Molecular Mechanism of Cyanidin-3-O-Glucoside Disassembling Aβ Fibril In Silico
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecule Preparation
2.2. Molecule Interaction
2.3. Analysis
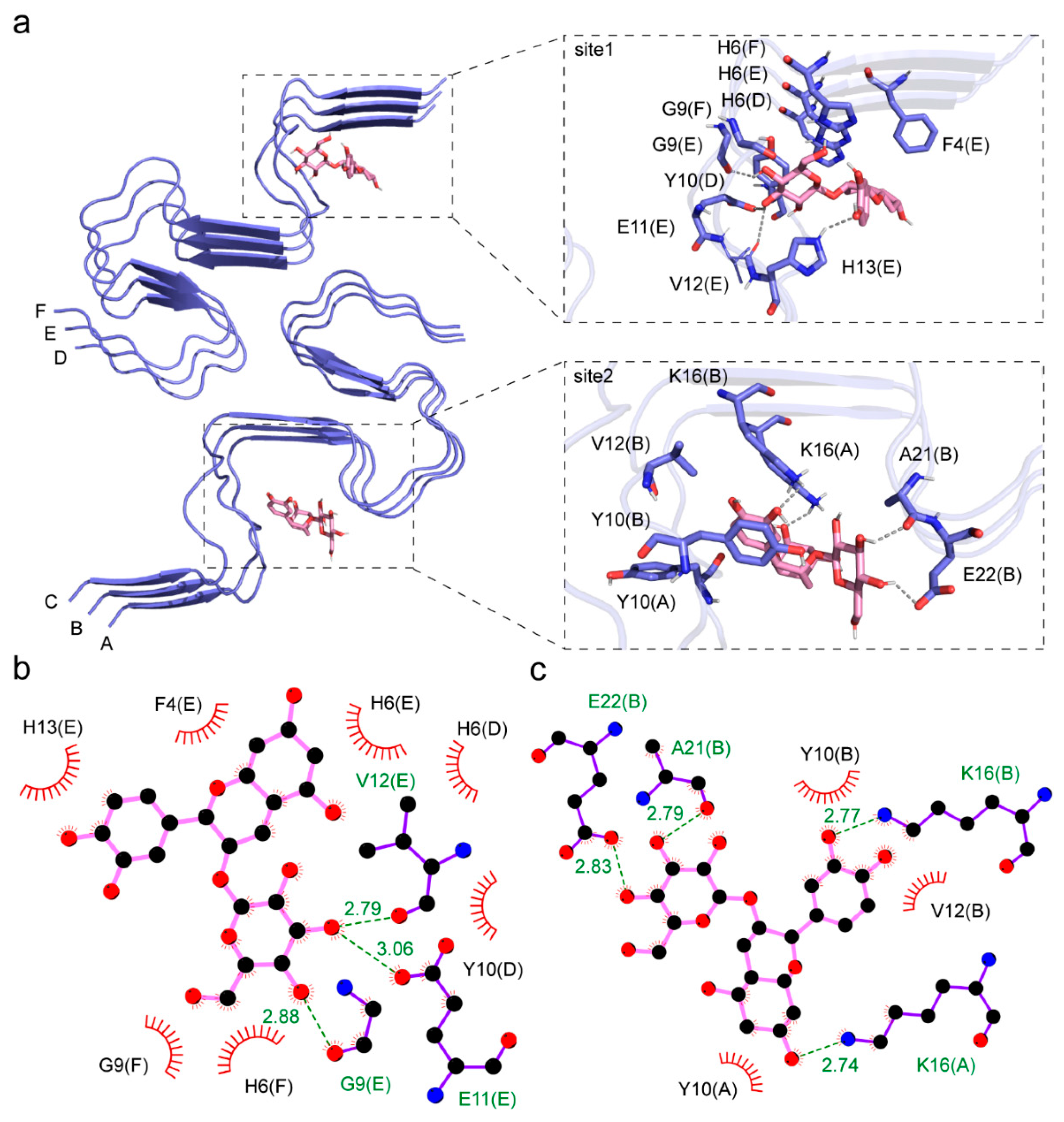
3. Results
3.1. Simulated Single Aβ peptide Solution Structure
3.2. Interaction between Aβ Fibril and Cy-3G
3.3. Interaction between a Single Aβ Peptide and Cy-3G
3.4. Preferential Binding of Cy-3G to a Single Aβ Peptide over the Fibrillar Polymorph
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dartigues, J.F. Alzheimer’s disease: A global challenge for the 21st century. Lancet Neurol. 2009, 8, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Servick, K. Doubts persist for claimed Alzheimer’s drug. Science 2019, 366, 1298. [Google Scholar] [CrossRef]
- Mullane, K.; Williams, M. Alzheimer’s disease beyond amyloid: Can the repetitive failures of amyloid-targeted therapeutics inform future approaches to dementia drug discovery? Biochem. Pharmacol. 2020, 177, 113945. [Google Scholar] [CrossRef] [PubMed]
- Sevigny, J.; Chiao, P.; Bussiere, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Benek, O.; Korabecny, J.; Soukup, O. A Perspective on Multi-target Drugs for Alzheimer’s Disease. Trends Pharmacol. Sci. 2020, 41, 434–445. [Google Scholar] [CrossRef]
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Prim. 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Campora, M.; Francesconi, V.; Schenone, S.; Tasso, B.; Tonelli, M. Journey on Naphthoquinone and Anthraquinone Derivatives: New Insights in Alzheimer’s Disease. Pharmaceuticals 2021, 14, 33. [Google Scholar] [CrossRef]
- De Strooper, B.; Vassar, R.; Golde, T. The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 99–107. [Google Scholar] [CrossRef]
- Huang, Y.; Potter, R.; Sigurdson, W.; Santacruz, A.; Shih, S.; Ju, Y.E.; Kasten, T.; Morris, J.C.; Mintun, M.; Duntley, S.; et al. Effects of age and amyloid deposition on Abeta dynamics in the human central nervous system. Arch. Neurol. 2012, 69, 51–58. [Google Scholar] [CrossRef]
- Iwatsubo, T.; Odaka, A.; Suzuki, N.; Mizusawa, H.; Nukina, N.; Ihara, Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: Evidence that an initially deposited species is A beta 42(43). Neuron 1994, 13, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.L.; Papayannopoulos, I.A.; Styles, J.; Bobin, S.A.; Lin, Y.Y.; Biemann, K.; Iqbal, K. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer’s disease. Arch. Biochem. Biophys. 1993, 301, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Kodali, R.; Williams, A.D.; Chemuru, S.; Wetzel, R. Abeta(1–40) forms five distinct amyloid structures whose beta-sheet contents and fibril stabilities are correlated. J. Mol. Biol. 2010, 401, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Agopian, A.; Guo, Z. Structural origin of polymorphism of Alzheimer’s amyloid beta-fibrils. Biochem. J. 2012, 447, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Petkova, A.T.; Leapman, R.D.; Guo, Z.; Yau, W.M.; Mattson, M.P.; Tycko, R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science 2005, 307, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Katzmarski, N.; Ziegler-Waldkirch, S.; Scheffler, N.; Witt, C.; Abou-Ajram, C.; Nuscher, B.; Prinz, M.; Haass, C.; Meyer-Luehmann, M. Abeta oligomers trigger and accelerate Abeta seeding. Brain Pathol. 