A Structure—Activity Relationship Study of the Inhibition of α-Amylase by Benzoic Acid and Its Derivatives
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Assessment of the In Vitro Inhibition of α-Amylase
2.3. Molecular Docking
2.4. Statistical Analysis
3. Results and Discussion
3.1. In Vitro Inhibitory Activity of Phenolic Acids towards α-Amylase
3.2. Effect of Hydroxylation on the Inhibition of α-Amylase
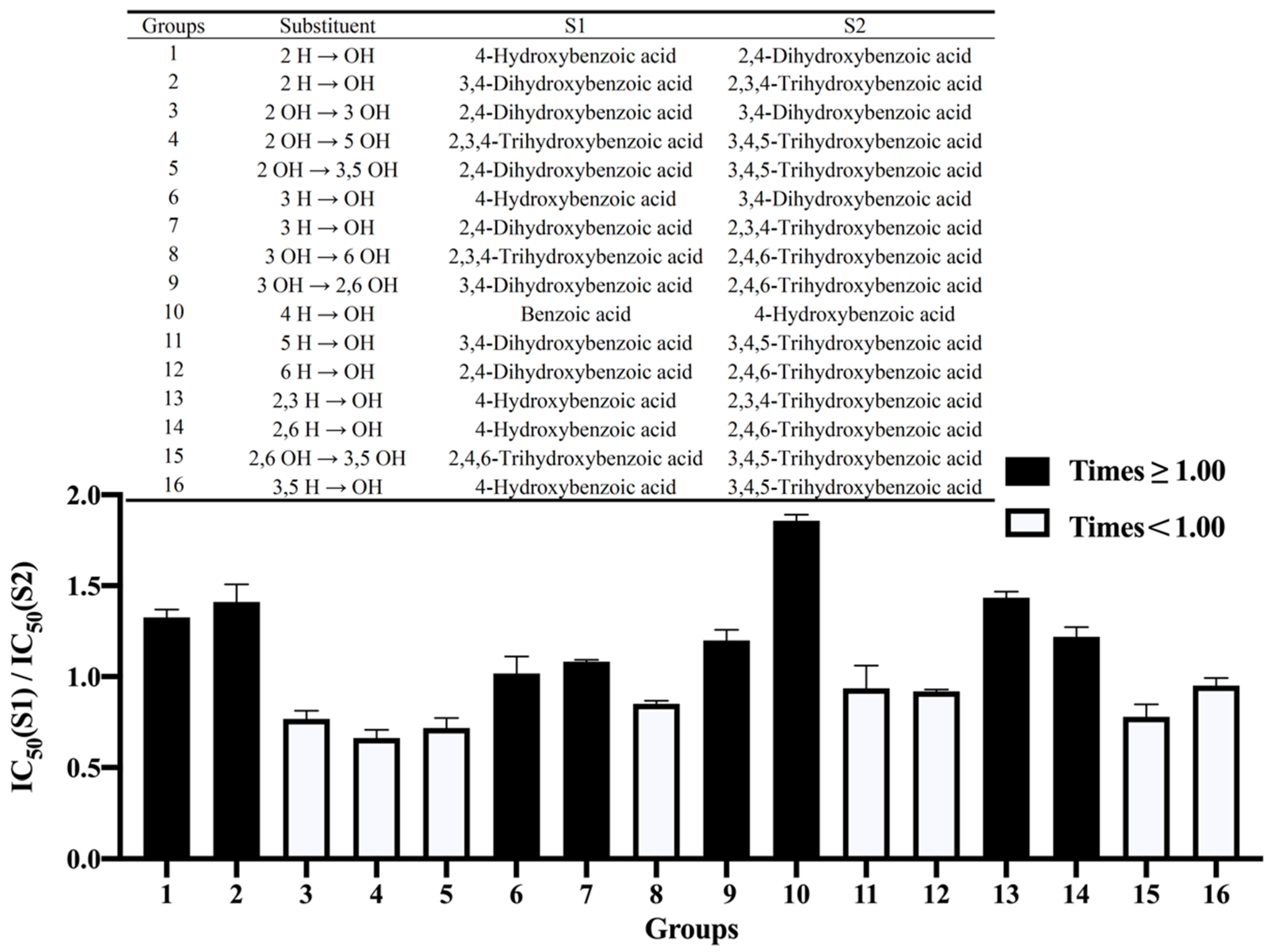
3.2.1. The Number of Hydroxyl Groups
3.2.2. The Position of the Hydroxyl Groups
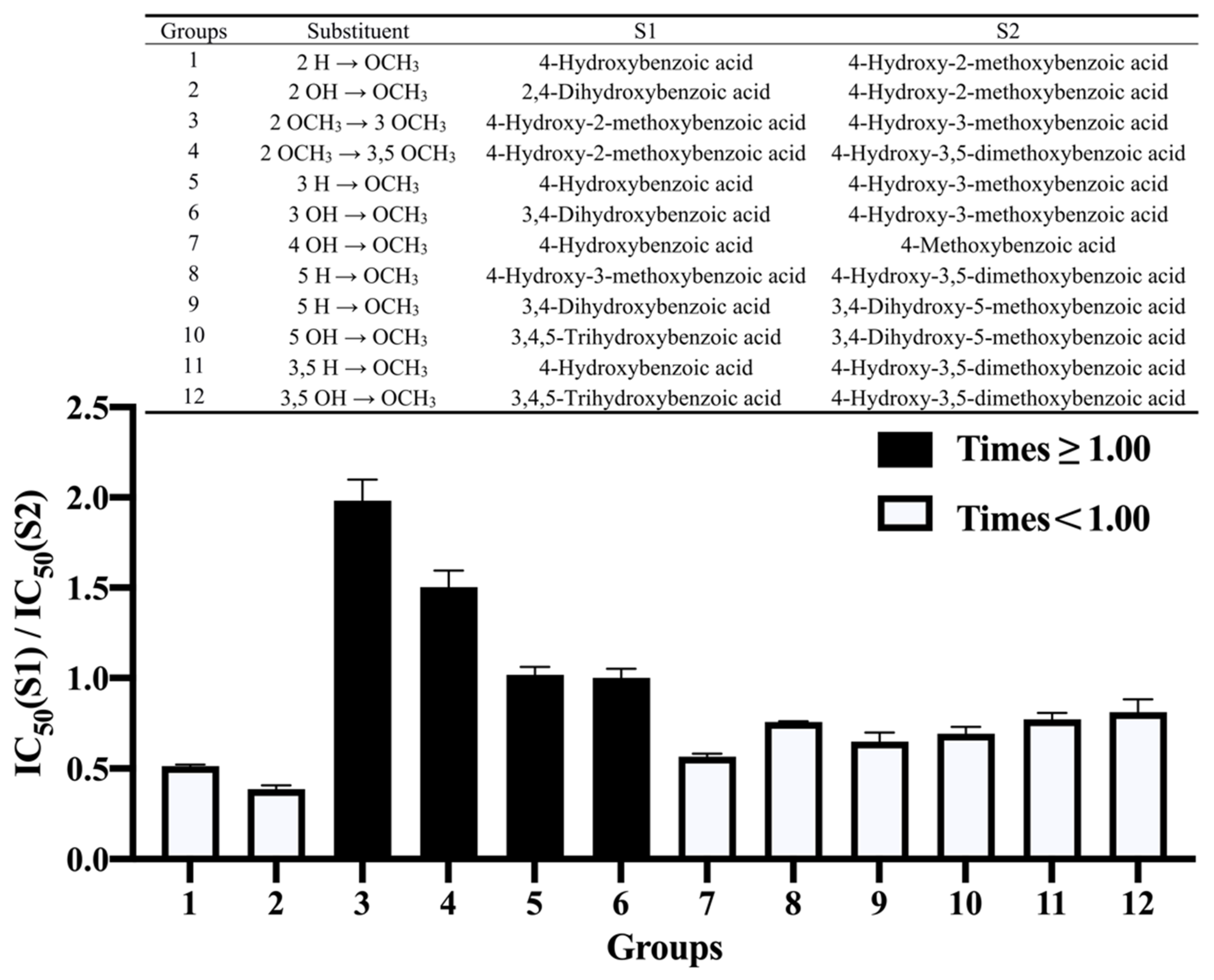
3.3. Effect of Methoxylation
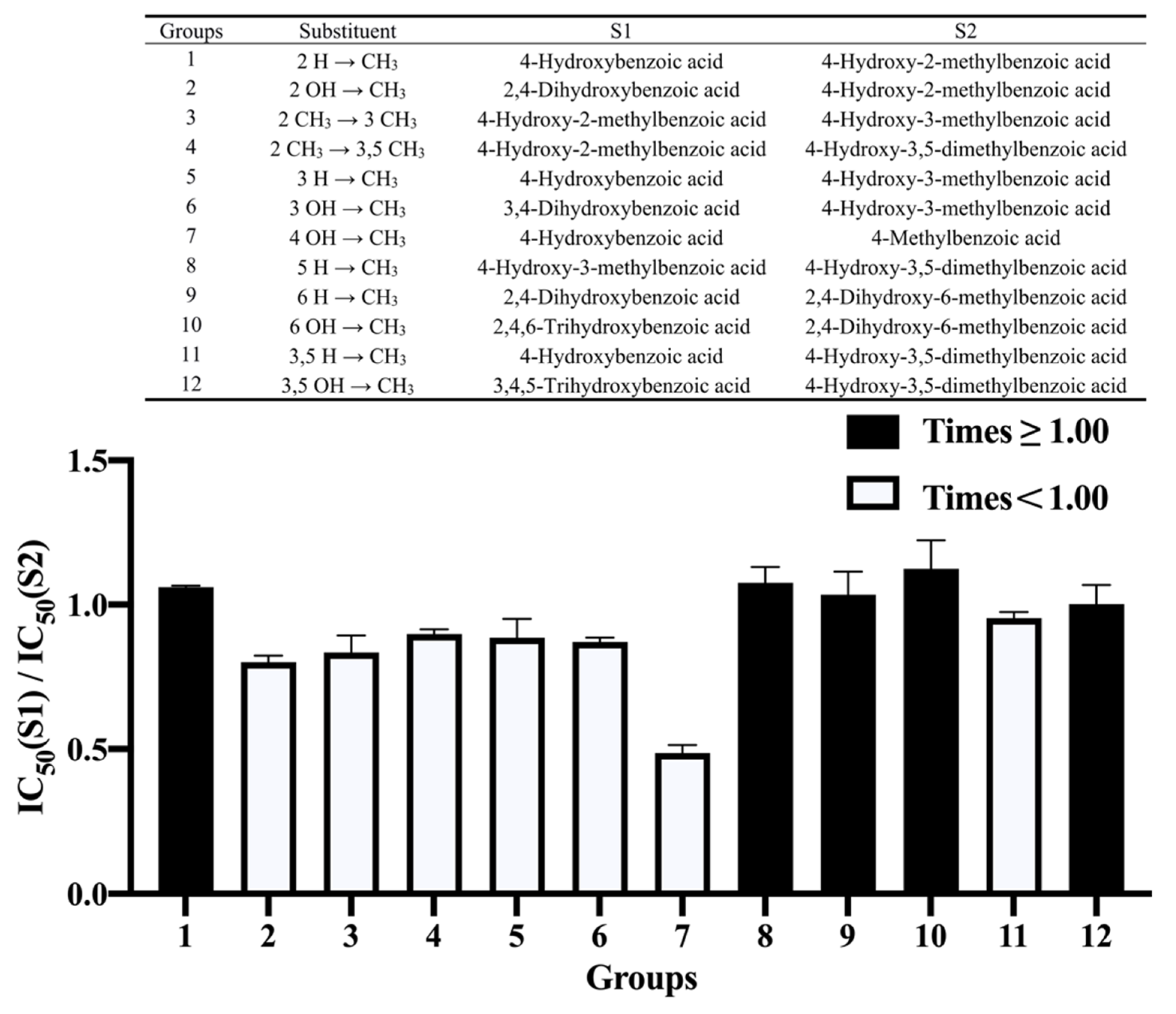
3.4. Effect of Methylation
3.5. Relationship between the Properties of the Phenolic Acids and Their IC50 Values
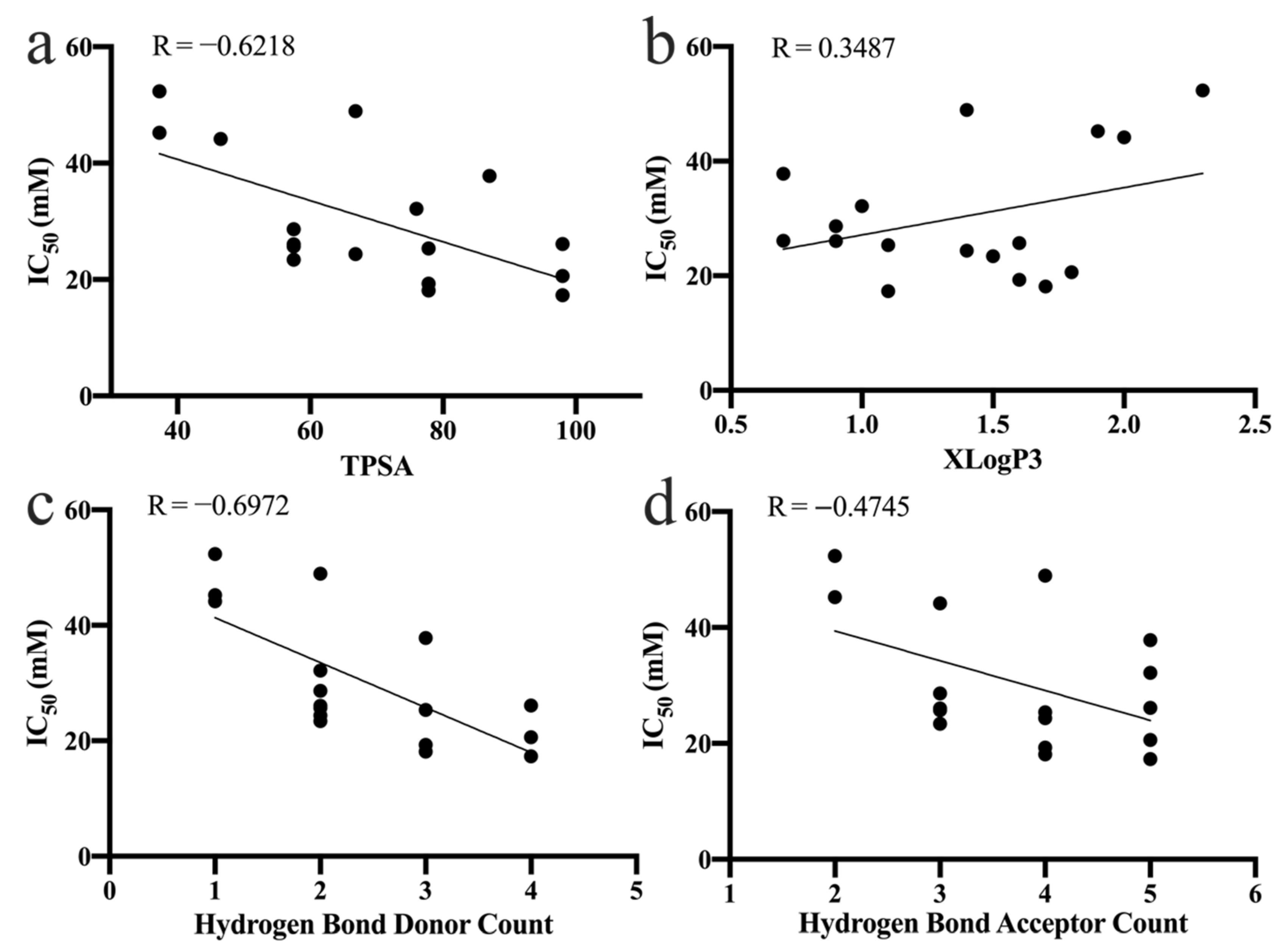
3.5.1. Topological Polar Surface Area
3.5.2. Partition Coefficient
3.5.3. Number of Hydrogen Bond Donors and Acceptors
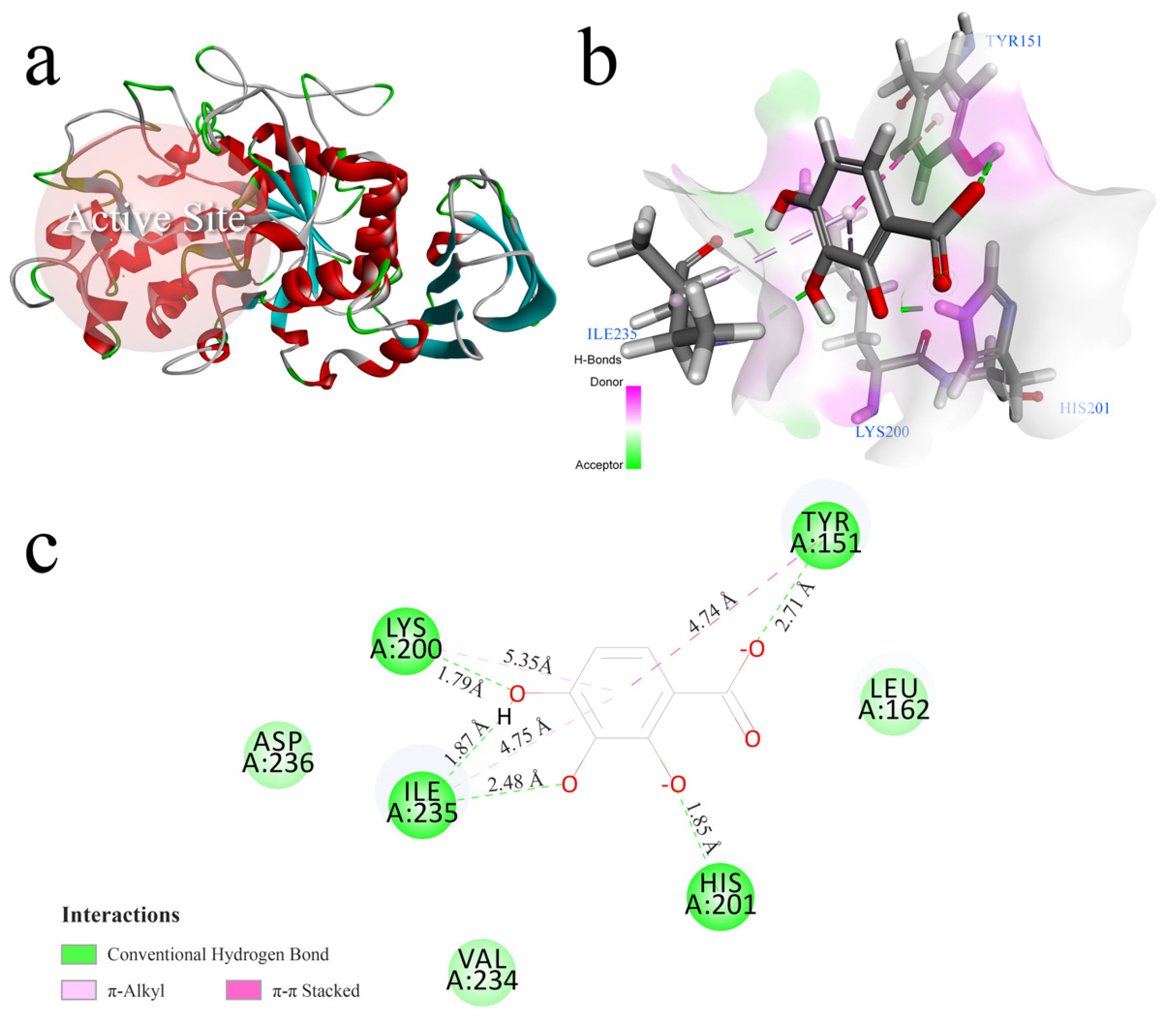
3.6. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Kaur, N.; Kumar, V.; Nayak, S.K.; Wadhwa, P.; Kaur, P.; Sahu, S.K. Alpha-amylase as Molecular Target for Treatment of Diabetes Mellitus: A Comprehensive Review. Chem. Biol. Drug. Des. 2021, 98, 539–560. