An Ethanol Extract of Coptidis rhizoma Induces Apoptotic Cell Death in Induced Pluripotent Stem Cells and Suppresses Teratoma Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of ECR
2.2. Cells and Reagents
2.3. RNA Isolation, Library Preparation, and Whole Transcriptome Profiling Using High-Throughput Sequencing
2.4. Cell Viability Assay in Monolayer and Spheroid Culture
2.5. Detection of Apoptotic Cell Death by AO/EB and DAPI Staining
2.6. Immunofluorescence Analysis for the γ-H2AX Foci
2.7. Measurement of Intracellular Reactive Oxygen Species (ROS)
2.8. Detection of Mitochondrial Membrane Potential (MMP)
2.9. Western Blot Analysis
2.10. Assessment of Caspase Activity
2.11. Analysis of Selective Elimination of iPSC in the Mixed Population with iPSC-Diff
2.12. In Ovo Teratoma Formation Assay
2.13. Ultra High-Performance Liquid Chromatography (UHPLC) Analysis
2.14. Statistical Analysis
3. Results and Discussion
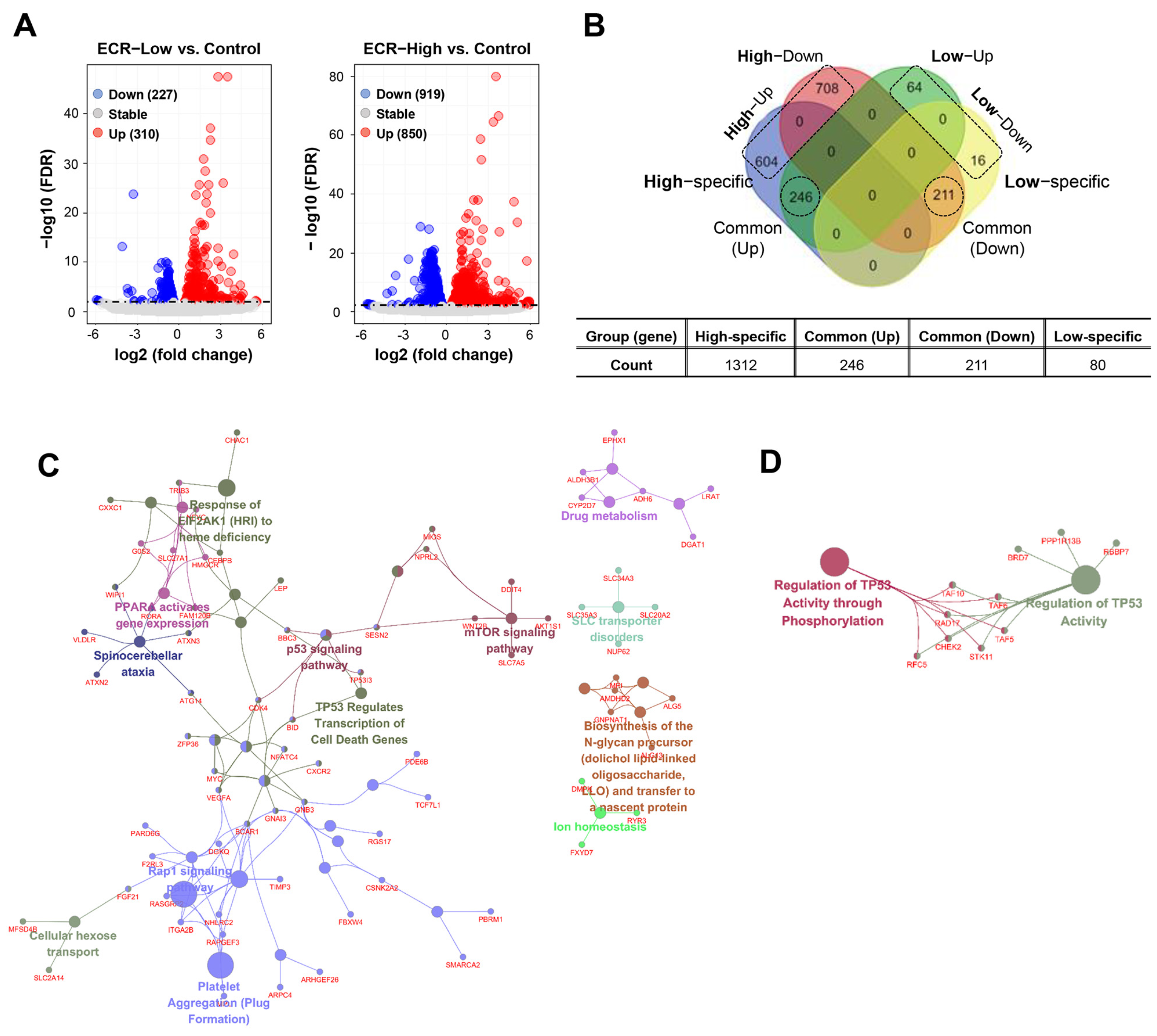
3.1. Identification of Functional Clusters of ECR-Treated iPSCs
3.2. The ECR Showed Cytotoxicity in iPSCs in Two-Dimensional (2D) and Three-Dimensional (3D) Cultures and Induced Apoptotic Cell Death
3.3. The ECR Induced Intracellular ROS Generation, Mitochondrial Damage, and Caspase-3/-9 Activation in iPSCs
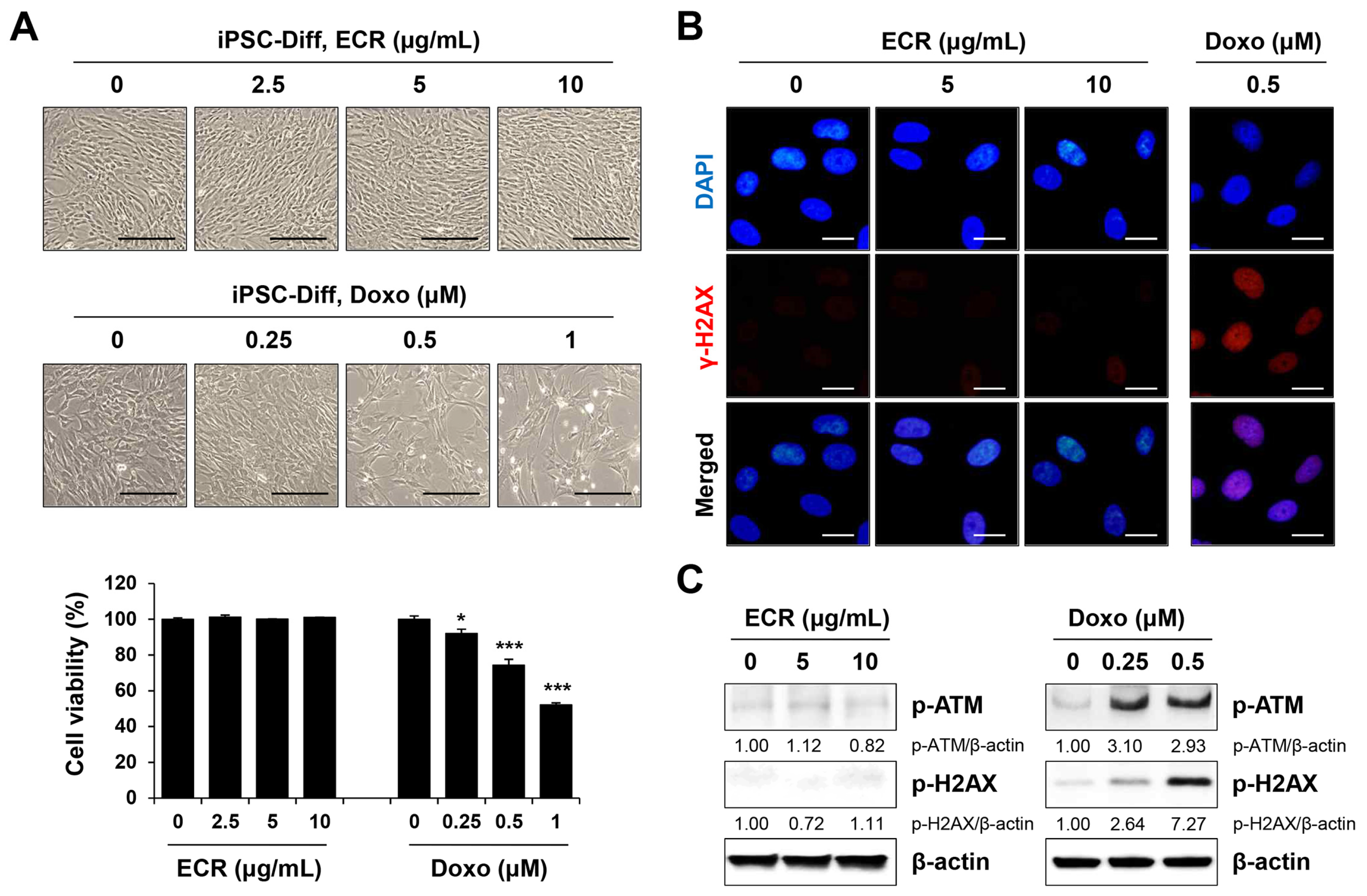
3.4. The ECR Did Not Induce Cell Death or Genotoxicity in iPSC-Diff
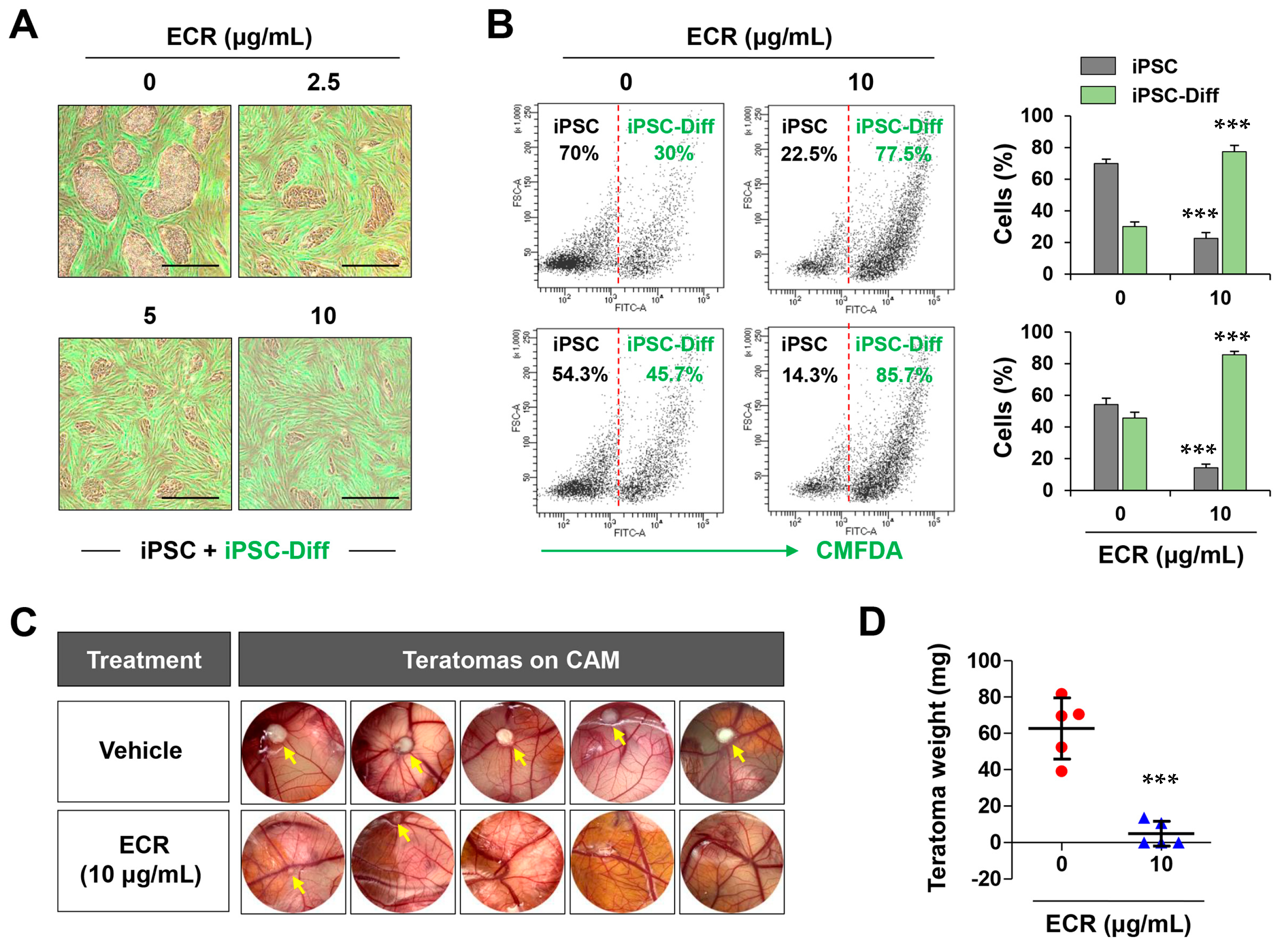
3.5. The ECR Selectively Eliminated iPSCs but Not iPSC-Diff in a Mixed Cell Population and Suppressed in Ovo Teratoma Formation
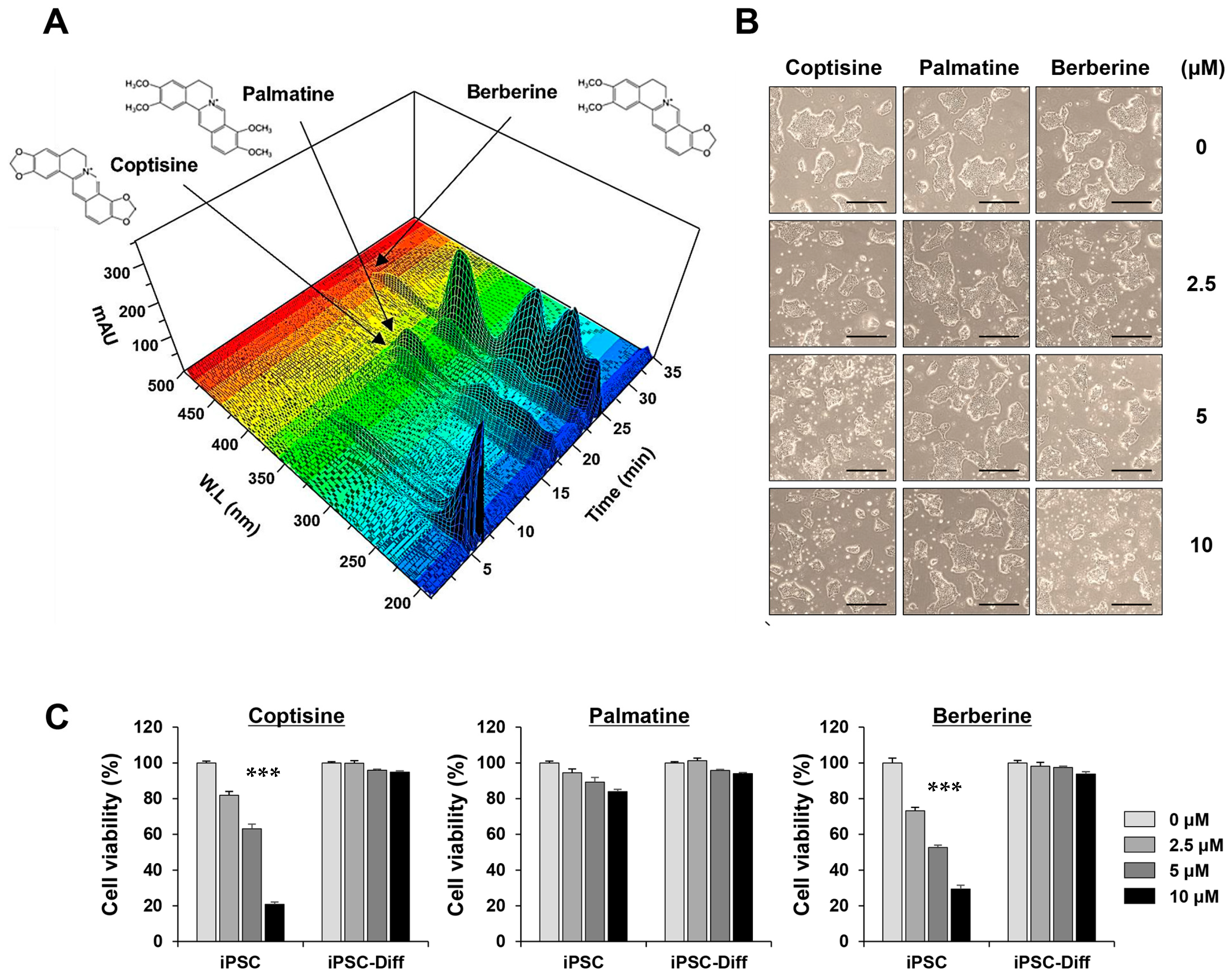
3.6. Coptisine and Berberine in the ECR Exhibited Selective Cytotoxicity in iPSCs but Not in iPSC-Diff
