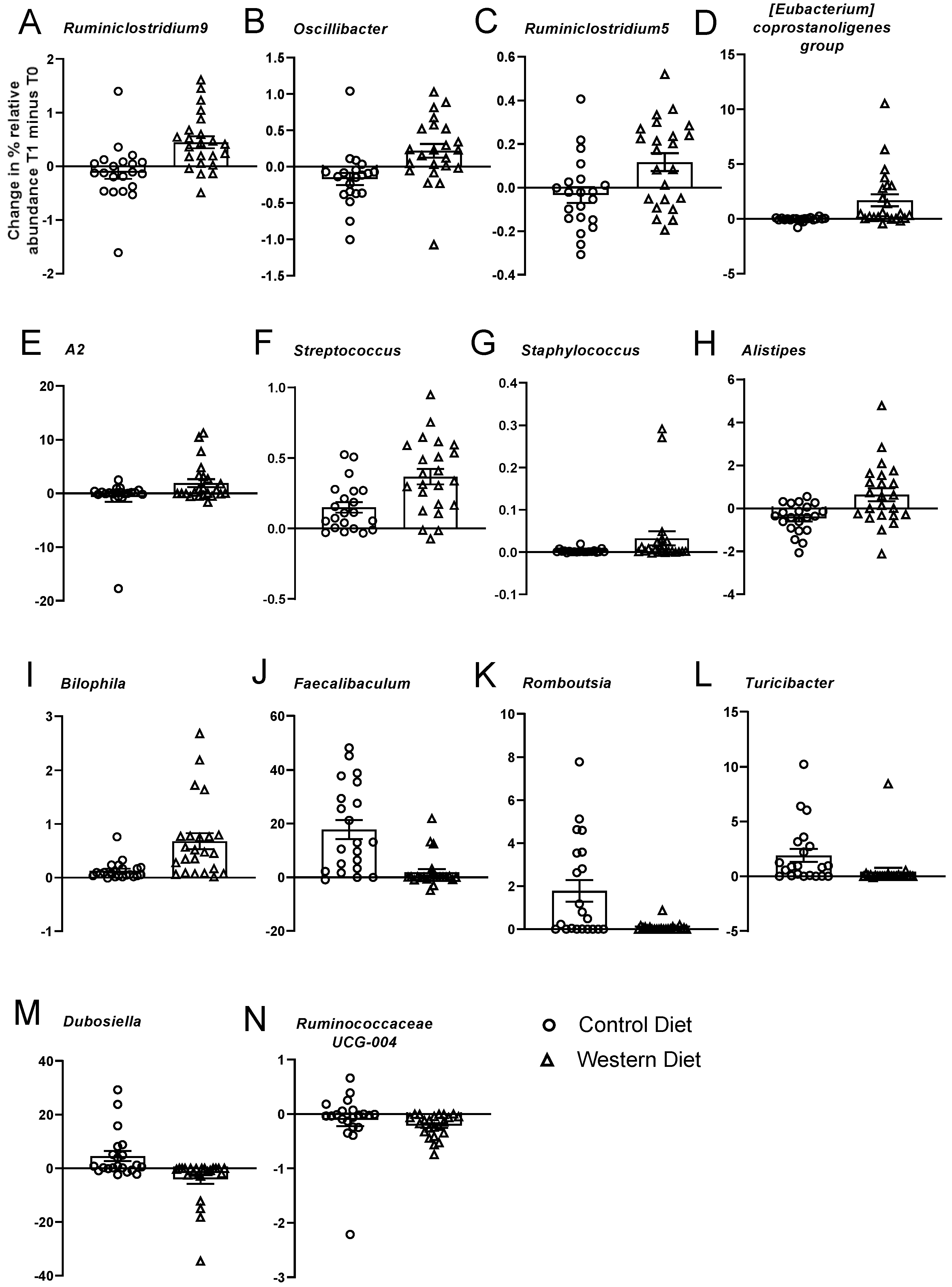
3.4. Effects of Diet on Microbiota
3.5. Effects of Genotype
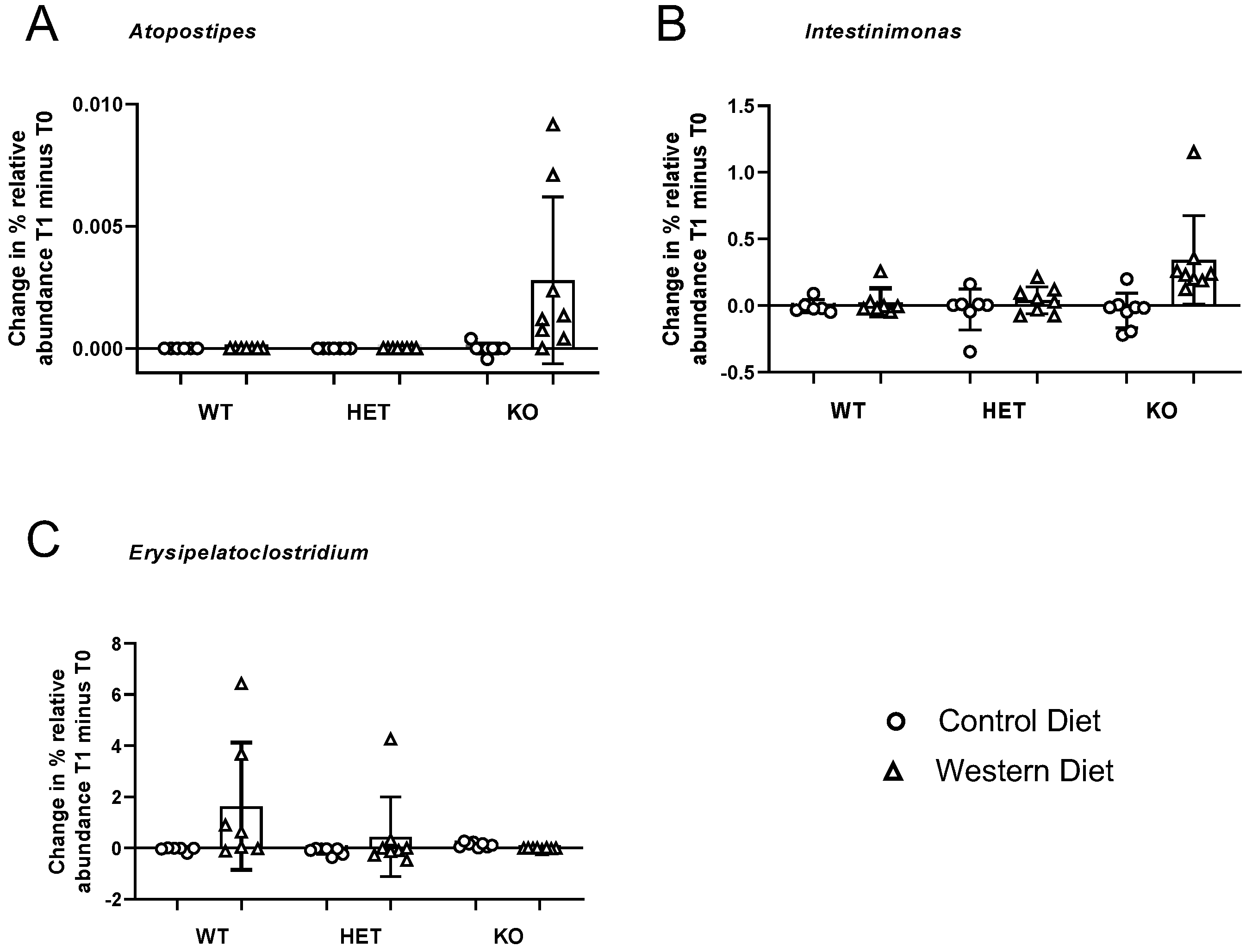
3.6. Diet × Genotype Interactions
3.7. Associations between the Effects of the Western Diet on the Gut Microbiota and Cellular and Molecular Changes
4. Discussion
4.1. Summary
4.2. Effects of the Western Diet on Gut Microbiota
4.3. Exploratory Correlations between Microbiota Changes and Physiological Changes in Mice Challenged with a Western Diet
4.4. Distinct Microbiota Effects of Western Diet on Microbiota of Mice with Differential SERT Gene Expression
4.5. Genetic SERT Deficiency and Gut Microbiota
4.6. Effects of Time
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AhR | Aryl hydrocarbon receptors |
| ANOVA | Analysis of variance |
| ARRIVE | Animal Research: Reporting of In Vivo Experiments |
| ASVs | Amplicon sequence variants |
| DADA2 | Divisive Amplicon Denoising Algorithm 2 |
| FDR | False discovery rate |
| HET | Heterozygous mice with partial genetic deficit of SERT, Sert+/− |
| KO | Knockout mice with complete genetic deficit of SERT, Sert−/− |
| mRNA | Messenger ribonucleic acid |
| OECD | Organisation for Economic Co-operation and Development |
| OTU | Operational taxonomic unit |
| PCR | Polymerase chain reaction |
| Pparg1b | Perixome-proliferator-activated-receptor-cofactor-1b |
| Ppargc1b | Perixome-proliferator-activated-receptor-cofactor-1b gene |
| QIIME2 | Quantitative Insights Into Microbial Ecology 2 |
| SEM | Standard error of the mean |
| SERT | Serotonin transporter |
| SLC6A4 | Solute Carrier Family 6 Member 4 gene, Sert gene (s for short allele, l for long allele) |
| TLR4 | Toll-like-Receptor |
| Tlr4 | Toll-like-Receptor 4 gene |
| V4 16S rRNA | V4 region of 16S ribosomal RNA |
| WT | Wild-type mice with normal expression of SERT, Sert+/+ |
References
- Lesch, K.-P.; Bengel, D.; Heils, A.; Sabol, S.Z.; Greenberg, B.D.; Petri, S.; Benjamin, J.; Müller, C.R.; Hamer, D.H.; Murphy, D.L. Association of Anxiety-Related Traits with a Polymorphism in the Serotonin Transporter Gene Regulatory Region. Science 1996, 274, 1527–1531. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, B.D.; Li, Q.; Lucas, F.R.; Hu, S.; Sirota, L.A.; Benjamin, J.; Lesch, K.-P.; Hamer, D.; Murphy, D.L. Association between the serotonin transporter promoter polymorphism and personality traits in a primarily female population sample. Am. J. Med. Genet. 2000, 96, 202–216. [Google Scholar] [CrossRef]
- Caspi, A.; Hariri, A.R.; Holmes, A.; Uher, R.; Moffitt, T.E.; Domingue, B.W.; Liu, H.; Okbay, A.; Belsky, D.W.; Rocha, T.B.-M.; et al. Genetic Sensitivity to the Environment: The Case of the Serotonin Transporter Gene and Its Implications for Studying Complex Diseases and Traits. Am. J. Psychiatry 2010, 167, 509–527. [Google Scholar] [CrossRef]
- Iordanidou, M.; Tavridou, A.; Petridis, I.; Arvanitidis, K.I.; Christakidis, D.; Vargemezis, V.; Manolopoulos, V.G. The serotonin transporter promoter polymorphism (5-HTTLPR) is associated with type 2 diabetes. Clin. Chim. Acta 2010, 411, 167–171. [Google Scholar] [CrossRef]
