Study of the Relationship between Mucosal Immunity and Commensal Microbiota: A Bibliometric Analysis
Abstract
1. Introduction
2. Method
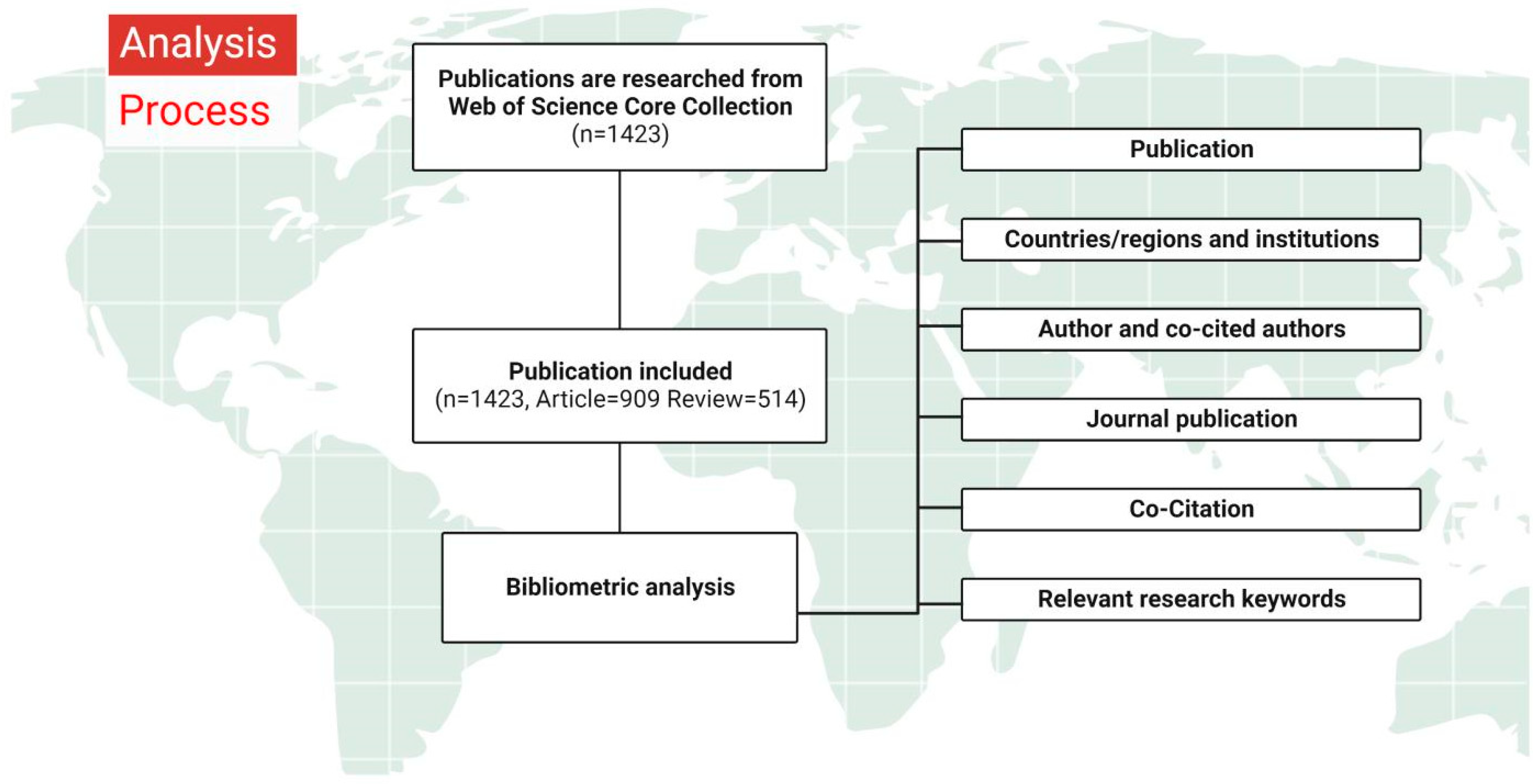
2.1. Data Collection
2.2. Research Strategy
2.3. Data Analysis and Visualization
3. Results
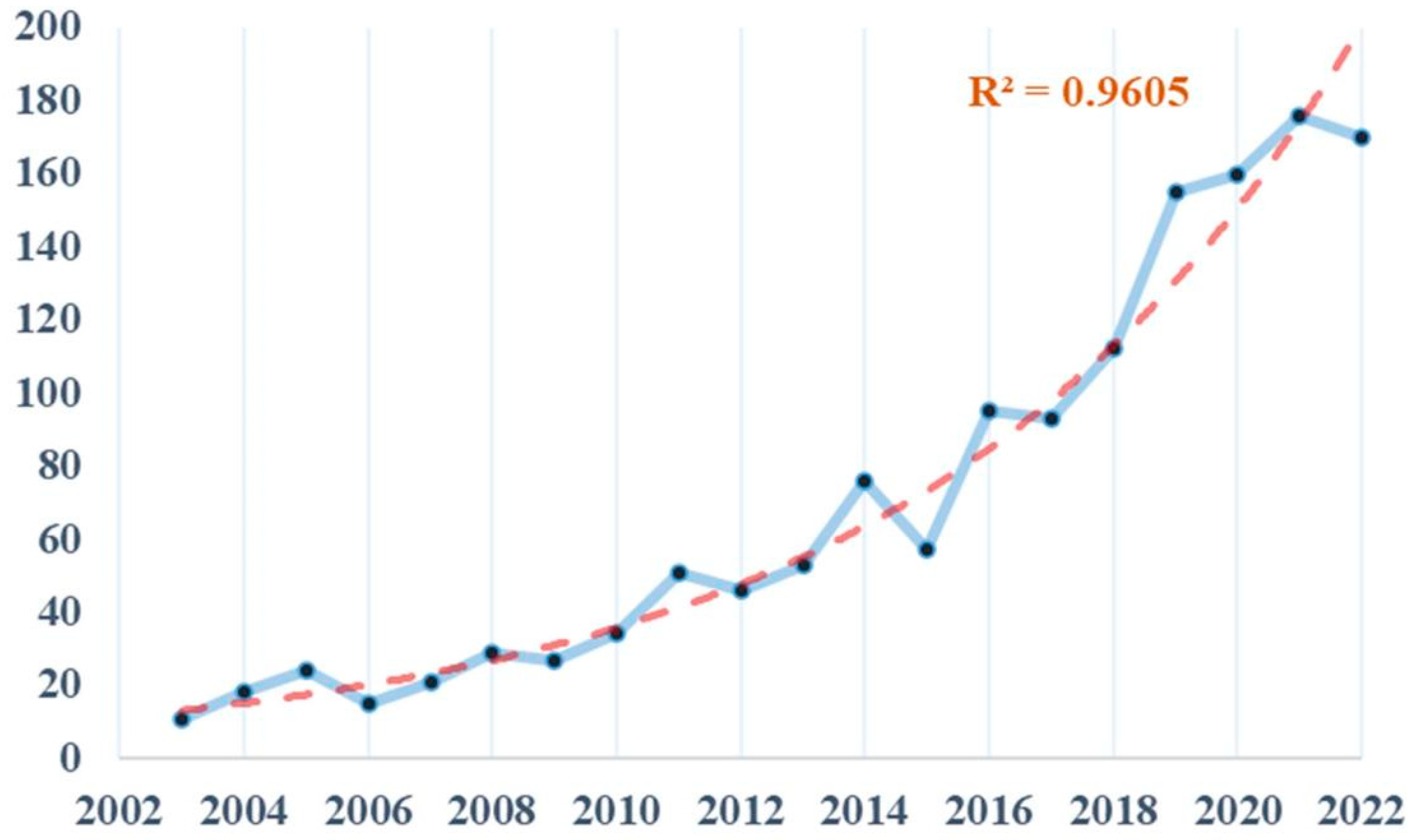
3.1. Basic Information about the Publication
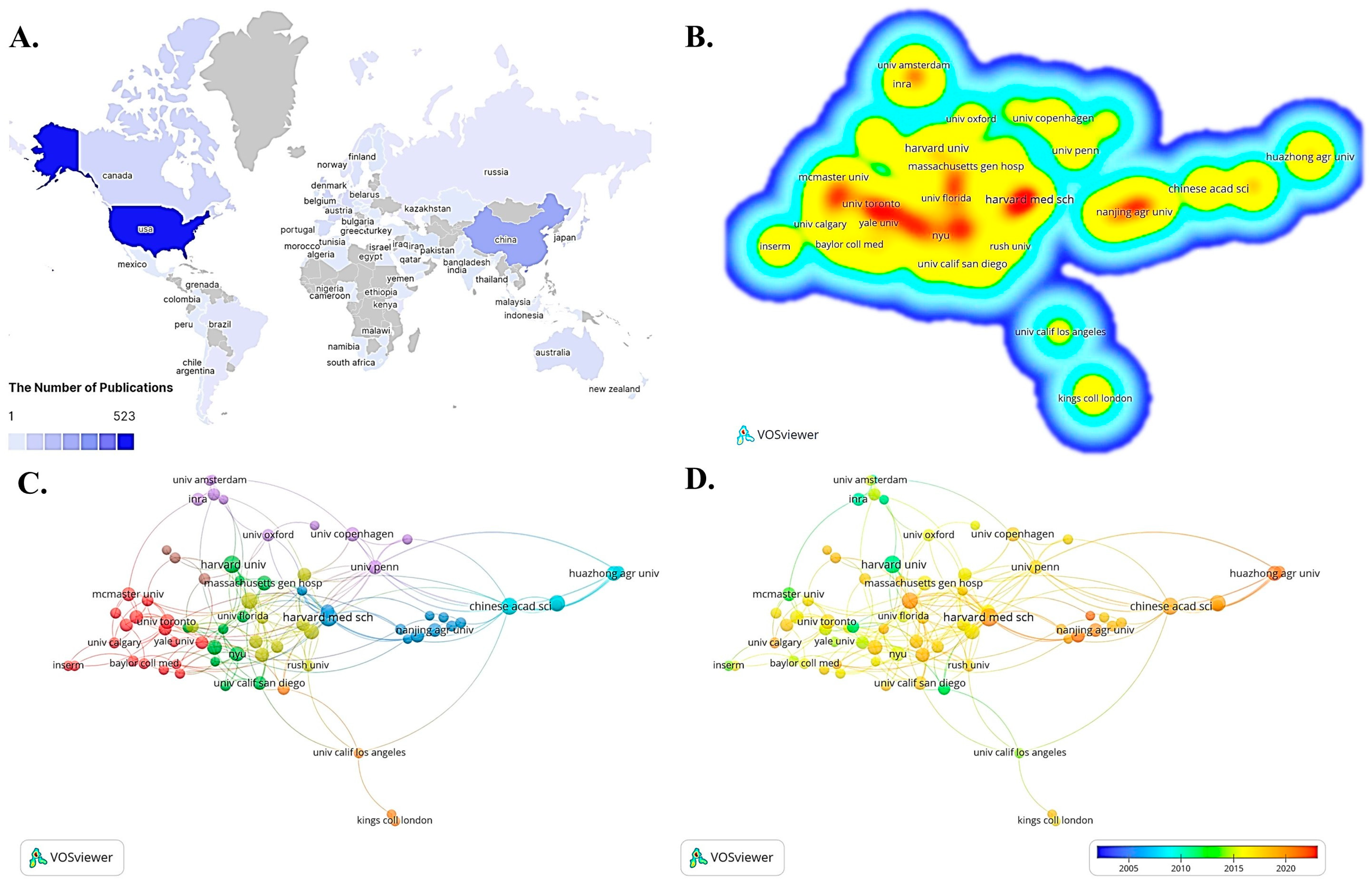
3.2. Distribution and Cooperation between Countries/Regions and Institutions
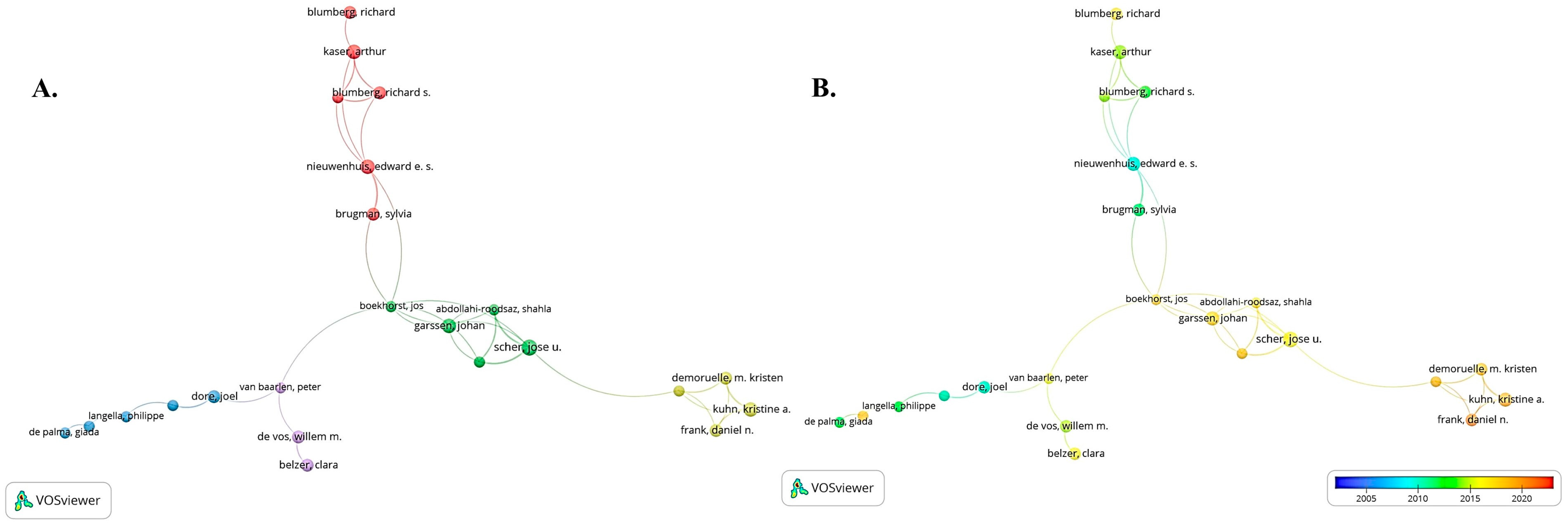
3.3. Conditions about the Author and Co-Cited Authors
3.4. Status of Journal Publication
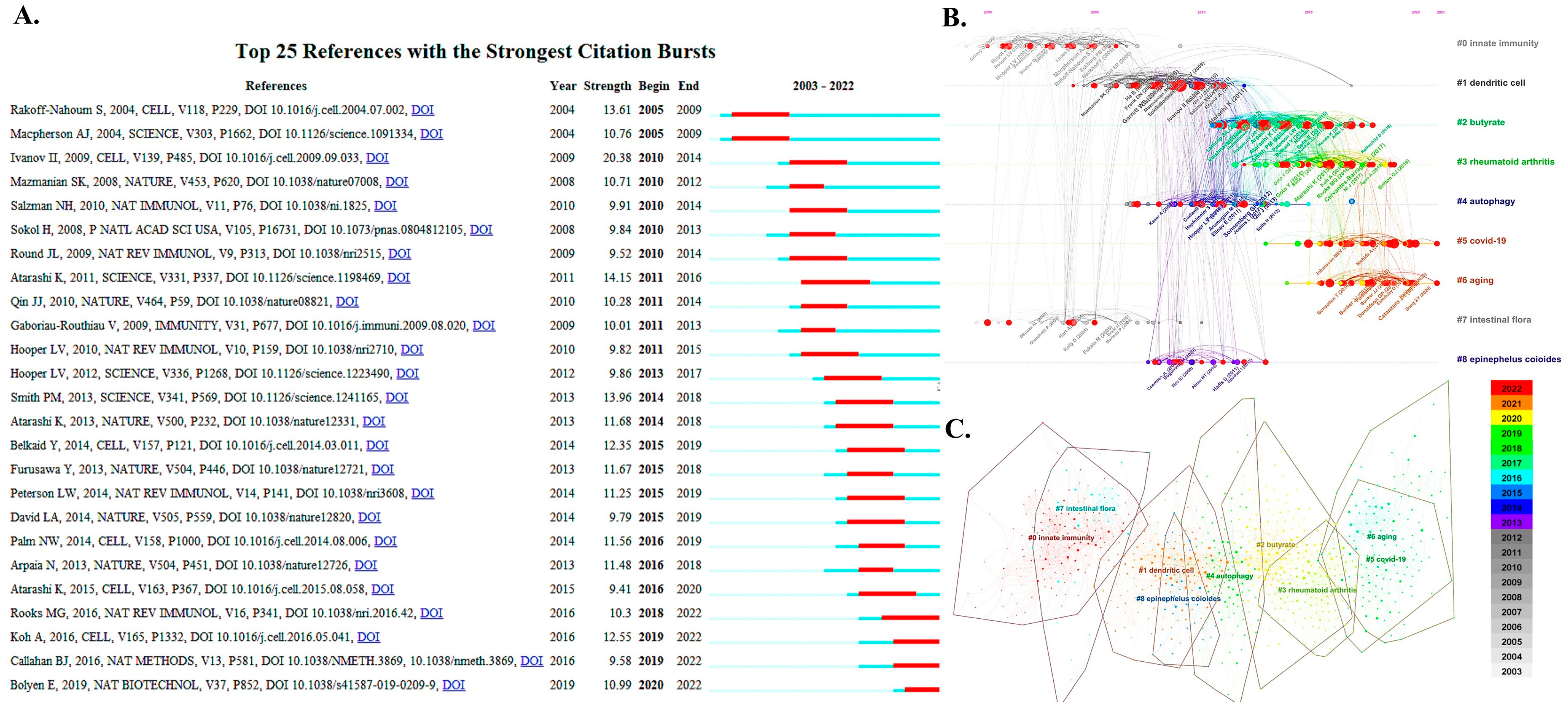
3.5. Co-Citation for References and References with Citation Burst
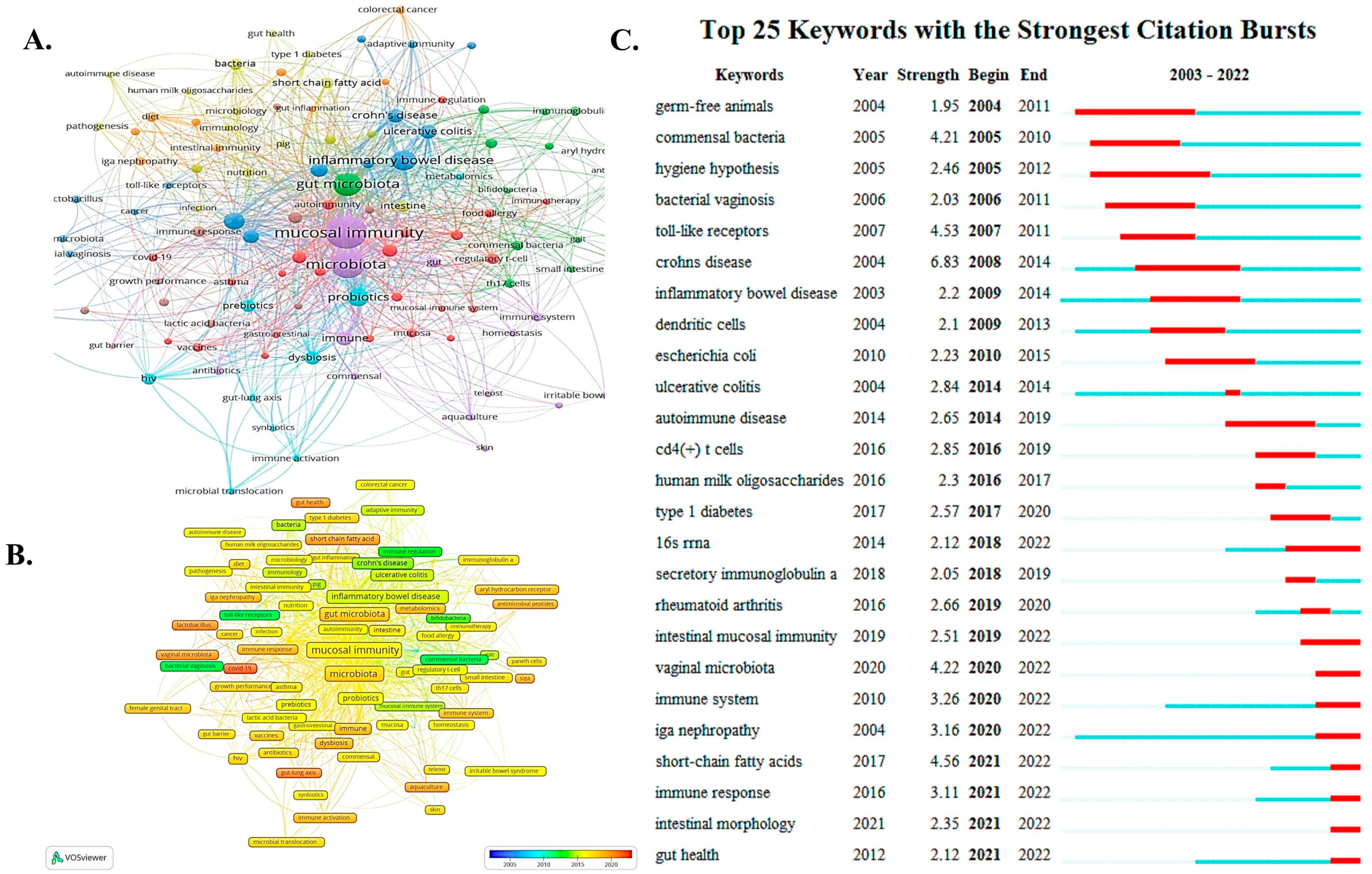
3.6. Summary from Relevant Research Keywords
4. Discussion
4.1. General Condition
4.2. Basic Knowledge Structure
4.3. Current Hotspots Analysis and Field Development Prediction
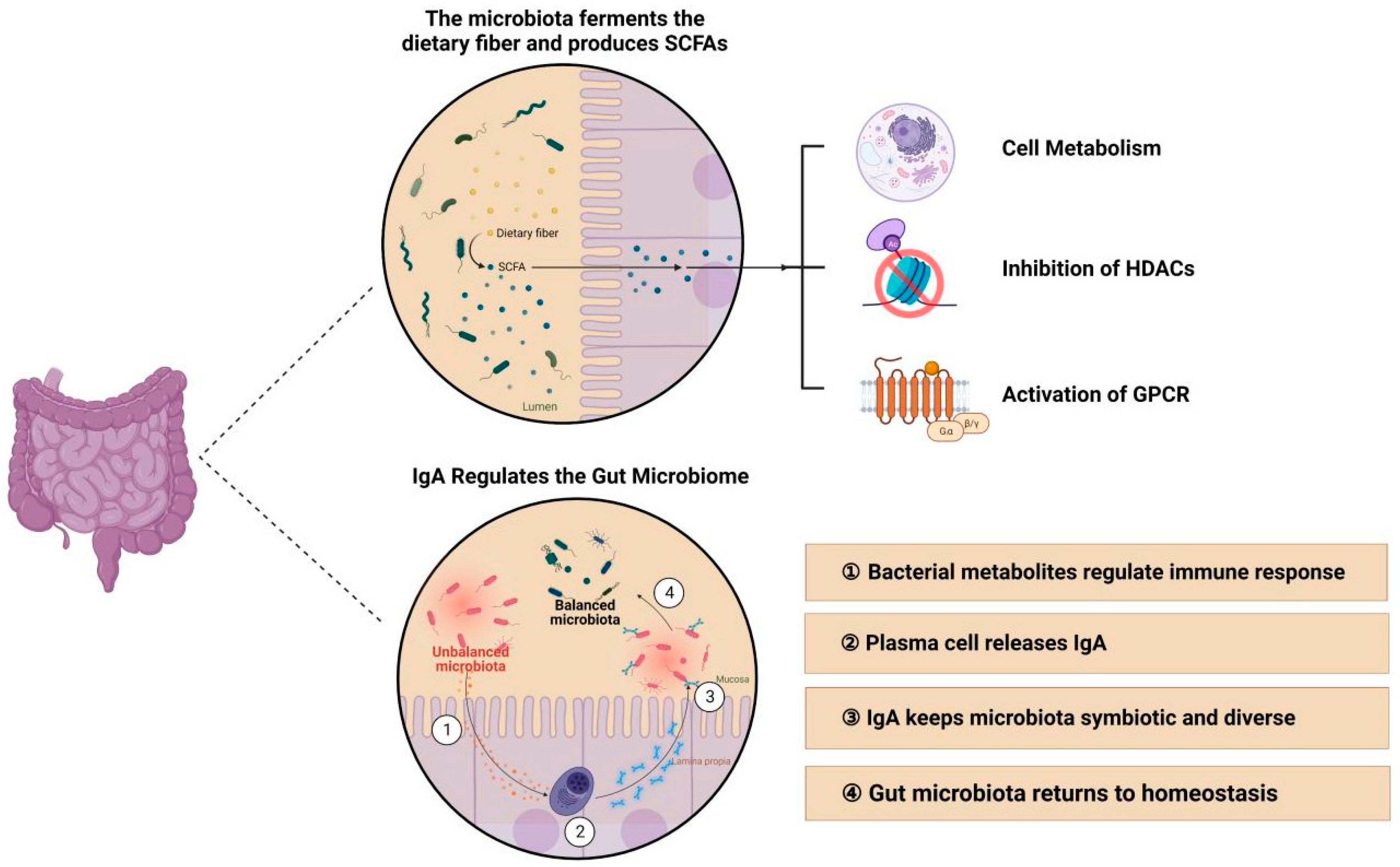
4.3.1. Intestinal Microbiota may Regulate Mucosal Immunity through Short-Chain Fatty Acids and Immunoglobulin A
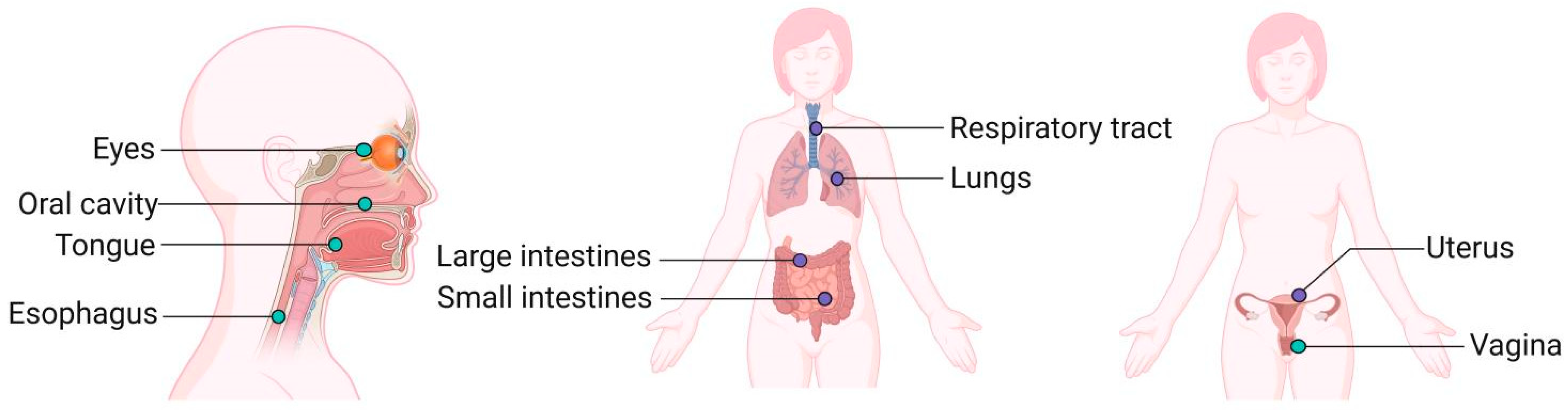
4.3.2. The Influence of Microbiota in Other Parts of the Body on Mucosal Immunity
4.3.3. The Relationship between COVID-19 and Mucosal Immunity and Microbiota (Potential Applications in Diseases)
4.4. Benefits and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Clemente, J.C.; Manasson, J.; Scher, J.U. The role of the gut microbiome in systemic inflammatory disease. BMJ 2018, 360, j5145. [Google Scholar] [CrossRef]
- Russell, M.W.; Moldoveanu, Z.; Ogra, P.L.; Mestecky, J. Mucosal Immunity in COVID-19: A Neglected but Critical Aspect of SARS-CoV-2 Infection. Front. Immunol. 2020, 11, 611337. [Google Scholar] [CrossRef]
- Perez-Lopez, A.; Behnsen, J.; Nuccio, S.P.; Raffatellu, M. Mucosal immunity to pathogenic intestinal bacteria. Nat. Rev. Immunol. 2016, 16, 135–148. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Saulnier, D.M.; Pot, B.; Versalovic, J. How can probiotics and prebiotics impact mucosal immunity? Gut Microbes 2010, 1, 293–300. [Google Scholar] [CrossRef]
- Fernandez, M.I.; Pedron, T.; Tournebize, R.; Olivo-Marin, J.C.; Sansonetti, P.J.; Phalipon, A. Anti-inflammatory role for intracellular dimeric immunoglobulin a by neutralization of lipopolysaccharide in epithelial cells. Immunity 2003, 18, 739–749. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Wei, H.; Lian, Z.X.; Sun, R.; Tian, Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014, 211, 2397–2410. [Google Scholar] [CrossRef]
