The Effect of Antibiotics Treatment on the Maternal Immune Response and Gut Microbiome in Pregnant and Non-Pregnant Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Set-Up
2.2. Animals
2.3. Antibiotic Treatment
2.4. Isolation and Staining of T Cells and Dendritic Cells from the Spleen, MLN and PP
2.5. Staining of Blood Monocytes
2.6. Flow Cytometry
2.7. Microbiota Measurement
2.8. Universal Bacterial Quantitative Polymerase Reaction
2.9. Statistics
3. Results
3.1. Weight Change of Pregnant and Non-Pregnant Mice following AB Treatment
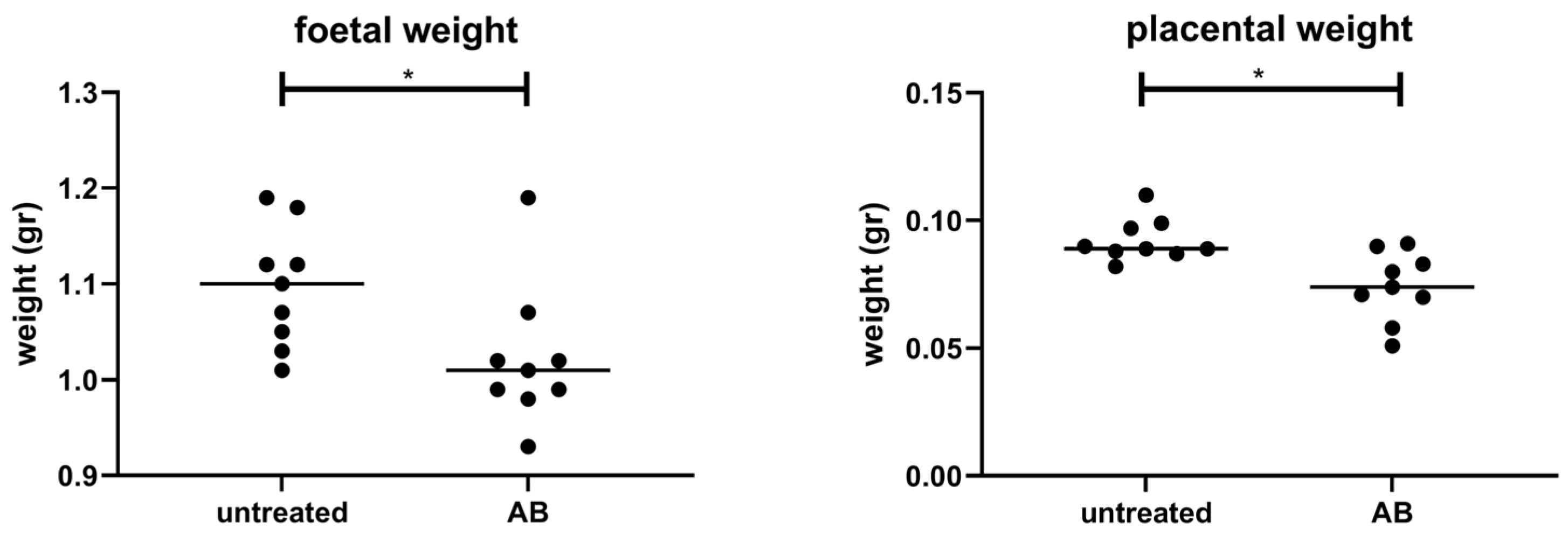
3.2. Weight of Fetuses and Placentas following AB Treatment of Pregnant Mice
3.3. Effect of AB Treatment on Immune Cell Populations
3.3.1. Effect of AB Treatment on Th Cell Subsets in the Spleens, PP and MLN
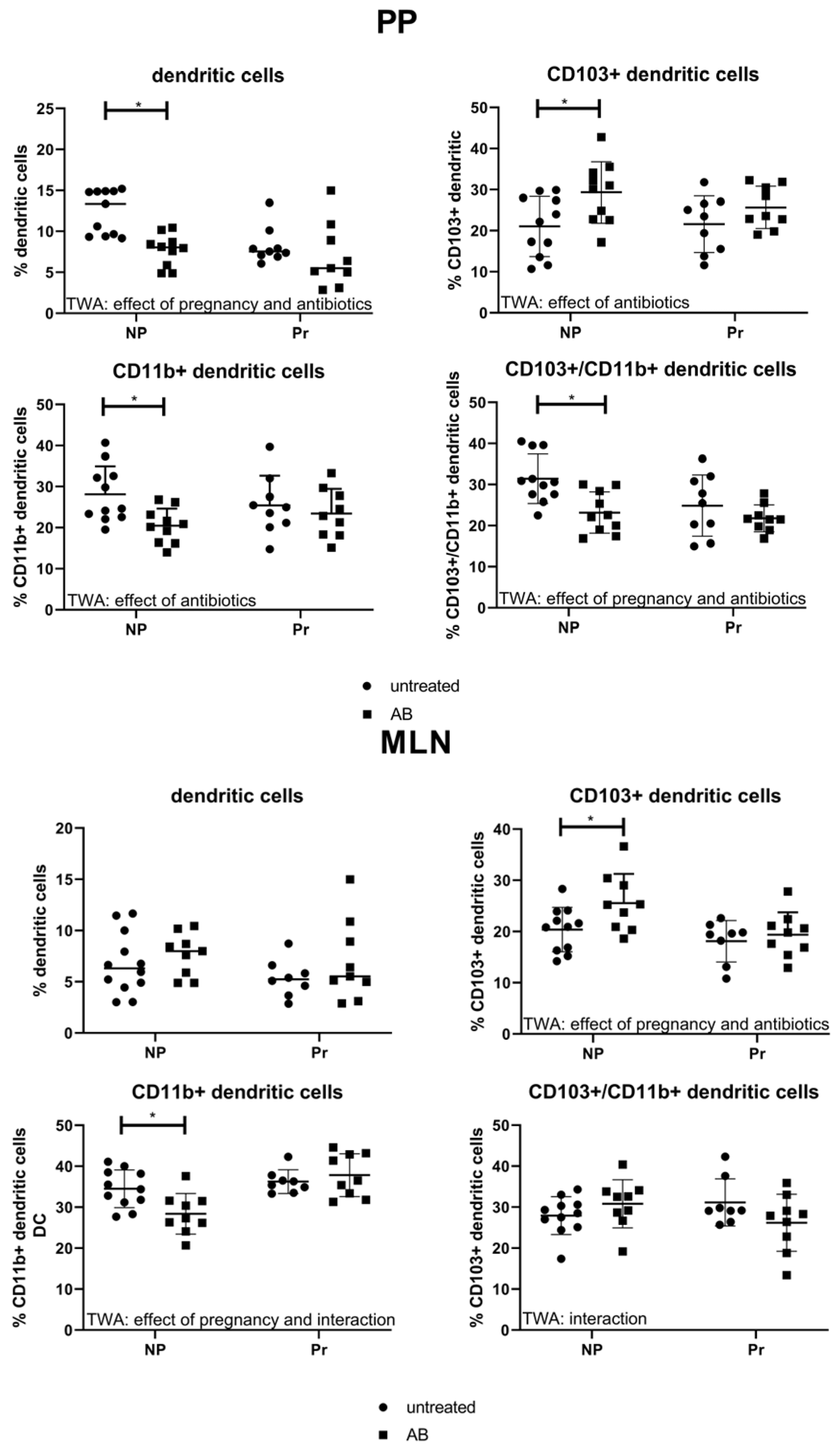
3.3.2. Effect of AB Treatment on Dendritic Cell Populations in the PP and MLN
3.3.3. Effect of AB Treatment on Blood Leukocyte Populations
Effect of AB Treatment on Peripheral Monocyte Subpopulations and Activation Status
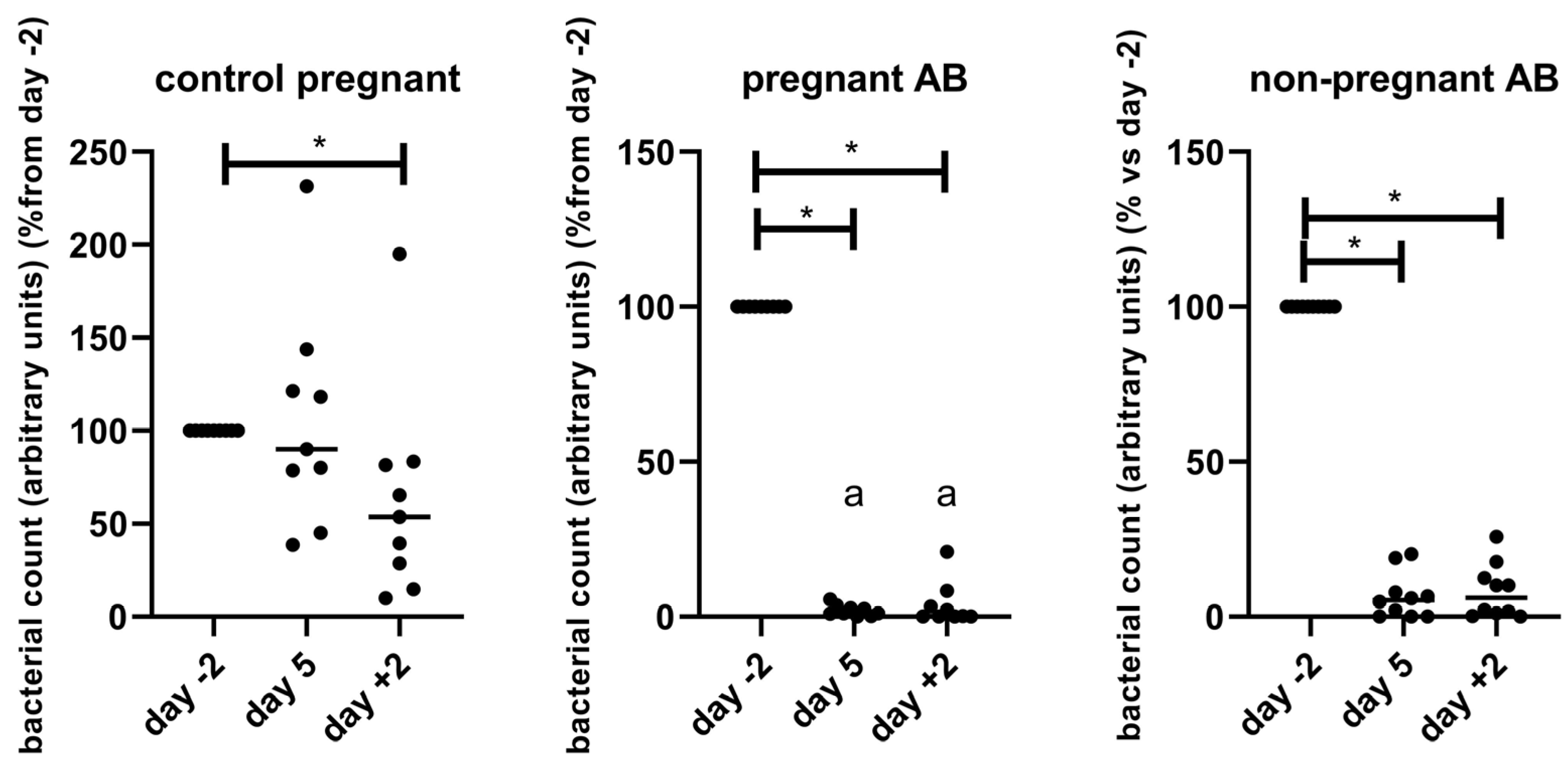
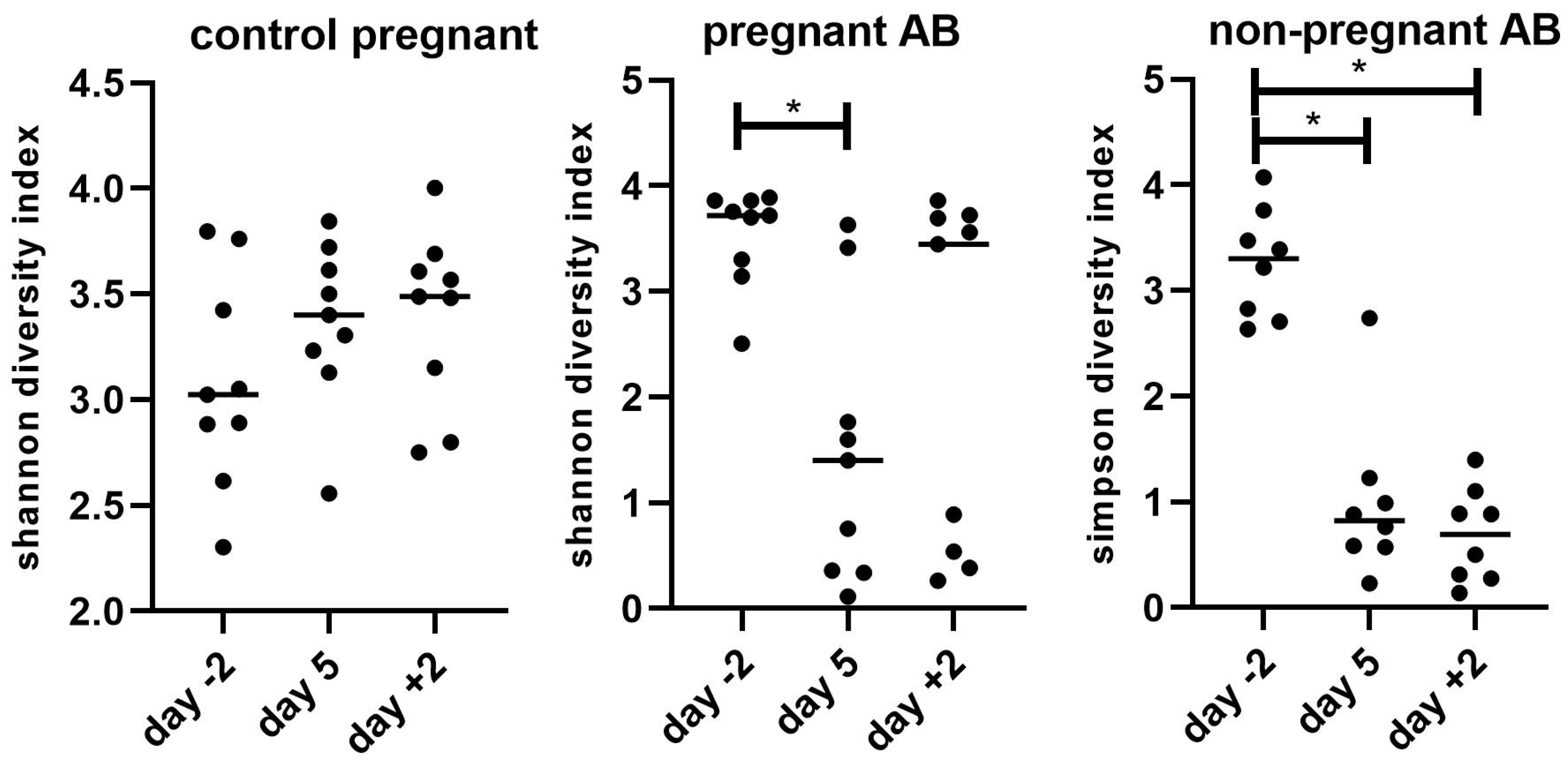
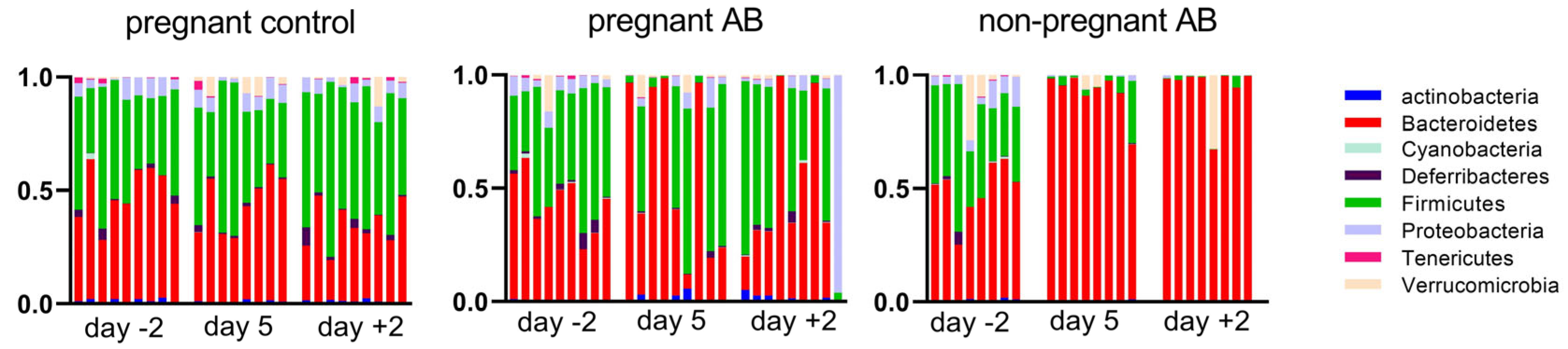
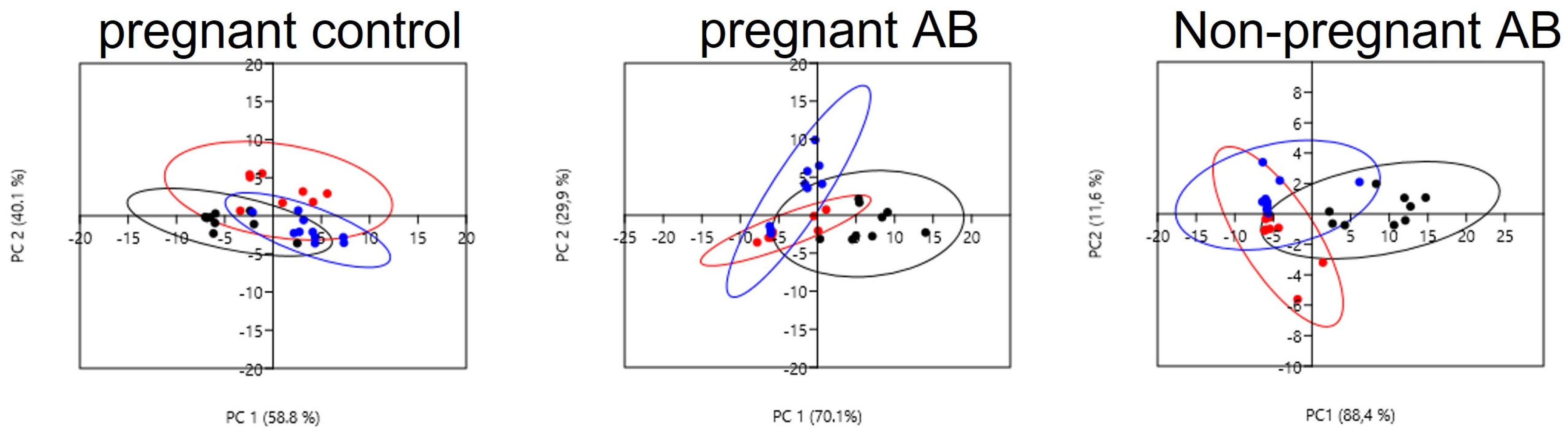
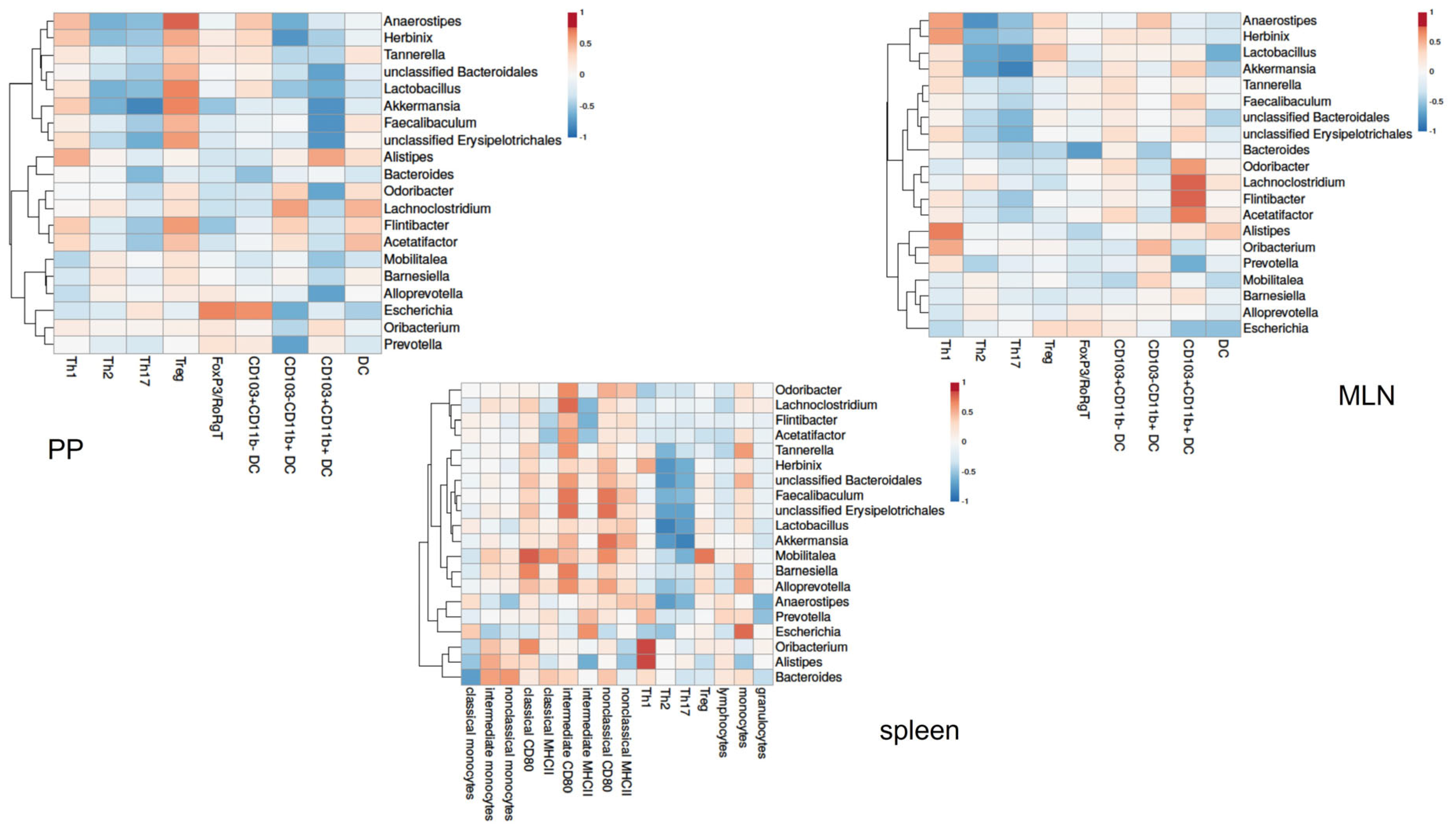