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, M. The Pathobiology of Diabetic Complications: A Unifying Mechanism. Diabetes 2005, 54, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Afkarian, M.; Zelnick, L.R.; Hall, Y.N.; Heagerty, P.J.; Tuttle, K.; Weiss, N.S.; De Boer, I.H. Clinical Manifestations of Kidney Disease among US Adults with Diabetes, 1988–2014. JAMA 2016, 316, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Gregg, E.W.; Li, Y.; Wang, J.; Rios Burrows, N.; Ali, M.K.; Rolka, D.; Williams, D.E.; Geiss, L. Changes in Diabetes-Related Complications in the United States, 1990–2010. N. Engl. J. Med. 2014, 370, 1514–1523. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kishore, L.; Kaur, N. Diabetic Peripheral Neuropathy: Current Perspective and Future Directions. Pharmacol. Res. 2014, 80, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Han, K.; Choi, K.S.; Ko, S.-H.; Rhee, E.; Park, C.-Y.; Park, J.-Y.; Lee, K.-U.; Ko, K.-S. Trends in Diabetic Retinopathy and Related Medical Practices among Type 2 Diabetes Patients: Results from the National Insurance Service Survey 2006–2013. J. Diabetes Investig. 2018, 9, 173–178. [Google Scholar] [CrossRef]
- Danaei, G.; Finucane, M.M.; Lu, Y.; Singh, G.M.; Cowan, M.J.; Paciorek, C.J.; Lin, J.K.; Farzadfar, F.; Khang, Y.-H.; Stevens, G.A. National, Regional, and Global Trends in Fasting Plasma Glucose and Diabetes Prevalence since 1980: Systematic Analysis of Health Examination Surveys and Epidemiological Studies with 370 Country-Years and 2·7 Million Participants. Lancet 2011, 378, 31–40. [Google Scholar] [CrossRef]