- Sencio, V.; Gallerand, A.; Gomes Machado, M.; Deruyter, L.; Heumel, S.; Soulard, D.; Barthelemy, J.; Cuinat, C.; Vieira, A.T.; Barthelemy, A.; et al. Influenza Virus Infection Impairs the Gut’s Barrier Properties and Favors Secondary Enteric Bacterial Infection through Reduced Production of Short-Chain Fatty Acids. Infect. Immun. 2021, 89, e0073420. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef]
- Yang, C.Y.; Zhang, F.Y.; Wang, I.J. Probiotics’ Efficacy in Preventing Asthmatic Allergic Reaction Induced by Air Particles: An Animal Study. Nutrients 2022, 14, 5219. [Google Scholar] [CrossRef] [PubMed]
- Karim, A.; Muhammad, T.; Iqbal, M.S.; Qaisar, R. A multistrain probiotic improves handgrip strength and functional capacity in patients with COPD: A randomized controlled trial. Arch. Gerontol. Geriatr. 2022, 102, 104721. [Google Scholar] [CrossRef] [PubMed]
- Jamalkandi, S.A.; Ahmadi, A.; Ahrari, I.; Salimian, J.; Karimi, M.; Ghanei, M. Oral and nasal probiotic administration for the prevention and alleviation of allergic diseases, asthma and chronic obstructive pulmonary disease. Nutr. Res. Rev. 2021, 34, 1–16. [Google Scholar] [CrossRef]
- Lu, W.; Fang, Z.; Liu, X.; Li, L.; Zhang, P.; Zhao, J.; Zhang, H.; Chen, W. The Potential Role of Probiotics in Protection against Influenza a Virus Infection in Mice. Foods 2021, 10, 902. [Google Scholar] [CrossRef] [PubMed]
- Reino-Gelardo, S.; Palop-Cervera, M.; Aparisi-Valero, N.; Espinosa-San Miguel, I.; Lozano-Rodríguez, N.; Llop-Furquet, G.; Sanchis-Artero, L.; Cortés-Castell, E.; Rizo-Baeza, M.; Cortés-Rizo, X. Effect of an Immune-Boosting, Antioxidant and Anti-Inflammatory Food Supplement in Hospitalized COVID-19 Patients: A Prospective Randomized Pilot Study. Nutrients 2023, 15, 1736. [Google Scholar] [CrossRef]
- Shi, K.; Zhang, Q.; Zhang, Y.; Bi, Y.; Zeng, X.; Wang, X. Association between probiotic therapy and the risk of hepatocellular carcinoma in patients with hepatitis B-related cirrhosis. Front. Cell Infect. Microbiol. 2023, 12, 1104399. [Google Scholar] [CrossRef]
- Yu, Z.; Shen, X.; Wang, A.; Hu, C.; Chen, J. The gut microbiome: A line of defense against tuberculosis development. Front. Cell Infect. Microbiol. 2023, 13, 1149679. [Google Scholar] [CrossRef]
- Liu, R.T.; Walsh, R.F.L.; Sheehan, A.E. Prebiotics and probiotics for depression and anxiety: A systematic review and meta-analysis of controlled clinical trials. Neurosci. Biobehav. Rev. 2019, 102, 13–23. [Google Scholar] [CrossRef]