2020, 30, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Pittman, J.M.; Zerweck, J.; Venkata, B.S.; Moore, P.C.; Sachleben, J.R.; Meredith, S.C. Beta-Amyloid aggregation and heterogeneous nucleation. Protein Sci. 2019, 28, 1567–1581. [Google Scholar] [CrossRef] [PubMed]
- Freyssin, A.; Page, G.; Fauconneau, B.; Rioux Bilan, A. Natural polyphenols effects on protein aggregates in Alzheimer’s and Parkinson’s prion-like diseases. Neural Regen. Res. 2018, 13, 955–961. [Google Scholar] [CrossRef]
- Jalili-Baleh, L.; Babaei, E.; Abdpour, S.; Nasir Abbas Bukhari, S.; Foroumadi, A.; Ramazani, A.; Sharifzadeh, M.; Abdollahi, M.; Khoobi, M. A review on flavonoid-based scaffolds as multi-target-directed ligands (MTDLs) for Alzheimer’s disease. Eur. J. Med. Chem. 2018, 152, 570–589. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.; Khan, M.; Shah, S.A.; Saeed, K.; Kim, M.O. Natural Antioxidant Anthocyanins-A Hidden Therapeutic Candidate in Metabolic Disorders with Major Focus in Neurodegeneration. Nutrients 2019, 11, 1195. [Google Scholar] [CrossRef]
- Tena, N.; Martin, J.; Asuero, A.G. State of the Art of Anthocyanins: Antioxidant Activity, Sources, Bioavailability, and Therapeutic Effect in Human Health. Antioxidants 2020, 9, 451. [Google Scholar] [CrossRef]
- Rupasinghe, H.P.V.; Arumuggam, N.; Amararathna, M.; De Silva, A.B.K.H. The potential health benefits of haskap (Lonicera caerulea L.): Role of cyanidin-3-O-glucoside. J. Funct. Foods 2018, 44, 24–39. [Google Scholar] [CrossRef]
- Olivas-Aguirre, F.J.; Rodrigo-Garcia, J.; Martinez-Ruiz, N.D.; Cardenas-Robles, A.I.; Mendoza-Diaz, S.O.; Alvarez-Parrilla, E.; Gonzalez-Aguilar, G.A.; de la Rosa, L.A.; Ramos-Jimenez, A.; Wall-Medrano, A. Cyanidin-3-O-glucoside: Physical-Chemistry, Foodomics and Health Effects. Molecules 2016, 21, 1264. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-s.; Li, B.; Li, D.-n.; Wang, Y.-h.; Lin, Y.; Meng, X.-j.; Sun, X.-y.; Liu, N. Cyanidin-3-O-glucoside attenuates amyloid-beta (1–40)-induced oxidative stress and apoptosis in SH-SY5Y cells through a Nrf2 mechanism. J. Funct. Foods 2017, 38, 474–485. [Google Scholar] [CrossRef]
- Shin, W.-H.; Park, S.-J.; Kim, E.-J. Protective effect of anthocyanins in middle cerebral artery occlusion and reperfusion model of cerebral ischemia in rats. Life Sci. 2006, 79, 130–137. [Google Scholar] [CrossRef]