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kimbrel, E.A.; Lanza, R. Current status of pluripotent stem cells: Moving the first therapies to the clinic. Nat. Rev. Drug Discov. 2015, 14, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, J.; Forget, B.G.; Gaines, P. Induced pluripotent stem cells in regenerative medicine: An argument for continued research on human embryonic stem cells. Regen. Med. 2009, 4, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Kalsan, M.; Kumar, N.; Saini, A.; Chandra, R. Induced pluripotent stem cells: Applications in regenerative medicine, disease modeling, and drug discovery. Front. Cell Dev. Biol. 2015, 3, 2. [Google Scholar] [CrossRef]
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int. J. Med. Sci. 2018, 15, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Trounson, A.; McDonald, C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell 2015, 17, 11–22. [Google Scholar] [CrossRef]
- Jeong, H.-C.; Cho, S.-J.; Lee, M.-O.; Cha, H.-J. Technical approaches to induce selective cell death of pluripotent stem cells. Cell. Mol. Life Sci. 2017, 74, 2601–2611. [Google Scholar] [CrossRef]
- Kuang, Y.; Miki, K.; Parr, C.J.C.; Hayashi, K.; Takei, I.; Li, J.; Iwasaki, M.; Nakagawa, M.; Yoshida, Y.; Saito, H. Efficient, Selective Removal of Human Pluripotent Stem Cells via Ecto-Alkaline Phosphatase-Mediated Aggregation of Synthetic Peptides. Cell Chem. Biol. 2017, 24, 685–694.e684. [Google Scholar] [CrossRef]
- Lee, M.-O.; Moon, S.H.; Jeong, H.-C.; Yi, J.-Y.; Lee, T.-H.; Shim, S.H.; Rhee, Y.-H.; Lee, S.-H.; Oh, S.-J.; Lee, M.-Y.; et al. Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc. Natl. Acad. Sci. USA 2013, 110, E3281–E3290. [Google Scholar] [CrossRef]