- Sharma, N.; Kang, D.-K.; Paik, H.-D.; Park, Y.-S. Beyond probiotics: A narrative review on an era of revolution. Food Sci. Biotechnol. 2022, 32, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Dong, D.; Dong, Z.; Zhang, Q.; Fang, H.; Wang, C.; Zhang, S.; Wu, S.; Dong, Y.; Wan, Y. Drug repositioning: A bibliometric analysis. Front. Pharmacol. 2022, 13, 974849. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Maniruzzaman, M. A global bibliometric and visualized analysis of bacteria-mediated cancer therapy. Drug. Discov. Today 2022, 27, 103297. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Wu, C.; Hu, S.; Liao, N.; Huang, Y.; Ding, H.; Li, R.; Li, Y. Research on neck dissection for oral squamous-cell carcinoma: A bibliometric analysis. Int. J. Oral. Sci. 2021, 13, 13. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; Deroos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J. Therese Uhr. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Salzman, N.H.; Hung, K.; Haribhai, D.; Chu, H.; Karlsson-Sjöberg, J.; Amir, E.; Teggatz, P.; Barman, M.; Hayward, M.; Eastwood, D.; et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010, 11, 76–83. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper T Cell Responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef]
- Hooper, L.V.; Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010, 10, 159–169. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; de Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Ando, M.; Kamada, N.; Nagano, Y.; Narushima, S.; Suda, W.; Imaoka, A.; Setoyama, H.; Nagamori, T.; et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 2015, 163, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.S.; Kriegel, M.A. Evolving concepts of host-pathobiont interactions in autoimmunity. Curr. Opin. Immunol. 2022, 80, 102265. [Google Scholar] [CrossRef]
- Kudelka, M.R.; Stowell, S.R.; Cummings, R.D.; Neish, A.S. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 597–617. [Google Scholar] [CrossRef]
- Seidelin, J.B.; Bahl, M.I.; Licht, T.R.; Mead, B.E.; Karp, J.M.; Johansen, J.V.; Riis, L.B.; Galera, M.R.; Woetmann, A.; Bjerrum, J.T. Acute Experimental Barrier Injury Triggers Ulcerative Colitis-Specific Innate Hyperresponsiveness and Ulcerative Colitis-Type Microbiome Changes in Humans. Cell Mol. Gastroenterol. Hepatol. 2021, 12, 1281–1296. [Google Scholar] [CrossRef]
- Rachid, R.; Stephen-Victor, E.; Chatila, T.A. The microbial origins of food allergy. J. Allergy Clin. Immunol. 2021, 147, 808–813. [Google Scholar] [CrossRef]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Cai, Y.; Zhu, G.; Liu, S.; Pan, Z.; Quintero, M.; Poole, C.J.; Lu, C.; Zhu, H.; Islam, B.; van Riggelen, J.; et al. Indispensable role of the Ubiquitin-fold modifier 1-specific E3 ligase in maintaining intestinal homeostasis and controlling gut inflammation. Cell Discov. 2019, 5, 7. [Google Scholar] [CrossRef]