- Dona, A.C.; Pages, G.; Gilbert, R.G.; Kuchel, P.W. Digestion of Starch: In Vivo and in Vitro Kinetic Models Used to Characterise Oligosaccharide or Glucose Release. Carbohydr. Polym. 2010, 80, 599–617. [Google Scholar] [CrossRef]
- Xiao, J.; Ni, X.; Kai, G.; Chen, X. A Review on Structure–Activity Relationship of Dietary Polyphenols Inhibiting α-Amylase. Crit. Rev. Food Sci. Nutr. 2013, 53, 497–506. [Google Scholar] [CrossRef]
- Gulati, V.; Harding, I.H.; Palombo, E.A. Enzyme Inhibitory and Antioxidant Activities of Traditional Medicinal Plants: Potential Application in the Management of Hyperglycemia. BMC Complement. Altern. Med. 2012, 12, 77. [Google Scholar] [CrossRef]
- Middleton, E.; Kandaswami, C.; Theoharides, T.C. The Effects of Plant Flavonoids on Mammalian Cells: Implications for Inflammation, Heart Disease, and Cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Sun, L.; Warren, F.J.; Gidley, M.J. Natural Products for Glycaemic Control: Polyphenols as Inhibitors of Alpha-Amylase. Trends Food Sci. Technol. 2019, 91, 262–273. [Google Scholar] [CrossRef]
- Zaharudin, N.; Salmeán, A.A.; Dragsted, L.O. Inhibitory Effects of Edible Seaweeds, Polyphenolics and Alginates on the Activities of Porcine Pancreatic α-Amylase. Food Chem. 2018, 245, 1196–1203. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Chang, S.K.C.; Zhang, Y. Comparison of α-Amylase, α-Glucosidase and Lipase Inhibitory Activity of the Phenolic Substances in Two Black Legumes of Different Genera. Food Chem. 2017, 214, 259–268. [Google Scholar] [CrossRef]