- Kim, A.; Lee, S.-Y.; Kim, B.-Y.; Chung, S.-K. Elimination of Teratogenic Human Induced Pluripotent Stem Cells by Bee Venom via Calcium-Calpain Pathway. Int. J. Mol. Sci. 2020, 21, 3265. [Google Scholar] [CrossRef]
- Kim, A.; Lee, S.-Y.; Seo, C.-S.; Chung, S.-K. Ethanol extract of Magnoliae cortex (EEMC) limits teratoma formation of pluripotent stem cells by selective elimination of undifferentiated cells through the p53-dependent mitochondrial apoptotic pathway. Phytomedicine 2020, 69, 153198. [Google Scholar] [CrossRef]
- Kim, A.; Lee, S.-Y.; Seo, C.-S.; Chung, S.-K. Prunellae Spica Extract Suppresses Teratoma Formation of Pluripotent Stem Cells through p53-Mediated Apoptosis. Nutrients 2020, 12, 721. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Lou, G.H.; Zeng, H.-R.; Hu, J.; Huang, Q.W.; Peng, W.; Yang, X.-B. Coptidis Rhizoma: A comprehensive review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. Pharm. Biol. 2019, 57, 193–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mu, W.; Shang, H.; Lin, J.; Lei, X. The antihyperglycemic effects of Rhizoma Coptidis and mechanism of actions: A review of systematic reviews and pharmacological research. BioMed Res. Int. 2014, 2014, 798093. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xiong, Y.-Y.; Wu, H.-Z.; Xiong, W.-C.; Liu, B.; Xie, Z.-T.; Xiao, W.-P.; Huang, B.-S.; Yang, Y.-F. Active Ingredients and Mechanism of Action of Rhizoma Coptidis against Type 2 Diabetes Based on Network-Pharmacology and Bioinformatics. Curr. Med. Sci. 2020, 40, 257–264. [Google Scholar] [CrossRef]