3.4. Gut Microbiota Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Veenstra van Nieuwenhoven, A.L.; Heineman, M.J.; Faas, M.M. The immunology of successful pregnancy. Hum. Reprod. Update 2003, 9, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Tsukaguchi, N.; Hasegawa, T.; Michimata, T.; Tsuda, H.; Narita, N. Distribution of Th1, Th2, and Th0 and the Th1/Th2 cell ratios in human peripheral and endometrial T cells. Am. J. Reprod. Immunol. 1999, 42, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Sacks, G.P.; Studena, K.; Sargent, I.L.; Redman, C.W.G. Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. Am. J. Obstet. Gynecol. 1998, 179, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Santner-Nanan, B.; Peek, M.J.; Khanam, R.; Richarts, L.; Zhu, E.; Fazekas de St Groth, B.; Nanan, R. Systemic increase in the ratio between Foxp3+ and IL-17-producing CD4+ T cells in healthy pregnancy but not in preeclampsia. J. Immunol. 2009, 183, 7023–7030. [Google Scholar] [CrossRef] [PubMed]
- Melgert, B.N.; Spaans, F.; Borghuis, T.; Klok, P.A.; Groen, B.; Bolt, A.; de Vos, P.; van Pampus, M.G.; Wong, T.Y.; van Goor, H.; et al. Pregnancy and preeclampsia affect monocyte subsets in humans and rats. PLoS ONE 2012, 7, e45229. [Google Scholar] [CrossRef]
- Faas, M.M.; de Vos, P. Uterine NK cells and macrophages in pregnancy. Placenta 2017, 56, 44–52. [Google Scholar] [CrossRef]