- Phan, M.A.T.; Wang, J.; Tang, J.; Lee, Y.Z.; Ng, K. Evaluation of α-Glucosidase Inhibition Potential of Some Flavonoids from Epimedium Brevicornum. LWT Food Sci. Technol. 2013, 53, 492–498. [Google Scholar] [CrossRef]
- Vieth, M.; Hirst, J.D.; Kolinski, A.; Brooks, C.L., III. Assessing Energy Functions for Flexible Docking. J. Comput. Chem. 1998, 19, 1612–1622. [Google Scholar] [CrossRef]
- Vieth, M.; Hirst, J.D.; Dominy, B.N.; Daigler, H.; Brooks, C.L., III. Assessing Search Strategies for Flexible Docking. J. Comput. Chem. 1998, 19, 1623–1631. [Google Scholar] [CrossRef]
- Miao, M.; Jiang, B.; Jiang, H.; Zhang, T.; Li, X. Interaction Mechanism between Green Tea Extract and Human α-Amylase for Reducing Starch Digestion. Food Chem. 2015, 186, 20–25. [Google Scholar] [CrossRef]
- Gaillard, T. Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J. Chem. Inf. Model. 2018, 58, 1697–1706. [Google Scholar] [CrossRef]
- Nielsen, J.E.; Borchert, T.V.; Vriend, G. The Determinants of α-Amylase PH–Activity Profiles. Protein Eng. Des. Sel. 2001, 14, 505–512. [Google Scholar] [CrossRef][Green Version]