- Giannaccini, G.; Betti, L.; Palego, L.; Marsili, A.; Santini, F.; Pelosini, C.; Fabbrini, L.; Schmid, L.; Giusti, L.; Maffei, M.; et al. The expression of platelet serotonin transporter (SERT) in human obesity. BMC Neurosci. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Gemma, C.; García, S.I.; Gianotti, T.F.; Dieuzeide, G.; Roussos, A.; Tonietti, M.; Trifone, L.; Kanevsky, D.; González, C.D.; et al. Short Allele of Serotonin Transporter Gene Promoter Is a Risk Factor for Obesity in Adolescents*. Obesity 2007, 15, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Fuemmeler, B.F.; Agurs-Collins, T.D.; McClernon, F.J.; Kollins, S.H.; Kail, M.E.; Bergen, A.W.; Ashley-Koch, A.E. Genes Implicated in Serotonergic and Dopaminergic Functioning Predict BMI Categories. Obesity 2008, 16, 348–355. [Google Scholar] [CrossRef]
- Jarrell, H.; Hoffman, J.B.; Kaplan, J.R.; Berga, S.; Kinkead, B.; Wilson, M.E. Polymorphisms in the serotonin reuptake transporter gene modify the consequences of social status on metabolic health in female rhesus monkeys. Physiol. Behav. 2008, 93, 807–819. [Google Scholar] [CrossRef]
- Kautzky-Willer, A.; Harreiter, J.; Pacini, G. Sex and Gender Differences in Risk, Pathophysiology and Complications of Type 2 Diabetes Mellitus. Endocr. Rev. 2016, 37, 278–316. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Matyka, D.; Jurczak, A.; Szkup, M.; Samochowiec, A.; Grzywacz, A.; Huszla, S.; Grochans, E. The influence of the serotonergic system on the personality and quality of life of postmenopausal women. Clin. Interv. Aging 2017, 12, 963–970. [Google Scholar] [CrossRef]
- Khabazkhoob, M.; Emamian, M.H.; Hashemi, H.; Shariati, M.; Fotouhi, A. Prevalence of Overweight and Obesity in the Middle-age Population: A Priority for the Health System. Iran. J. Public Health 2017, 46, 827–834. [Google Scholar] [PubMed]
- Batsis, J.A.; Zagaria, A.B. Addressing Obesity in Aging Patients. Med. Clin. N. Am. 2018, 102, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Yabut, J.M.; Crane, J.D.; Green, A.E.; Keating, D.J.; Khan, W.I.; Steinberg, G.R. Emerging Roles for Serotonin in Regulating Metabolism: New Implications for an Ancient Molecule. Endocr. Rev. 2019, 40, 1092–1107. [Google Scholar] [CrossRef]
- Koopman, K.E.; Booij, J.; Fliers, E.; Serlie, M.J.; la Fleur, S.E. Diet-induced changes in the Lean Brain: Hypercaloric high-fat-high-sugar snacking decreases serotonin transporters in the human hypothalamic region. Mol. Metab. 2013, 2, 417–422. [Google Scholar] [CrossRef]
- Nam, S.B.; Kim, K.; Kim, B.S.; Im, H.-J.; Lee, S.H.; Kim, S.-J.; Kim, I.J.; Pak, K. The Effect of Obesity on the Availabilities of Dopamine and Serotonin Transporters. Sci. Rep. 2018, 8, 4924. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, R.I.; Koopman, K.E.; Booij, J.; Ackermans, M.T.; Unmehopa, U.A.; Fliers, E.; La Fleur, S.E.; Serlie, M.J. Serotonin Transporter Binding in the Diencephalon Is Reduced in Insulin-Resistant Obese Humans. Neuroendocrinology 2017, 105, 141–149. [Google Scholar] [CrossRef]
- Kakiuchi, T.; Tsukada, H.; Fukumoto, D.; Nishiyama, S. Effects of aging on serotonin transporter availability and its response to fluvoxamine in the living brain: PET study with [11C](+)McN5652 and [11C](-)McN5652 in conscious monkeys. Synapse 2001, 40, 170–179. [Google Scholar] [CrossRef]