- Faas, M. Uterine natural killer cells and successful pregnancy: From mouse experiments to human physiology. Explor. Immunol. 2022, 2, 518–539. [Google Scholar] [CrossRef]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Backhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef]
- Faas, M.M.; Liu, Y.; Borghuis, T.; van Loo-Bouwman, C.A.; Harmsen, H.; de Vos, P. Microbiota Induced Changes in the Immune Response in Pregnant Mice. Front. Immunol. 2019, 10, 2976. [Google Scholar] [CrossRef]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef]
- Richards, J.L.; Yap, Y.A.; McLeod, K.H.; Mackay, C.R.; Mariño, E. Dietary metabolites and the gut microbiota: An alternative approach to control inflammatory and autoimmune diseases. Clin. Transl. Immunol. 2016, 5, e82. [Google Scholar] [CrossRef]
- Jung, C.; Hugot, J.P.; Barreau, F. Peyer’s Patches: The Immune Sensors of the Intestine. Int. J. Inflam. 2010, 2010, 823710. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Smith, K. Mesenteric lymph nodes at the center of immune anatomy. J. Exp. Med. 2006, 203, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Elderman, M.; Hugenholtz, F.; Belzer, C.; Boekschoten, M.; de Haan, B.; de Vos, P.; Faas, M. Changes in intestinal gene expression and microbiota composition during late pregnancy are mouse strain dependent. Sci. Rep. 2018, 8, 10001. [Google Scholar] [CrossRef] [PubMed]