- Mishra, P.J.; Ragunath, C.; Ramasubbu, N. The Mechanism of Salivary Amylase Hydrolysis: Role of Residues at Subsite S2′. Biochem. Biophys. Res. Commun. 2002, 292, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Haser, R.; Payan, F. Structure and Molecular Model Refinement of Pig Pancreatic α-Amylase at 2·1 Å Resolution. J. Mol. Biol. 1993, 231, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yang, W.; Sun, W.; Chen, S.; Liu, D.; Kong, X.; Tian, J.; Ye, X. Inhibition of Porcine Pancreatic α-Amylase Activity by Chlorogenic Acid. J. Funct. Foods 2020, 64, 103587. [Google Scholar] [CrossRef]
- Dunn, G.E.; Kung, F.-L. Effect of Intramolecular Hydrogen Bonding on Ionization Constants of Substituted Salicylic Acids. Can. J. Chem. 1966, 44, 1261–1269. [Google Scholar] [CrossRef]
- Mosher, M. Organic Chemistry. (Morrison, Robert Thornton; Boyd, Robert Neilson). ACS Publ. 1992, 69, A305. [Google Scholar]
- Sun, L.; Wang, Y.; Miao, M. Inhibition of α-Amylase by Polyphenolic Compounds: Substrate Digestion, Binding Interactions and Nutritional Intervention. Trends Food Sci. Technol. 2020, 104, 190–207. [Google Scholar] [CrossRef]
- Lo Piparo, E.; Scheib, H.; Frei, N.; Williamson, G.; Grigorov, M.; Chou, C.J. Flavonoids for Controlling Starch Digestion: Structural Requirements for Inhibiting Human α-Amylase. J. Med. Chem. 2008, 51, 3555–3561. [Google Scholar] [CrossRef]
- Miao, M.; Jiang, H.; Jiang, B.; Li, Y.; Cui, S.W.; Zhang, T. Structure Elucidation of Catechins for Modulation of Starch Digestion. LWT-Food Sci. Technol. 2014, 57, 188–193. [Google Scholar] [CrossRef]