- He, L.; Zhong, Z.; Chen, M.; Liang, Q.; Wang, Y.; Tan, W. Current Advances in Coptidis Rhizoma for Gastrointestinal and Other Cancers. Front. Pharmacol. 2021, 12, 775084. [Google Scholar] [CrossRef]
- Meng, F.-C.; Wu, Z.-F.; Yin, Z.-Q.; Lin, L.-G.; Wang, R.; Zhang, Q.-W. Coptidis rhizoma and its main bioactive components: Recent advances in chemical investigation, quality evaluation and pharmacological activity. Chin. Med. 2018, 13, 13. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Park, J.-H.; Jeong, S.; Kim, B.-Y.; Kang, Y.-K.; Xu, Y.; Chung, S.-K. K120R mutation inactivates p53 by creating an aberrant splice site leading to nonsense-mediated mRNA decay. Oncogene 2019, 38, 1597–1610. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, N.; Miyamoto, K.; Okita, K.; Tangoku, A.; Hayashi, H.; Yosino, S.; Abe, T.; Morioka, T.; Hazama, S.; Oka, M. Inhibitory effect of Coptidis Rhizoma and berberine on the proliferation of human esophageal cancer cell lines. Cancer Lett. 2000, 148, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.-J.; Noh, J.-W.; Lee, B.-C. Mechanisms and Effect of Coptidis Rhizoma on Obesity-Induced Inflammation: In Silico and In Vivo Approaches. Int. J. Mol. Sci. 2021, 22, 8075. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Feng, S.; Zhang, X.; Zhao, W.; Feng, J.; Ma, C.; Wang, R.; Song, W.; Cheng, J. Biological Response Profiling Reveals the Functional Differences of Main Alkaloids in Rhizoma Coptidis. Molecules 2021, 26, 7389. [Google Scholar] [CrossRef]
- Kim, A.; Lee, S.-Y.; Chung, S.-K. Caffeic acid selectively eliminates teratogenic human-induced pluripotent stem cells via apoptotic cell death. Phytomedicine 2022, 102, 154144. [Google Scholar] [CrossRef]