- Tochitani, S.; Ikeno, T.; Ito, T.; Sakurai, A.; Yamauchi, T.; Matsuzaki, H. Administration of Non-Absorbable Antibiotics to Pregnant Mice to Perturb the Maternal Gut Microbiota Is Associated with Alterations in Offspring Behavior. PLoS ONE 2016, 11, e0138293. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Zaharia, M.; Bolosky, W.; Curtis, K.; Fox, A.; Patterson, D.; Shenker, S.; Stoica, I.; Karp, R.M.; Sittler, T. Faster and more accurate sequence alignment with SNAP. arXiv 2011, arXiv:1111.5572. [Google Scholar]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Ladirat, S.E.; Schols, H.A.; Nauta, A.; Schoterman, M.H.; Keijser, B.J.; Montijn, R.C.; Gruppen, H.; Schuren, F.H.J. High-throughput analysis of the impact of antibiotics on the human intestinal microbiota composition. J. Microbiol. Methods 2013, 92, 387–397. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.K.; Ryan, P.D. Past: Paleontological statistics software package for educatoin and data analysis. Paleontol. Electron. 2001, 4, 4. [Google Scholar]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Guerrieri, D.; Tateosian, N.L.; Maffia, P.C.; Reiteri, R.M.; Amiano, N.O.; Costa, M.J.; Villalonga, X.; Sanchez, M.L.; Estein, S.M.; Garcia, V.E.; et al. Serine leucocyte proteinase inhibitor-treated monocyte inhibits human CD4(+) lymphocyte proliferation. Immunology 2011, 133, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Farina, C.; Theil, D.; Semlinger, B.; Hohlfeld, R.; Meinl, E. Distinct responses of monocytes to Toll-like receptor ligands and inflammatory cytokines. Int. Immunol. 2004, 16, 799–809. [Google Scholar] [CrossRef]
- Benner, M.; Lopez-Rincon, A.; Thijssen, S.; Garssen, J.; Ferwerda, G.; Joosten, I.; van der Molen, R.G.; Hogenkamp, A. Antibiotic Intervention Affects Maternal Immunity during Gestation in Mice. Front. Immunol. 2021, 12, 685742. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. Mucosal Immunology. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef]
- Liang, J.; Huang, H.I.; Benzatti, F.P.; Karlsson, A.B.; Zhang, J.J.; Youssef, N.; Ma, A.; Hale, L.P.; Hammer, G.E. Inflammatory Th1 and Th17 in the Intestine Are Each Driven by Functionally Specialized Dendritic Cells with Distinct Requirements for MyD88. Cell Rep. 2016, 17, 1330–1343. [Google Scholar] [CrossRef] [PubMed]
- Blaschitz, C.; Raffatellu, M. Th17 cytokines and the gut mucosal barrier. J. Clin. Immunol. 2010, 30, 196–203. [Google Scholar] [CrossRef]
- Feng, Y.; Huang, Y.; Wang, Y.; Wang, P.; Song, H.; Wang, F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS ONE 2019, 14, e0218384. [Google Scholar] [CrossRef]