- Yamamoto, M.; Suhara, T.; Okubo, Y.; Ichimiya, T.; Sudo, Y.; Inoue, M.; Takano, A.; Yasuno, F.; Yoshikawa, K.; Tanada, S. Age-related decline of serotonin transporters in living human brain of healthy males. Life Sci. 2002, 71, 751–757. [Google Scholar] [CrossRef]
- Rodríguez, J.J.; Noristani, H.N.; Verkhratsky, A. The serotonergic system in ageing and Alzheimer’s disease. Prog. Neurobiol. 2012, 99, 15–41. [Google Scholar] [CrossRef]
- Herrera-Pérez, J.J.; Fernández-Guasti, A.; Martínez-Mota, L. Brain SERT Expression of Male Rats Is Reduced by Aging and Increased by Testosterone Restitution. Neurosci. J. 2013, 2013, 201909. [Google Scholar] [CrossRef]
- Morita, Y.; Iwamoto, I.; Mizuma, N.; Kuwahata, T.; Matsuo, T.; Yoshinaga, M.; Douchi, T. Precedence of the shift of body-fat distribution over the change in body composition after menopause. J. Obstet. Gynaecol. Res. 2006, 32, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Karakelides, H.; Irving, B.A.; Short, K.R.; O’Brien, P.; Nair, K.S. Age, Obesity, and Sex Effects on Insulin Sensitivity and Skeletal Muscle Mitochondrial Function. Diabetes 2010, 59, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Boemi, M.; Furlan, G.; Luconi, M.P. Glucose Metabolism, Insulin, and Aging. In Molecular Basis of Nutrition and Aging; Academic Press: Cambridge, MA, USA, 2016; pp. 393–409. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef]
- Zheng, L.J.; Lin, L.; Zhong, J.; Zhang, Z.; Ye, Y.B.; Zhang, X.Y.; Wang, Y.F.; Zhang, H.; Liu, Y.; Lu, G.M.; et al. Gut dysbiosis-influence on amygdala-based functional activity in patients with end stage renal disease: A preliminary study. Brain Imaging Behav. 2020, 14, 2731–2744. [Google Scholar] [CrossRef]
- Lin, H.; An, Y.; Hao, F.; Wang, Y.; Tang, H. Correlations of Fecal Metabonomic and Microbiomic Changes Induced by High-fat Diet in the Pre-Obesity State. Sci. Rep. 2016, 6, 21618. [Google Scholar] [CrossRef]
- Walker, A.; Pfitzner, B.; Neschen, S.; Kahle, M.; Harir, M.; Lucio, M.; Moritz, F.; Tziotis, D.; Witting, M.; Rothballer, M.; et al. Distinct signatures of host–microbial meta-metabolome and gut microbiome in two C57BL/6 strains under high-fat diet. ISME J. 2014, 8, 2380–2396. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef]
- Agus, A.; Denizot, J.; Thévenot, J.; Martinez-Medina, M.; Massier, S.; Sauvanet, P.; Bernalier-Donadille, A.; Denis, S.; Hofman, P.; Bonnet, R.; et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci. Rep. 2016, 6, 19032. [Google Scholar] [CrossRef]
- Cabral, D.J.; Wurster, J.I.; Korry, B.J.; Penumutchu, S.; Belenky, P. Consumption of a Western-Style Diet Modulates the Response of the Murine Gut Microbiome to Ciprofloxacin. mSystems 2020, 5, e00317-20. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Denizot, J.; Dreux, N.; Robin, F.; Billard, E.; Bonnet, R.; Darfeuille-Michaud, A.; Barnich, N. Western diet induces dysbiosis with increased E. coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 2014, 63, 116–124. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jazwinski, S.M. The Gut Microbiota and Healthy Aging: A Mini-Review. Gerontology 2018, 64, 513–520. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Singhal, M.; Turturice, B.A.; Manzella, C.R.; Ranjan, R.; Metwally, A.A.; Theorell, J.; Huang, Y.; Alrefai, W.A.; Dudeja, P.K.; Finn, P.W.; et al. Serotonin Transporter Deficiency is Associated with Dysbiosis and Changes in Metabolic Function of the Mouse Intestinal Microbiome. Sci. Rep. 2019, 9, 2138. [Google Scholar] [CrossRef]
- Sun, W.; Guo, Y.; Zhang, S.; Chen, Z.; Wu, K.; Liu, Q.; Liu, K.; Wen, L.; Wei, Y.; Wang, B.; et al. Fecal Microbiota Transplantation Can Alleviate Gastrointestinal Transit in Rats with High-Fat Diet-Induced Obesity via Regulation of Serotonin Biosynthesis. BioMed Res. Int. 2018, 2018, 8308671. [Google Scholar] [CrossRef]
- Bi, Z.; Zhang, S.; Meng, Y.; Feng, Y.; Wang, Y.; Wang, E.; Pan, X.; Zhu, R.; Fan, H.; Pang, S.; et al. Female serotonin transporter-knockout rat: A potential model of irritable bowel syndrome. FASEB J. 2021, 35, e21701. [Google Scholar] [CrossRef]
- Fung, T.C.; Vuong, H.E.; Luna, C.D.G.; Pronovost, G.N.; Aleksandrova, A.A.; Riley, N.G.; Vavilina, A.; McGinn, J.; Rendon, T.; Forrest, L.R.; et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 2019, 4, 2064–2073. [Google Scholar] [CrossRef]
- Di Ye, D.; Xu, H.; Xia, H.; Zhang, C.; Tang, Q.; Bi, F. Targeting SERT promotes tryptophan metabolism: Mechanisms and implications in colon cancer treatment. J. Exp. Clin. Cancer Res. 2021, 40, 1–17. [Google Scholar] [CrossRef]
- Veniaminova, E.; Cespuglio, R.; Chernukha, I.; Schmitt-Boehrer, A.G.; Morozov, S.; Kalueff, A.V.; Kuznetsova, O.; Anthony, D.C.; Lesch, K.-P.; Strekalova, T. Metabolic, Molecular, and Behavioral Effects of Western Diet in Serotonin Transporter-Deficient Mice: Rescue by Heterozygosity? Front. Neurosci. 2020, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- Strekalova, T.; Costa-Nunes, J.P.; Veniaminova, E.; Kubatiev, A.; Lesch, K.-P.; Chekhonin, V.P.; Evans, M.C.; Steinbusch, H.W. Insulin receptor sensitizer, dicholine succinate, prevents both Toll-like receptor 4 (TLR4) upregulation and affective changes induced by a high-cholesterol diet in mice. J. Affect. Disord. 2016, 196, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Strekalova, T.; Evans, M.; Costa-Nunes, J.; Bachurin, S.; Couch, Y.; Steinbusch, H.M.W.; Köhler, S.E.; Lesch, P.; Anthony, D.C. Tlr4 upregulation in the brain accompanies depression- and anxiety-like behaviors induced by a high-cholesterol diet. Brain Behav. Immun. 2015, 48, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Veniaminova, E.; Cespuglio, R.; Cheung, C.W.; Umriukhin, A.; Markova, N.; Shevtsova, E.; Lesch, K.-P.; Anthony, D.C.; Strekalova, T. Autism-Like Behaviours and Memory Deficits Result from a Western Diet in Mice. Neural Plast. 2017, 2017, 9498247. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Genet. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Lathi, L.; Shetty, S. Tools for Microbiome Analysis in R. 2017. Available online: http://microbiome.github.com/microbiome (accessed on 8 June 2023).