- Kawamura-Konishi, Y.; Watanabe, N.; Saito, M.; Nakajima, N.; Sakaki, T.; Katayama, T.; Enomoto, T. Isolation of a New Phlorotannin, a Potent Inhibitor of Carbohydrate-Hydrolyzing Enzymes, from the Brown Alga Sargassum Patens. J. Agric. Food Chem. 2012, 60, 5565–5570. [Google Scholar] [CrossRef]
- Prasanna, S.; Doerksen, R.J. Topological Polar Surface Area: A Useful Descriptor in 2D-QSAR. Curr. Med. Chem. 2009, 16, 21–41. [Google Scholar] [CrossRef]
- Benoit, J.M.; Gilmour, C.C.; Mason, R. Aspects of Bioavailability of Mercury for Methylation in Pure Cultures of Desulfobulbus Propionicus (1pr3). Appl. Environ. Microbiol. 2001, 67, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Benoit, J.M.; Gilmour, C.C.; Mason, R.P. The Influence of Sulfide on Solid-Phase Mercury Bioavailability for Methylation by Pure Cultures of Desulfobulbus Propionicus (1pr3). Environ. Sci. Technol. 2001, 35, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Miao, M. Dietary Polyphenols Modulate Starch Digestion and Glycaemic Level: A Review. Crit. Rev. Food Sci. Nutr. 2020, 60, 541–555. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, E.A.; Janeček, Š.; Svensson, B. Relationship of Sequence and Structure to Specificity in the α-Amylase Family of Enzymes. Biochim. Biophys. Acta BBA-Protein Struct. Mol. Enzymol. 2001, 1546, 1–20. [Google Scholar] [CrossRef]