- Correa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Nakashima, A.; Shima, T.; Ito, M. Th1/Th2/Th17 and regulatory T-cell paradigm in pregnancy. Am. J. Reprod. Immunol. 2010, 63, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, H.; Way, S.S. Immunological Basis for Recurrent Fetal Loss and Pregnancy Complications. Annu. Rev. Pathol. 2019, 14, 185–210. [Google Scholar] [CrossRef]
- Sykes, L.; MacIntyre, D.A.; Yap, X.J.; Teoh, T.G.; Bennett, P.R. The Th1:th2 dichotomy of pregnancy and preterm labour. Mediat. Inflamm. 2012, 2012, 967629. [Google Scholar] [CrossRef]
- Makhseed, M.; Raghupathy, R.; Azizieh, F.; Omu, A.; Al-Shamali, E.; Ashkanani, L. Th1 and Th2 cytokine profiles in recurrent aborters with successful pregnancy and with subsequent abortions. Hum. Reprod. 2001, 16, 2219–2226. [Google Scholar] [CrossRef]
- Saito, S.; Umekaga, H.; Sakamoto, Y.; Sakai, M.; Tanebe, K.; Sasaki, Y.; Morikawa, H. Increased T-Helper-1-Type immunity and decreased T-helper-2-type immunity in patients with preeclampsia. Am. J. Reprod. Immunol. 1999, 41, 297–306. [Google Scholar] [CrossRef]
- Raghupathy, R.; Al-Azemi, M.; Azizieh, F. Intrauterine growth restriction: Cytokine profiles of trophoblast antigen-stimulated maternal lymphocytes. Clin. Dev. Immunol. 2012, 2012, 734865. [Google Scholar] [CrossRef]
- Tomar, N.; Uldbjerg, C.S.; Bech, B.H.; Burgner, D.P.; Pedersen, L.H.; Miller, J.E. Prenatal antibiotic exposure and birth weight. Pediatr. Obes. 2022, 17, e12831. [Google Scholar] [CrossRef]