- Huelsenbeck, S.C.; Schorr, A.; Roos, W.P.; Huelsenbeck, J.; Henninger, C.; Kaina, B.; Fritz, G. Rac1 protein signaling is required for DNA damage response stimulated by topoisomerase II poisons. J. Biol. Chem. 2012, 287, 38590–38599. [Google Scholar] [CrossRef]
- Banáth, J.P.; Klokov, D.; MacPhail, S.H.; Banuelos, C.A.; Olive, P.L. Residual γH2AX foci as an indication of lethal DNA lesions. BMC Cancer 2010, 10, 4. [Google Scholar] [CrossRef]
- Mariotti, L.G.; Pirovano, G.; Savage, K.I.; Ghita, M.; Ottolenghi, A.; Prise, K.M.; Schettino, G. Use of the γ-H2AX Assay to Investigate DNA Repair Dynamics Following Multiple Radiation Exposures. PLoS ONE 2013, 8, e79541. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, C.; Kim, M.Y.; Ji, S.Y.; Hwangbo, H.; Lee, H.; Hong, S.H.; Han, M.H.; Jeong, J.-W.; Kim, G.-Y.; et al. ROS-Mediated Anti-Tumor Effect of Coptidis Rhizoma against Human Hepatocellular Carcinoma Hep3B Cells and Xenografts. Int. J. Mol. Sci. 2021, 22, 4797. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The Anti-Cancer Mechanisms of Berberine: A Review. Cancer Manag. Res. 2020, 12, 695–702. [Google Scholar] [CrossRef]
- Xiong, R.-G.; Huang, S.-Y.; Wu, S.-X.; Zhou, D.-D.; Yang, Z.-J.; Saimaiti, A.; Zhao, C.-N.; Shang, A.; Zhang, Y.-J.; Gan, R.-Y.; et al. Anticancer Effects and Mechanisms of Berberine from Medicinal Herbs: An Update Review. Molecules 2022, 27, 4523. [Google Scholar] [CrossRef] [PubMed]