- Xu, Z.; Parra, D.; Gómez, D.; Salinas, I.; Zhang, Y.A.; von Gersdorff Jørgensen, L.; Heinecke, R.D.; Buchmann, K.; LaPatra, S.; Sunyer, J.O. Teleost skin, an ancient mucosal surface that elicits gut-like immune response. Proc. Natl. Acad. Sci. USA 2013, 110, 13097–13102. [Google Scholar] [CrossRef]
- Vakili, F.; Roosta, Z.; Safari, R.; Raeisi, M.; Hossain, M.S.; Guerreiro, I.; Akbazadeh, A.; Hoseinifar, S.H. Effects of dietary nutmeg (Myristica fragrans) seed meals on growth, non-specific immune indices, antioxidant status, gene expression analysis, and cold stress tolerance in zebrafish (Danio rerio). Front. Nutr. 2023, 9, 1038748. [Google Scholar] [CrossRef] [PubMed]
- Papotto, P.H.; Yilmaz, B.; Pimenta, G.; Mensurado, S.; Cunha, C.; Fiala, G.J.; da Costa, D.G.; Gonçalves-Sousa, N.; Chan, B.H.; Blankenhaus, B.; et al. Maternal γδ T cells shape offspring pulmonary type 2 immunity in a microbiota-dependent manner. Cell Rep. 2023, 42, 112074. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Fuhrer, T.; Morgenthaler, D.; Krupka, N.; Wang, D.; Spari, D.; Candinas, D.; Misselwitz, B.; Beldi, G.; Sauer, U.; et al. Plasticity of the adult human small intestinal stoma microbiota. Cell Host Microbe 2022, 30, 1773–1787.e6. [Google Scholar] [CrossRef]
- Ramani, K.; Cormack, T.; Cartwright, A.N.; Alami, A.; Parameswaran, P.; Abdou, M.; Wang, I.; Hilliard-Barth, K.; Argueta, S.; Raghunathan, D.; et al. Regulation of Peripheral Inflammation by a Non-Viable, Non-Colonizing Strain of Commensal Bacteria. Front. Immunol. 2022, 13, 768076. [Google Scholar] [CrossRef]
- Stoidis, C.N.; Misiakos, E.P.; Patapis, P.; Fotiadis, C.I.; Spyropoulos, B.G. Potential benefits of pro- and prebiotics on intestinal mucosal immunity and intestinal barrier in short bowel syndrome. Nutr. Res. Rev. 2011, 24, 21–30. [Google Scholar] [CrossRef]
- Sam, Q.H.; Ling, H.; Yew, W.S.; Tan, Z.; Ravikumar, S.; Chang, M.W.; Chai, L.Y.A. The Divergent Immunomodulatory Effects of Short Chain Fatty Acids and Medium Chain Fatty Acids. Int. J. Mol. Sci. 2021, 22, 6453. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.H.; Kim, M.; Yun, C.H. Regulation of Gastrointestinal Immunity by Metabolites. Nutrients 2021, 13, 167. [Google Scholar] [CrossRef]