- Vidal, A.C.; Murphy, S.K.; Murtha, A.P.; Schildkraut, J.M.; Soubry, A.; Huang, Z.; Neelon, S.E.B.; Fuemmeler, B.; Iversen, E.; Wang, F.; et al. Associations between antibiotic exposure during pregnancy, birth weight and aberrant methylation at imprinted genes among offspring. Int. J. Obes. 2013, 37, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Ding, J.; Di, X.; Sun, W.; Chen, H.; Zhang, H. Association between prenatal antibiotics exposure and measures of fetal growth: A repeated-measure study. Ecotoxicol. Environ. Saf. 2022, 244, 114041. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, P.; Liu, M.; Zheng, H.; He, Y.; Chen, M.X.; Tang, W.; Yue, X.; Huang, Y.; Zhuang, L.; et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut 2020, 69, 513–522. [Google Scholar] [CrossRef] [PubMed]




| T Cell Subset | Antibodies |
|---|---|
| Th1 | CD3+CD4+Tbet+ |
| Th2 | CD3+CD4+Gata3+ |
| Th17 | CD3+CD4+RoRgT+ |
| Treg | CD3+CD4+FoxP3+ |
| FoxP3+/RoRgT+ | CD3+CD4+FoxP3+RoRgT+ |
| Dendritic Cell Subsets | Antibodies |
|---|---|
| Dendritic cell population | MHCII+CD64-CD11c+ |
| CD103+ dendritic cells | MHCII+CD64-CD11c+CD103+ |
| CD11b+ dendritic cells | MHCII+CD64-CD11c+CD11b+ |
| CD103+CD11b+ dendritic cells | MHCII+CD64-CD11c+CD103+CD11b+ |
| Monocyte Subsets | Antibodies |
|---|---|