- Andres-Lacueva, C.; Shukitt-Hale, B.; Galli, R.L.; Jauregui, O.; Lamuela-Raventos, R.M.; Joseph, J.A. Anthocyanins in aged blueberry-fed rats are found centrally and may enhance memory. Nutr. Neurosci. 2005, 8, 111–120. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, F.; Wang, W.; Sang, J.; Jia, L.; Li, L.; Lu, F. Cyanidin-3-O-glucoside inhibits Abeta40 fibrillogenesis, disintegrates preformed fibrils, and reduces amyloid cytotoxicity. Food Funct. 2020, 11, 2573–2587. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.; Zhang, W.; Ding, Y.; Zhao, T.; Zhang, M.; Mao, G.; Feng, W.; Wu, X.; Yang, L. Bioaccessibility and biotransformation of anthocyanin monomers following in vitro simulated gastric-intestinal digestion and in vivo metabolism in rats. Food Funct. 2019, 10, 6052–6061. [Google Scholar] [CrossRef]
- Walti, M.A.; Ravotti, F.; Arai, H.; Glabe, C.G.; Wall, J.S.; Bockmann, A.; Guntert, P.; Meier, B.H.; Riek, R. Atomic-resolution structure of a disease-relevant Abeta(1–42) amyloid fibril. Proc. Natl. Acad. Sci. USA 2016, 113, E4976–E4984. [Google Scholar] [CrossRef]
- Gao, J.; Yu, P.; Liang, H.; Fu, J.; Luo, Z.; Yang, D. The wPDI Redox Cycle Coupled Conformational Change of the Repetitive Domain of the HMW-GS 1Dx5-A Computational Study. Molecules 2020, 25, 4393. [Google Scholar] [CrossRef]
- Yu, P.; Zhou, F.; Yang, D. Curdlan conformation change during its hydrolysis by multi-domain beta-1,3-glucanases. Food Chem. 2019, 287, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.Y.; Kang, S.S.; Lee, S.K.; Han, B.H. Polyphenolic Biflavonoids Inhibit Amyloid-Beta Fibrillation and Disaggregate Preformed Amyloid-Beta Fibrils. Biomol. Ther. 2020, 28, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Baumketner, A.; Bernstein, S.L.; Wyttenbach, T.; Bitan, G.; Teplow, D.B.; Bowers, M.T.; Shea, J.E. Amyloid β-protein monomer structure: A computational and experimental study. J. Protein Sci. 2006, 15, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Lazo, N.D.; Grant, M.A.; Condron, M.C.; Rigby, A.C.; Teplow, D.B. On the nucleation of amyloid β-protein monomer folding. J. Protein Sci. 2005, 14, 1581–1596. [Google Scholar] [CrossRef] [PubMed]


| Aβ Polymorph | CDOCKER Interaction Energy (kJ/mol) | Number of Hydrogen Bonds | Number of Amino Acid Residues Involved in Hydrophobic Interactions |
|---|---|---|---|
| Single peptide | −43.29 | 4 | 11 |
| Fibril site1 | −181.31 | 4 | 9 |
| Fibril site2 | −197.72 | 4 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Fu, J.; Gao, X.; Yang, D. Molecular Mechanism of Cyanidin-3-O-Glucoside Disassembling Aβ Fibril In Silico. Nutrients 2023, 15, 109. https://doi.org/10.3390/nu15010109
Gao J, Fu J, Gao X, Yang D. Molecular Mechanism of Cyanidin-3-O-Glucoside Disassembling Aβ Fibril In Silico. Nutrients. 2023; 15(1):109. https://doi.org/10.3390/nu15010109
Chicago/Turabian StyleGao, Jihui, Jiahui Fu, Xiaoyu Gao, and Dong Yang. 2023. "Molecular Mechanism of Cyanidin-3-O-Glucoside Disassembling Aβ Fibril In Silico" Nutrients 15, no. 1: 109. https://doi.org/10.3390/nu15010109
APA StyleGao, J., Fu, J., Gao, X., & Yang, D. (2023). Molecular Mechanism of Cyanidin-3-O-Glucoside Disassembling Aβ Fibril In Silico. Nutrients, 15(1), 109. https://doi.org/10.3390/nu15010109