- Szopinska-Tokov, J.; Dam, S.; Naaijen, J.; Konstanti, P.; Rommelse, N.; Belzer, C.; Buitelaar, J.; Franke, B.; Bloemendaal, M.; Aarts, E.; et al. Investigating the Gut Microbiota Composition of Individuals with Attention-Deficit/Hyperactivity Disorder and Association with Symptoms. Microorganisms 2020, 8, 406. [Google Scholar] [CrossRef] [PubMed]
- Kloke, J.D.; Mckean, J.W. Rfit: Rank-based Estimation for Linear Models Rank-regression. R J. 2012, 4, 57–64. [Google Scholar] [CrossRef]
- Hou, D.; Zhao, Q.; Yousaf, L.; Xue, Y.; Shen, Q. Beneficial effects of mung bean seed coat on the prevention of high-fat diet-induced obesity and the modulation of gut microbiota in mice. Eur. J. Nutr. 2020, 60, 2029–2045. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Li, M.; Li, J.; Han, X.; Zhu, H.; Yu, G.; Cheng, J. Medium-, long- and medium-chain-type structured lipids ameliorate high-fat diet-induced atherosclerosis by regulating inflammation, adipogenesis, and gut microbiota in ApoE−/− mice. Food Funct. 2020, 11, 5142–5155. [Google Scholar] [CrossRef]
- Zhuang, P.; Zhang, Y.; Shou, Q.; Li, H.; Zhu, Y.; He, L.; Chen, J.; Jiao, J. Eicosapentaenoic and Docosahexaenoic Acids Differentially Alter Gut Microbiome and Reverse High-Fat Diet–Induced Insulin Resistance. Mol. Nutr. Food Res. 2020, 64, e1900946. [Google Scholar] [CrossRef]
- Gong, S.; Ye, T.; Wang, M.; Wang, M.; Li, Y.; Ma, L.; Yang, Y.; Wang, Y.; Zhao, X.; Liu, L.; et al. Traditional Chinese Medicine Formula Kang Shuai Lao Pian Improves Obesity, Gut Dysbiosis, and Fecal Metabolic Disorders in High-Fat Diet-Fed Mice. Front. Pharmacol. 2020, 11, 297. [Google Scholar] [CrossRef]
- Ijaz, M.U.; Ahmed, M.I.; Zou, X.; Hussain, M.; Zhang, M.; Zhao, F.; Xu, X.; Zhou, G.; Li, C. Beef, Casein, and Soy Proteins Differentially Affect Lipid Metabolism, Triglycerides Accumulation and Gut Microbiota of High-Fat Diet-Fed C57BL/6J Mice. Front. Microbiol. 2018, 9, 2200. [Google Scholar] [CrossRef]
- Kim, Y.; Hwang, S.W.; Kim, S.; Lee, Y.-S.; Kim, T.-Y.; Lee, S.-H.; Kim, S.J.; Yoo, H.J.; Na Kim, E.; Kweon, M.-N. Dietary cellulose prevents gut inflammation by modulating lipid metabolism and gut microbiota. Gut Microbes 2020, 11, 944–961. [Google Scholar] [CrossRef]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; Qin, H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 2019, 60, 175–184. [Google Scholar] [CrossRef]
- Zhang, Q.; Fan, X.; Ye, R.; Hu, Y.; Zheng, T.; Shi, R.; Cheng, W.; Lv, X.; Chen, L.; Liang, P. The Effect of Simvastatin on Gut Microbiota and Lipid Metabolism in Hyperlipidemic Rats Induced by a High-Fat Diet. Front. Pharmacol. 2020, 11, 522. [Google Scholar] [CrossRef]
- Zhou, D.; Pan, Q.; Xin, F.-Z.; Zhang, R.-N.; He, C.-X.; Chen, G.-Y.; Liu, C.; Chen, Y.-W.; Fan, J.-G. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J. Gastroenterol. 2017, 23, 60–75. [Google Scholar] [CrossRef]
- Zeng, H.; Larson, K.J.; Cheng, W.-H.; Bukowski, M.; Safratowich, B.D.; Liu, Z.; Hakkak, R. Advanced liver steatosis accompanies an increase in hepatic inflammation, colonic, secondary bile acids and Lactobacillaceae/Lachnospiraceae bacteria in C57BL/6 mice fed a high-fat diet. J. Nutr. Biochem. 2020, 78, 108336. [Google Scholar] [CrossRef]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef]
- Tan, R.; Jin, M.; Shao, Y.; Yin, J.; Li, H.; Chen, T.; Shi, D.; Zhou, S.; Li, J.; Yang, D. High-sugar, high-fat, and high-protein diets promote antibiotic resistance gene spreading in the mouse intestinal microbiota. Gut Microbes 2022, 14, 2022442. [Google Scholar] [CrossRef]