| Classical monocytes | CD11b+Ly6G-Ly6C++CD43dim |
| Intermediate monocytes | CD11b+Ly6G-Ly6C+CD43dim |
| Non-classical monocytes | CD11b+Ly6G-Ly6CdimCD43+ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faas, M.M.; Liu, Y.; Wekema, L.; Weiss, G.A.; van Loo-Bouwman, C.A.; Silva Lagos, L. The Effect of Antibiotics Treatment on the Maternal Immune Response and Gut Microbiome in Pregnant and Non-Pregnant Mice. Nutrients 2023, 15, 2723. https://doi.org/10.3390/nu15122723
Faas MM, Liu Y, Wekema L, Weiss GA, van Loo-Bouwman CA, Silva Lagos L. The Effect of Antibiotics Treatment on the Maternal Immune Response and Gut Microbiome in Pregnant and Non-Pregnant Mice. Nutrients. 2023; 15(12):2723. https://doi.org/10.3390/nu15122723
Chicago/Turabian StyleFaas, Marijke M., Yuanrui Liu, Lieske Wekema, Gisela A. Weiss, Carolien A. van Loo-Bouwman, and Luis Silva Lagos. 2023. "The Effect of Antibiotics Treatment on the Maternal Immune Response and Gut Microbiome in Pregnant and Non-Pregnant Mice" Nutrients 15, no. 12: 2723. https://doi.org/10.3390/nu15122723
APA StyleFaas, M. M., Liu, Y., Wekema, L., Weiss, G. A., van Loo-Bouwman, C. A., & Silva Lagos, L. (2023). The Effect of Antibiotics Treatment on the Maternal Immune Response and Gut Microbiome in Pregnant and Non-Pregnant Mice. Nutrients, 15(12), 2723. https://doi.org/10.3390/nu15122723