- Kühlbrandt, W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Sivandzade, F.; Bhalerao, A.; Cucullo, L. Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe. Bio-Protoc. 2019, 9, e3128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Campbell, T.; Perry, B.; Beaurepaire, C.; Qin, L. Hypoglycemic and insulin-sensitizing effects of berberine in high-fat diet- and streptozotocin-induced diabetic rats. Metabolism 2011, 60, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Zhao, Y.; Dong, F.; Yan, Z.; Zheng, W.; Fan, J.; Sun, G. Meta-analysis of the effect and safety of berberine in the treatment of type 2 diabetes mellitus, hyperlipemia and hypertension. J. Ethnopharmacol. 2015, 161, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Xiao, Y.; Yi, L.; Li, L.; Wang, M.; Tian, C.; Ma, H.; He, K.; Wang, Y.; Han, B.; et al. Coptisine from Rhizoma Coptidis Suppresses HCT-116 Cells-related Tumor Growth in vitro and in vivo. Sci. Rep. 2017, 7, srep38524. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.-L.; Jia, W.-H.; Zhang, L.; Xu, C.-Y.; Chen, X.; Yin, L.; Wang, N.-Q.; Fang, L.-H.; Qiang, G.-F.; Yang, X.-Y.; et al. Glucose consumption assay discovers coptisine with beneficial effect on diabetic mice. Eur. J. Pharmacol. 2019, 859, 172523. [Google Scholar] [CrossRef]
- Long, J.; Song, J.; Zhong, L.; Liao, Y.; Liu, L.; Li, X. Palmatine: A review of its pharmacology, toxicity and pharmacokinetics. Biochimie 2019, 162, 176–184. [Google Scholar] [CrossRef]


| Parameter | Analytical Conditions | ||
|---|---|---|---|
| Column temperature | 30 °C | ||
| UV | DAD at 350 nm | ||
| Spectra range | 190 to 500 nm | ||
| Injection volume | 2 µL | ||
| Flow rate | 0.5 mL/min | ||
| Column | Phenomenex Luna C18 (4.6 × 250 mm, 5 µm) | ||
| Sample reconstruction | 2 mg/mL in MeOH | ||
| Mobile phase | Time (min) | A (%) (0.1% formic acid in water) | B (%) (Acetonitrile) |
| 0 | 72 | 28 | |