- Natividad, J.M.; Lamas, B.; Pham, H.P.; Michel, M.-L.; Rainteau, D.; Bridonneau, C.; da Costa, G.; Van Hylckama Vlieg, J.; Sovran, B.; Chamignon, C.; et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018, 9, 2802. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; Nilaweera, K.; Ross, P.R.; Shanahan, F.; O’toole, P.W.; Cotter, P.D. The gut microbiota and its relationship to diet and obesity: New insights. Gut Microbes 2012, 3, 186–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Fan, X.-Y.; Guo, W.-L.; Cao, Y.-J.; Lin, Y.-C.; Cheng, W.-J.; Chen, L.-J.; Rao, P.-F.; Ni, L.; Lv, X.-C. The protective mechanisms of macroalgae Laminaria japonica consumption against lipid metabolism disorders in high-fat diet-induced hyperlipidemic rats. Food Funct. 2020, 11, 3256–3270. [Google Scholar] [CrossRef]
- Wei, W.; Jiang, W.; Tian, Z.; Wu, H.; Ning, H.; Yan, G.; Zhang, Z.; Li, Z.; Dong, F.; Sun, Y.; et al. Fecal g. Streptococcus and g. Eubacterium_coprostanoligenes_group combined with sphingosine to modulate the serum dyslipidemia in high-fat diet mice. Clin. Nutr. 2021, 40, 4234–4245. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Mo, W.; Zheng, C.; Li, W.; Tang, J.; Wu, X. Alleviating effects of noni fruit polysaccharide on hepatic oxidative stress and inflammation in rats under a high-fat diet and its possible mechanisms. Food Funct. 2020, 11, 2953–2968. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Alderete, T.L.; Kim, J.S.; Millstein, J.; Gilliland, F.D.; Goran, M.I. High intake of dietary fructose in overweight/obese teenagers associated with depletion of Eubacterium and Streptococcus in gut microbiome. Gut Microbes 2019, 10, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, D.-Q.; Liu, J.-R.; Zhang, J.; Vaziri, N.D.; Zhuang, S.; Chen, H.; Feng, Y.-L.; Guo, Y.; Zhao, Y.-Y. Unilateral ureteral obstruction causes gut microbial dysbiosis and metabolome disorders contributing to tubulointerstitial fibrosis. Exp. Mol. Med. 2019, 51, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.-Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships Between Diet and Metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef]
- Huang, C.; Chen, J.; Wang, J.; Zhou, H.; Lu, Y.; Lou, L.; Zheng, J.; Tian, L.; Wang, X.; Cao, Z.; et al. Dysbiosis of Intestinal Microbiota and Decreased Antimicrobial Peptide Level in Paneth Cells during Hypertriglyceridemia-Related Acute Necrotizing Pancreatitis in Rats. Front. Microbiol. 2017, 8, 776. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wu, S.; Xia, Y.; Huang, J.; Ye, J.; Xuan, Z.; Li, P.; Du, B. Probiotic-fermented black tartary buckwheat alleviates hyperlipidemia and gut microbiota dysbiosis in rats fed with a high-fat diet. Food Funct. 2021, 12, 6045–6057. [Google Scholar] [CrossRef]
- Miao, M.; Wang, Q.; Wang, X.; Fan, C.; Yan, L.; Yue, Z.; Can, R.; Wenwen, H.; Xin, Z.; Yongmei, D.; et al. Inulin-type Fructans Protect Against High Fat- sucrose Diet-induced Gestational Diabetes Mice in Association with Gut Microbiota Regulation. Res. Sq. 2021, 1–26. [Google Scholar] [CrossRef]
- Li, L.; Guo, W.-L.; Zhang, W.; Xu, J.-X.; Qian, M.; Bai, W.-D.; Zhang, Y.-Y.; Rao, P.-F.; Ni, L.; Lv, X.-C. Grifola frondosa polysaccharides ameliorate lipid metabolic disorders and gut microbiota dysbiosis in high-fat diet fed rats. Food Funct. 2019, 10, 2560–2572. [Google Scholar] [CrossRef]
- Zhong, Y.; Nyman, M.; Fak, F. Modulation of gut microbiota in rats fed high-fat diets by processing. Mol. Nutr. Food Res. 2015, 59, 2066–2076. [Google Scholar] [CrossRef]
- Ai, X.; Wu, C.; Yin, T.; Zhur, O.; Liu, C.; Yan, X.; Yi, C.; Liu, D.; Xiao, L.; Li, W.; et al. Antidiabetic Function of Lactobacillus fermentum MF423-Fermented Rice Bran and Its Effect on Gut Microbiota Structure in Type 2 Diabetic Mice. Front. Microbiol. 2021, 12, 682290. [Google Scholar] [CrossRef]
- Bai, Y.-F.; Wang, S.-W.; Wang, X.-X.; Weng, Y.-Y.; Fan, X.-Y.; Sheng, H.; Zhu, X.-T.; Lou, L.-J.; Zhang, F. The flavonoid-rich Quzhou Fructus Aurantii extract modulates gut microbiota and prevents obesity in high-fat diet-fed mice. Nutr. Diabetes 2019, 9, 30. [Google Scholar] [CrossRef]