- Chun, E.; Lavoie, S.; Fonseca-Pereira, D.; Bae, S.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Comeau, C.A.G.; Glickman, J.N.; Fuller, M.H.; et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity 2019, 51, 871–884.e6. [Google Scholar] [CrossRef]
- Goverse, G.; Molenaar, R.; Macia, L.; Tan, J.; Erkelens, M.N.; Konijn, T.; Knippenberg, M.; Cook, E.C.; Hanekamp, D.; Veldhoen, M.; et al. Diet-Derived Short Chain Fatty Acids Stimulate Intestinal Epithelial Cells To Induce Mucosal Tolerogenic Dendritic Cells. J. Immunol. 2017, 198, 2172–2181. [Google Scholar] [CrossRef]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef]
- Liu, E.G.; Zhang, B.; Martin, V.; Anthonypillai, J.; Kraft, M.; Grishin, A.; Grishina, G.; Catanzaro, J.R.; Chinthrajah, S.; Sindher, T.; et al. Food-specific immunoglobulin A does not correlate with natural tolerance to peanut or egg allergens. Sci. Transl. Med. 2022, 14, eabq0599. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Chen, X.; Cheng, H.; Zhang, T.; Li, Z. Advances in IgA glycosylation and its correlation with diseases. Front. Chem. 2022, 10, 974854. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Vogelzang, A.; Maruya, M.; Miyajima, M.; Murata, M.; Son, A.; Kuwahara, T.; Tsuruyama, T.; Yamada, S.; Matsuura, M.; et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 2018, 215, 2019–2034. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K. Diversified IgA–Bacteria Interaction in Gut Homeostasis. Adv. Exp. Med. Biol. 2020, 1254, 105–116. [Google Scholar] [PubMed]
- Pietrzak, B.; Tomela, K.; Olejnik-Schmidt, A.; Mackiewicz, A.; Schmidt, M. Secretory IgA in Intestinal Mucosal Secretions as an Adaptive Barrier against Microbial Cells. Int. J. Mol. Sci. 2020, 21, 9254. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; McKinley, S.A.; Shi, F.; Wang, S.; Mucha, P.J.; Harit, D.; Forest, M.G.; Lai, S.K. Modeling of Virion Collisions in Cervicovaginal Mucus Reveals Limits on Agglutination as the Protective Mechanism of Secretory Immunoglobulin A. PLoS ONE 2015, 10, e0131351. [Google Scholar] [CrossRef]
- de Fays, C.; Carlier, F.M.; Gohy, S.; Pilette, C. Secretory Immunoglobulin A Immunity in Chronic Obstructive Respiratory Diseases. Cells 2022, 11, 1324. [Google Scholar] [CrossRef]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the Respiratory Microbiome in Healthy Nonsmokers and Smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef] [PubMed]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the Upper Respiratory Tract Microbiotas as the Source of the Lung and Gastric Microbiotas in Healthy Individuals. mBio 2015, 6, e00037. [Google Scholar] [CrossRef] [PubMed]