- Karonen, M.; Oraviita, M.; Mueller-Harvey, I.; Salminen, J.-P.; Green, R.J. Binding of an Oligomeric Ellagitannin Series to Bovine Serum Albumin (BSA): Analysis by Isothermal Titration Calorimetry (ITC). J. Agric. Food Chem. 2015, 63, 10647–10654. [Google Scholar] [CrossRef]
- Martinez-Gonzalez, A.I.; Díaz-Sánchez, Á.G.; de la Rosa, L.A.; Vargas-Requena, C.L.; Bustos-Jaimes, I. Polyphenolic Compounds and Digestive Enzymes: In Vitro Non-Covalent Interactions. Molecules 2017, 22, 669. [Google Scholar] [CrossRef]
- Martinez-Gonzalez, A.I.; Díaz-Sánchez, Á.G.; de la Rosa, L.A.; Bustos-Jaimes, I.; Alvarez-Parrilla, E. Inhibition of α-Amylase by Flavonoids: Structure Activity Relationship (SAR). Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 206, 437–447. [Google Scholar] [CrossRef]
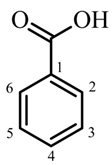
| Basic Structure | Compound No. | Substitution | IC50 Values against α-Amylase | |||
|---|---|---|---|---|---|---|
| OH | OCH3 | CH3 | mM | mg/mL | ||
 | 1 | 45.25 ± 2.15 | 5.525 ± 0.263 | |||
| 2 | 4 | 25.72 ± 1.33 | 3.552 ± 0.184 | |||
| 3 | 2,4 | 19.29 ± 1.18 | 2.973 ± 0.182 | |||
| 4 | 3,4 | 25.36 ± 1.23 | 3.908 ± 0.190 | |||
| 5 | 2,3,4 | 17.30 ± 0.73 | 2.943 ± 0.124 | |||
| 6 | 2,4,6 | 20.62 ± 0.53 | 3.508 ± 0.090 | |||
| 7 | 3,4,5 | 26.12 ± 1.89 | 4.444 ± 0.332 | |||
| 8 | 4 | 52.35 ± 3.31 | 7.127 ± 0.450 | |||