- Oubraim, S.; Wang, R.; Hausknecht, K.A.; Shen, R.-Y.; Haj-Dahmane, S. Tonic Endocannabinoid Signaling Gates Synaptic Plasticity in Dorsal Raphe Nucleus Serotonin Neurons through Peroxisome Proliferator-Activated Receptors. Front. Pharmacol. 2021, 12, 691219. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K. The role of the dorsal raphé nucleus in reward-seeking behavior. Front. Integr. Neurosci. 2013, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.-Y.; Shi, H.-T.; Gong, Z.-X.; Li, J.-H.; Song, K.; Huang, J.-N.; Yi, P.-F.; Shen, H.-Q.; Fu, B.-D. Baicalin Protects against APEC-Induced Colibacillosis by Regulating Gut Microbiota in Chickens. SSRN Electron. J. 2020, 1–20. [Google Scholar] [CrossRef]
- Fujita, Y.; Matsuoka, H.; Hirooka, K. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007, 66, 829–839. [Google Scholar] [CrossRef]
- Crescenzo, R.; Spagnuolo, M.S.; Cancelliere, R.; Iannotta, L.; Mazzoli, A.; Gatto, C.; Iossa, S.; Cigliano, L. Effect of Initial Aging and High-Fat/High-Fructose Diet on Mitochondrial Bioenergetics and Oxidative Status in Rat Brain. Mol. Neurobiol. 2019, 56, 7651–7663. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, A.; Spagnuolo, M.S.; Gatto, C.; Nazzaro, M.; Cancelliere, R.; Crescenzo, R.; Iossa, S.; Cigliano, L. Adipose Tissue and Brain Metabolic Responses to Western Diet—Is There a Similarity between the Two? Int. J. Mol. Sci. 2020, 21, 786. [Google Scholar] [CrossRef]
- Cho, S.; Hwang, O.; Park, S. Effect of Dietary Protein Levels on Composition of Odorous Compounds and Bacterial Ecology in Pig Manure. Asian Australas. J. Anim. Sci. 2015, 28, 1362–1370. [Google Scholar] [CrossRef]
- Cotta, M.A.; Whitehead, T.R.; Collins, M.D.; Lawson, P.A. Atopostipes suicloacale gen. nov., sp. nov., isolated from an underground swine manure storage pit. Anaerobe 2004, 10, 191–195. [Google Scholar] [CrossRef]
- Li, L.; Shi, M.; Salerno, S.; Tang, M.; Guo, F.; Liu, J.; Feng, Y.; Fu, M.; Huang, Q.; Ma, L.; et al. Microbial and metabolomic remodeling by a formula of Sichuan dark tea improves hyperlipidemia in apoE-deficient mice. PLoS ONE 2019, 14, e0219010. [Google Scholar] [CrossRef]
- Afouda, P.; Durand, G.A.; Lagier, J.-C.; Labas, N.; Cadoret, F.; Armstrong, N.; Raoult, D.; Dubourg, G. Noncontiguous finished genome sequence and description of Intestinimonas massiliensis sp. nov strain GD2T, the second Intestinimonas species cultured from the human gut. Microbiologyopen 2019, 8, e00621. [Google Scholar] [CrossRef]
- Lu, F.; Liu, F.; Zhou, Q.; Hu, X.; Zhang, Y. Effects of grape pomace and seed polyphenol extracts on the recovery of gut microbiota after antibiotic treatment in high-fat diet-fed mice. Food Sci. Nutr. 2019, 7, 2897–2906. [Google Scholar] [CrossRef]
- Hsu, C.-N.; Lin, I.-C.; Yu, H.-R.; Huang, L.-T.; Tiao, M.-M.; Tain, Y.-L. Maternal Tryptophan Supplementation Protects Adult Rat Offspring against Hypertension Programmed by Maternal Chronic Kidney Disease: Implication of Tryptophan-Metabolizing Microbiome and Aryl Hydrocarbon Receptor. Int. J. Mol. Sci. 2020, 21, 4552. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci. Rep. 2017, 7, 2594. [Google Scholar] [CrossRef] [PubMed]
- Kahalehili, H.M.; Newman, N.K.; Pennington, J.M.; Kolluri, S.K.; Kerkvliet, N.I.; Shulzhenko, N.; Morgun, A.; Ehrlich, A.K. Dietary Indole-3-Carbinol Activates AhR in the Gut, Alters Th17-Microbe Interactions, and Exacerbates Insulitis in NOD Mice. Front. Immunol. 2021, 11, 606441. [Google Scholar] [CrossRef]
- Bailén, M.; Bressa, C.; Martínez-López, S.; González-Soltero, R.; Lominchar, M.G.M.; Juan, C.S.; Larrosa, M. Microbiota Features Associated with a High-Fat/Low-Fiber Diet in Healthy Adults. Front. Nutr. 2020, 7, 583608. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Wang, S.; Lv, Z.; Zhao, W.; Li, S. Low molecular weight chitosan oligosaccharides (LMW-COSs) prevent obesity-related metabolic abnormalities in association with the modification of gut microbiota in high-fat diet (HFD)-fed mice. Food Funct. 2020, 11, 9947–9959. [Google Scholar] [CrossRef]