- Ipci, K.; Altıntoprak, N.; Muluk, N.B.; Senturk, M.; Cingi, C. The possible mechanisms of the human microbiome in allergic diseases. Eur. Arch. Otorhinolaryngol. 2017, 274, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, W.; Yuan, Y.; Zhu, W.; Shang, A. Vaginal microecological characteristics of women in different physiological and pathological period. Front. Cell Infect. Microbiol. 2022, 12, 959793. [Google Scholar] [CrossRef] [PubMed]
- Thurman, A.R.; Kimble, T.; Herold, B.; Mesquita, P.M.; Fichorova, R.N.; Dawood, H.Y.; Fashemi, T.; Chandra, N.; Rabe, L.; Cunningham, T.D.; et al. Bacterial Vaginosis and Subclinical Markers of Genital Tract Inflammation and Mucosal Immunity. AIDS Res. Hum. Retroviruses. 2015, 31, 1139–1152. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.F.; Nhan-Chang, C.L.; Sobel, J.D.; Workowski, K.; Conde-Agudelo, A.; Romero, R. Treatment of abnormal vaginal flora in early pregnancy with clindamycin for the prevention of spontaneous preterm birth: A systematic review and metaanalysis. Am. J. Obstet. Gynecol. 2011, 205, 177–190. [Google Scholar] [CrossRef]
- Leger, A.J.S.; Desai, J.V.; Drummond, R.A.; Kugadas, A.; Almaghrabi, F.; Silver, P.; Raychaudhuri, K.; Gadjeva, M.; Iwakura, Y.; Lionakis, M.S.; et al. An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal γδ T Cells. Immunity 2017, 47, 148–158.e5. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Han, Y.; Jia, Z.; Shi, J.; Wang, W.; He, K. The active lung microbiota landscape of COVID-19 patients. Bioimpacts 2022, 12, 139–146. [Google Scholar] [CrossRef]
- Gaibani, P.; Viciani, E.; Bartoletti, M.; Lewis, R.E.; Tonetti, T.; Lombardo, D.; Castagnetti, A.; Bovo, F.; Horna, C.S.; Ranieri, M.; et al. The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci. Rep. 2021, 11, 10103. [Google Scholar] [CrossRef]
- Baradaran Ghavami, S.; Pourhamzeh, M.; Farmani, M.; Raftar, S.K.A.; Shahrokh, S.; Shpichka, A.; Asadzadeh Aghdaei, H.; Hakemi-Vala, M.; Hossein-Khannazer, N.; Timashev, P.; et al. Cross-talk between immune system and microbiota in COVID-19. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 1281–1294. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Tilg, H. COVID-19 and the gastrointestinal tract: More than meets the eye. Gut 2020, 69, 973–974. [Google Scholar] [CrossRef] [PubMed]
| Country | Rank | The Number of Publications (% of 1423) | Organization | Rank | The Number of Publications (% of 1423) |
|---|---|---|---|---|---|
| USA | 1 | 523 (36.8) | Harvard Med Sch | 1 | 24 (1.7) |
| China | 2 | 282 (19.8) | Harvard Univ | 2 | 22 (1.5) |
| England | 3 | 94 (6.6) | Chinese Acad Sci | 3 | 21 (1.5) |