| 35 | 90 | 10 | |
| 50 | Washing and equilibrium | ||
| Constituent | Linear Range (µg/mL) | Regression Equation (a) | Correlation Coefficient, r2 | LOD (µg/mL) | LOQ (µg/mL) | |
|---|---|---|---|---|---|---|
| Slope | Intercept | |||||
| Coptisine | 2–200 | 24,405 | 8.067 | 0.9999 | 0.541 | 1.638 |
| Palmatine | 5–200 | 15,315 | 5.335 | 0.9999 | 0.890 | 2.696 |
| Berberine | 1–200 | 35,455 | 15.435 | 0.9999 | 0.263 | 0.797 |
| Compound | Content (mg/g) |
|---|---|
| Coptisine | 31.62 ± 0.09 |
| Palmatine | 48.56 ± 0.23 |
| Berberine | 84.01 ± 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, A.; Baek, S.-J.; Shin, S.; Lee, S.-Y.; Chung, S.-K. An Ethanol Extract of Coptidis rhizoma Induces Apoptotic Cell Death in Induced Pluripotent Stem Cells and Suppresses Teratoma Formation. Nutrients 2023, 15, 2364. https://doi.org/10.3390/nu15102364
Kim A, Baek S-J, Shin S, Lee S-Y, Chung S-K. An Ethanol Extract of Coptidis rhizoma Induces Apoptotic Cell Death in Induced Pluripotent Stem Cells and Suppresses Teratoma Formation. Nutrients. 2023; 15(10):2364. https://doi.org/10.3390/nu15102364
Chicago/Turabian StyleKim, Aeyung, Su-Jin Baek, Sarah Shin, Seo-Young Lee, and Sun-Ku Chung. 2023. "An Ethanol Extract of Coptidis rhizoma Induces Apoptotic Cell Death in Induced Pluripotent Stem Cells and Suppresses Teratoma Formation" Nutrients 15, no. 10: 2364. https://doi.org/10.3390/nu15102364
APA StyleKim, A., Baek, S.-J., Shin, S., Lee, S.-Y., & Chung, S.-K. (2023). An Ethanol Extract of Coptidis rhizoma Induces Apoptotic Cell Death in Induced Pluripotent Stem Cells and Suppresses Teratoma Formation. Nutrients, 15(10), 2364. https://doi.org/10.3390/nu15102364