| 9 | 4 | 2 | 23.40 ± 1.11 | 3.560 ± 0.169 | ||
| 10 | 4 | 3 | 28.65 ± 0.76 | 4.359 ± 0.116 | ||
| 11 | 4 | 3,5 | 26.05 ± 1.00 | 4.328 ± 0.167 | ||
| 12 | 2,4 | 6 | 18.12 ± 1.30 | 3.047 ± 0.219 | ||
| 13 | 4 | 44.15 ± 1.39 | 6.717 ± 0.212 | |||
| 14 | 4 | 2 | 48.95 ± 2.51 | 8.230 ± 0.422 | ||
| 15 | 4 | 3 | 24.38 ± 0.71 | 4.099 ± 0.119 | ||
| 16 | 3,4 | 5 | 37.82 ± 1.59 | 6.965 ± 0.292 | ||
| 17 | 4 | 3,5 | 32.16 ± 0.88 | 6.372 ± 0.175 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, L.; Long, H.; Ren, F.; Li, Y.; Zhang, H. A Structure—Activity Relationship Study of the Inhibition of α-Amylase by Benzoic Acid and Its Derivatives. Nutrients 2022, 14, 1931. https://doi.org/10.3390/nu14091931
Guan L, Long H, Ren F, Li Y, Zhang H. A Structure—Activity Relationship Study of the Inhibition of α-Amylase by Benzoic Acid and Its Derivatives. Nutrients. 2022; 14(9):1931. https://doi.org/10.3390/nu14091931
Chicago/Turabian StyleGuan, Lei, Haoyuan Long, Fazheng Ren, Yixuan Li, and Hao Zhang. 2022. "A Structure—Activity Relationship Study of the Inhibition of α-Amylase by Benzoic Acid and Its Derivatives" Nutrients 14, no. 9: 1931. https://doi.org/10.3390/nu14091931
APA StyleGuan, L., Long, H., Ren, F., Li, Y., & Zhang, H. (2022). A Structure—Activity Relationship Study of the Inhibition of α-Amylase by Benzoic Acid and Its Derivatives. Nutrients, 14(9), 1931. https://doi.org/10.3390/nu14091931