- Swarte, J.C.; Eelderink, C.; Douwes, R.M.; Said, M.Y.; Hu, S.; Post, A.; Westerhuis, R.; Bakker, S.J.; Harmsen, H.J. Effect of High versus Low Dairy Consumption on the Gut Microbiome: Results of a Randomized, Cross-Over Study. Nutrients 2020, 12, 2129. [Google Scholar] [CrossRef]
- Mandić, A.D.; Woting, A.; Jaenicke, T.; Sander, A.; Sabrowski, W.; Rolle-Kampcyk, U.; von Bergen, M.; Blaut, M. Clostridium ramosum regulates enterochromaffin cell development and serotonin release. Sci. Rep. 2019, 9, 1177. [Google Scholar] [CrossRef]
- Waclawiková, B.; El Aidy, S. Role of Microbiota and Tryptophan Metabolites in the Remote Effect of Intestinal Inflammation on Brain and Depression. Pharmaceuticals 2018, 11, 63. [Google Scholar] [CrossRef]

| Genus | Diet | Diet × Genotype | Direction of Effect | ||
|---|---|---|---|---|---|
| F | FDR p | F | FDR p | ||
| Atopostipes | 97.55 | 6.60 × 10−10 | 48.77 | 4.40 × 10−9 | ↑ Western in KO group |
| Intestinimonas | 22.06 | 0.0017 | 12.39 | 0.0049 | ↑ Western in KO group |
| Erysipelatoclostridium | 6.63 | 0.0771 | 8.94 | 0.0301 | ↑ Western in WT group |
| Ruminiclostridium.9 | 17.17 | 0.0063 | 1.37 | 0.7788 | ↑ Western |
| Oscillibacter | 15.30 | 0.0084 | 2.24 | 0.6131 | ↑ Western |
| Ruminococcaceae.uncultured | 9.35 | 0.0367 | 1.03 | 0.8702 | ↑ Western |
| Ruminiclostridium.5 | 9.25 | 0.0367 | 0.32 | 1 | ↑ Western |
| Coprostanoligenes.group | 10.47 | 0.0287 | 1.77 | 0.7185 | ↑ Western |
| Lachnospiraceae(f).A2 | 11.59 | 0.0240 | 5.86 | 0.1070 | ↑ Western |
| Peptococcaceae(f).uncultured | 9.39 | 0.0367 | 1.40 | 0.7788 | ↑ Western |
| Streptococcus | 10.93 | 0.0258 | 0.30 | 1 | ↑ Western |
| Staphylococcus | 9.23 | 0.0367 | 3.76 | 0.2767 | ↑ Western |
| Alistipes | 8.43 | 0.0442 | 0.62 | 0.9626 | ↑ Western |
| Bilophila | 12.60 | 0.0179 | 0.20 | 1 | ↑ Western |
| Faecalibaculum | 16.32 | 0.0069 | 0.03 | 1 | ↑ Control |
| Romboutsia | 14.11 | 0.0113 | 6.69 | 0.0888 | ↑ Control |
| RF39(f).uncultured | 10.98 | 0.0258 | 4.42 | 0.2145 | ↑ Control |
| Turicibacter | 21.84 | 0.0017 | 5.66 | 0.1070 | ↑ Control |
| Dubosiella | 8.89 | 0.0402 | 1.12 | 0.8292 | ↑ Control |
| Ruminococcaceae.UCG.014 | 8.56 | 0.0440 | 1.94 | 0.6574 | ↑ Control |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bloemendaal, M.; Veniaminova, E.; Anthony, D.C.; Gorlova, A.; Vlaming, P.; Khairetdinova, A.; Cespuglio, R.; Lesch, K.P.; Arias Vasquez, A.; Strekalova, T. Serotonin Transporter (SERT) Expression Modulates the Composition of the Western-Diet-Induced Microbiota in Aged Female Mice. Nutrients 2023, 15, 3048. https://doi.org/10.3390/nu15133048
Bloemendaal M, Veniaminova E, Anthony DC, Gorlova A, Vlaming P, Khairetdinova A, Cespuglio R, Lesch KP, Arias Vasquez A, Strekalova T. Serotonin Transporter (SERT) Expression Modulates the Composition of the Western-Diet-Induced Microbiota in Aged Female Mice. Nutrients. 2023; 15(13):3048. https://doi.org/10.3390/nu15133048
Chicago/Turabian StyleBloemendaal, Mirjam, Ekaterina Veniaminova, Daniel C. Anthony, Anna Gorlova, Priscilla Vlaming, Adel Khairetdinova, Raymond Cespuglio, Klaus Peter Lesch, Alejandro Arias Vasquez, and Tatyana Strekalova. 2023. "Serotonin Transporter (SERT) Expression Modulates the Composition of the Western-Diet-Induced Microbiota in Aged Female Mice" Nutrients 15, no. 13: 3048. https://doi.org/10.3390/nu15133048
APA StyleBloemendaal, M., Veniaminova, E., Anthony, D. C., Gorlova, A., Vlaming, P., Khairetdinova, A., Cespuglio, R., Lesch, K. P., Arias Vasquez, A., & Strekalova, T. (2023). Serotonin Transporter (SERT) Expression Modulates the Composition of the Western-Diet-Induced Microbiota in Aged Female Mice. Nutrients, 15(13), 3048. https://doi.org/10.3390/nu15133048