| Canada | 4 | 89 (6.3) | Univ Washington | 4 | 20 (1.4) |
| Japan | 5 | 79 (5.6) | China Agr Univ | 5 | 20 (1.4) |
| Germany | 6 | 78 (5.5) | Univ Toronto | 6 | 17 (1.2) |
| Italy | 7 | 73 (5.1) | Univ Tokyo | 7 | 17 (1.2) |
| France | 8 | 70 (4.9) | Univ Calif Davis | 8 | 17 (1.2) |
| Spain | 9 | 56 (3.9) | NYU | 9 | 17 (1.2) |
| Iran | 10 | 53 (3.7) | Huazhong Agr Univ | 10 | 17 (1.2) |
| Name | Rank | The Number of Publications | H-Index |
|---|---|---|---|
| Xu, Zhen | 1 | 13 | 16 |
| Hoseinifar, Seyed Hossein | 2 | 13 | 45 |
| Ding, Li-guo | 3 | 7 | 8 |
| Tlaskalova-hogenova, Helena | 4 | 7 | 31 |
| Salinas, Rene | 5 | 7 | 30 |
| Yu, Yong-yao | 6 | 6 | 14 |
| Sunyer, J. Oriol | 7 | 6 | 35 |
| Stepankova, Renata | 8 | 6 | 30 |
| Scher, Jose U. | 9 | 6 | 37 |
| Shanahan, Fergus | 10 | 6 | 100 |
| Name | Rank | The Number of Co-Cited Times | H-Index |
|---|---|---|---|
| Macpherson, Andrew J. | 1 | 292 | 72 |
| Hooper, Lora V. | 2 | 260 | 63 |
| Atarashi, Koji | 3 | 254 | 34 |
| Brandtzaeg, Per | 4 | 243 | 95 |
| Ivanov, I. I. | 5 | 230 | 10 |
| Turnbaugh, Peter J. | 6 | 187 | 52 |
| Round, June | 7 | 172 | 33 |
| Ley, Ruth E | 8 | 160 | 75 |
| Johansson, Malin E. V. | 9 | 149 | 48 |
| Sokol, Harry | 10 | 128 | 63 |
| Name of the Journal | Rank | The Number of Publications | IF (2021–2022) |
|---|---|---|---|
| Frontiers in Immunology | 1 | 98 | 8.786 |
| Frontiers in Microbiology | 2 | 34 | 6.064 |
| Plos One | 3 | 33 | 3.752 |
| Mucosal Immunology | 4 | 25 | 8.701 |
| Frontiers in Cellular and Infection Microbiology | 5 | 22 | 6.073 |
| Proceedings of the National Academy of Sciences of the United States of America | 6 | 21 | 12.779 |
| Current Opinion in Gastroenterology | 7 | 19 | 2.741 |
| Fish & Shellfish Immunology | 8 | 19 | 4.622 |
| Immunology | 9 | 18 | 7.215 |
| Microorganisms | 10 | 16 | 4.926 |
| International Journal of Molecular Sciences | 11 | 16 | 6.208 |
| Gut Microbes | 12 | 15 | 9.434 |
| Gastroenterology | 13 | 15 | 33.883 |
| Infection and Immunity | 14 | 14 | 3.609 |
| Nutrients | 15 | 13 | 6.706 |
| Title | Type | Rank | Year | Cited Times | Journal |
|---|---|---|---|---|---|
| Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria | A | 1 | 2009 | 133 | Cell |
| Induction of colonic regulatory T cells by indigenous Clostridium species | A | 2 | 2011 | 93 | Science |
| Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis | A | 3 | 2004 | 89 | Cell |
| QIIME allows analysis of high-throughput community sequencing data | L | 4 | 2010 | 82 | Nat Methods |
| The gut microbiota shapes intestinal immune responses during health and disease | R | 5 | 2009 | 80 | Nat Rev Immunol |
| Diversity of the human intestinal microbial flora | A | 6 | 2005 | 78 | Science |
| The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis | A | 7 | 2013 | 76 | Science |
| Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation | A | 8 | 2013 | 70 | Nature |
| Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases | A | 9 | 2007 | 70 | Proc Natl Acad Sci USA |
| Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells | A | 10 | 2013 | 68 | Nature |
| A human gut microbial gene catalogue established by metagenomic sequencing | A | 11 | 2010 | 68 | Nature |
| Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota | A | 12 | 2013 | 67 | Nature |
| Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria | A | 13 | 2004 | 61 | Science |
| Interactions between the microbiota and the immune system | R | 14 | 2012 | 61 | Science |
| An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system | A | 15 | 2004 | 60 | Cell |
| Rank | Keywords | Counts |
|---|---|---|
| 1 | mucosal immunity | 440 |
| 2 | gut microbiota | 182 |
| 3 | inflammatory bowel disease | 100 |
| 4 | Crohn’s disease | 44 |
| 5 | innate immunity | 40 |
| 6 | barrier function | 35 |
| 7 | ulcerative colitis | 34 |
| 8 | dendritic cells | 28 |
| 9 | short-chain fatty acids | 20 |
| 10 | commensal bacteria | 19 |
| 11 | intestinal epithelium | 18 |
| 12 | B cells | 14 |
| 13 | immune response | 14 |
| 14 | 16S rRNA | 14 |
| 15 | gastrointestinal tract | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wu, J.; Ran, D.; Ou, G.; Chen, Y.; Xu, H.; Deng, L.; Chen, X. Study of the Relationship between Mucosal Immunity and Commensal Microbiota: A Bibliometric Analysis. Nutrients 2023, 15, 2398. https://doi.org/10.3390/nu15102398
Wang S, Wu J, Ran D, Ou G, Chen Y, Xu H, Deng L, Chen X. Study of the Relationship between Mucosal Immunity and Commensal Microbiota: A Bibliometric Analysis. Nutrients. 2023; 15(10):2398. https://doi.org/10.3390/nu15102398
Chicago/Turabian StyleWang, Shiqi, Jialin Wu, Duo Ran, Guosen Ou, Yaokang Chen, Huachong Xu, Li Deng, and Xiaoyin Chen. 2023. "Study of the Relationship between Mucosal Immunity and Commensal Microbiota: A Bibliometric Analysis" Nutrients 15, no. 10: 2398. https://doi.org/10.3390/nu15102398
APA StyleWang, S., Wu, J., Ran, D., Ou, G., Chen, Y., Xu, H., Deng, L., & Chen, X. (2023). Study of the Relationship between Mucosal Immunity and Commensal Microbiota: A Bibliometric Analysis. Nutrients, 15(10), 2398. https://doi.org/10.3390/nu15102398






