Critical Role of Maternal Selenium Nutrition in Neurodevelopment: Effects on Offspring Behavior and Neuroinflammatory Profile
Abstract
:1. Introduction
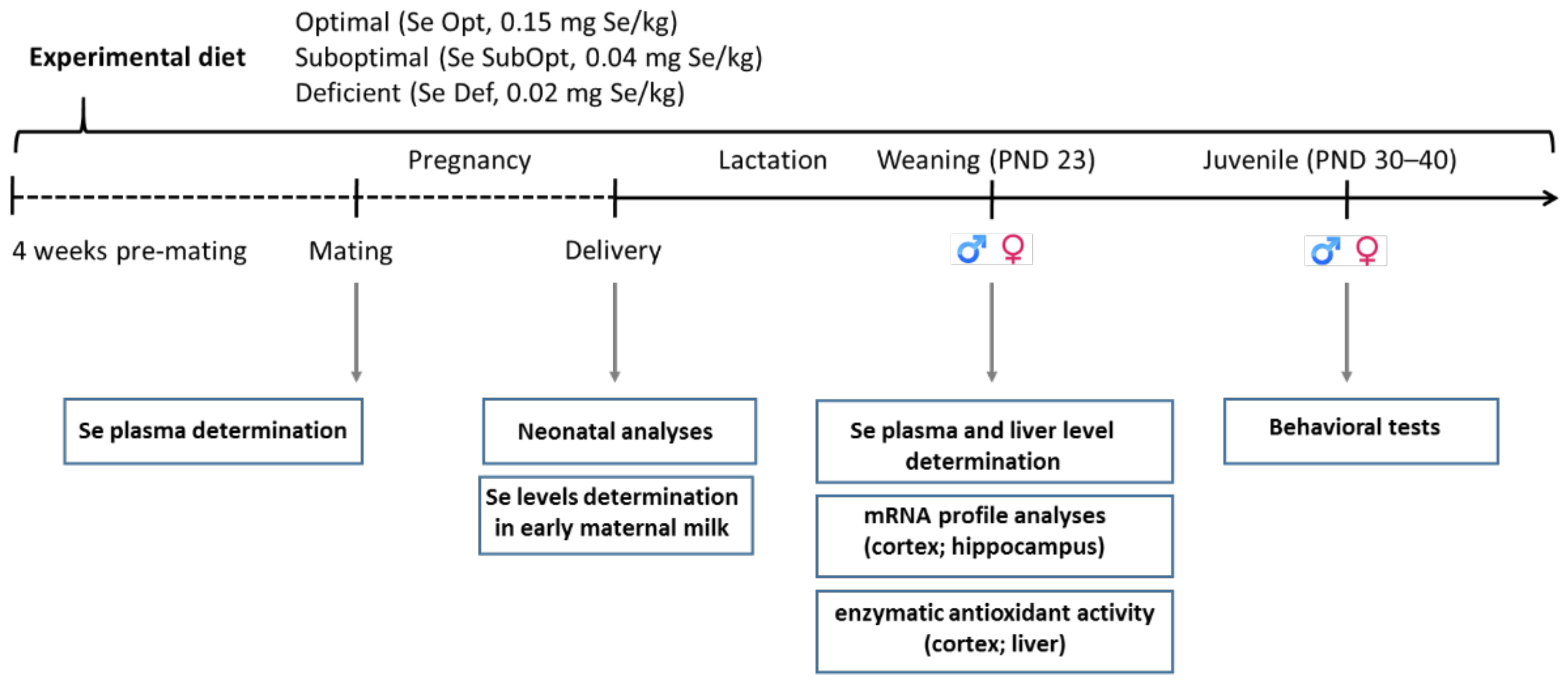
2. Materials and Methods
2.1. Animals
2.2. Diets
2.3. Plasma and Milk Sampling
2.4. Brain and Liver Sampling
2.5. Selenium Determination
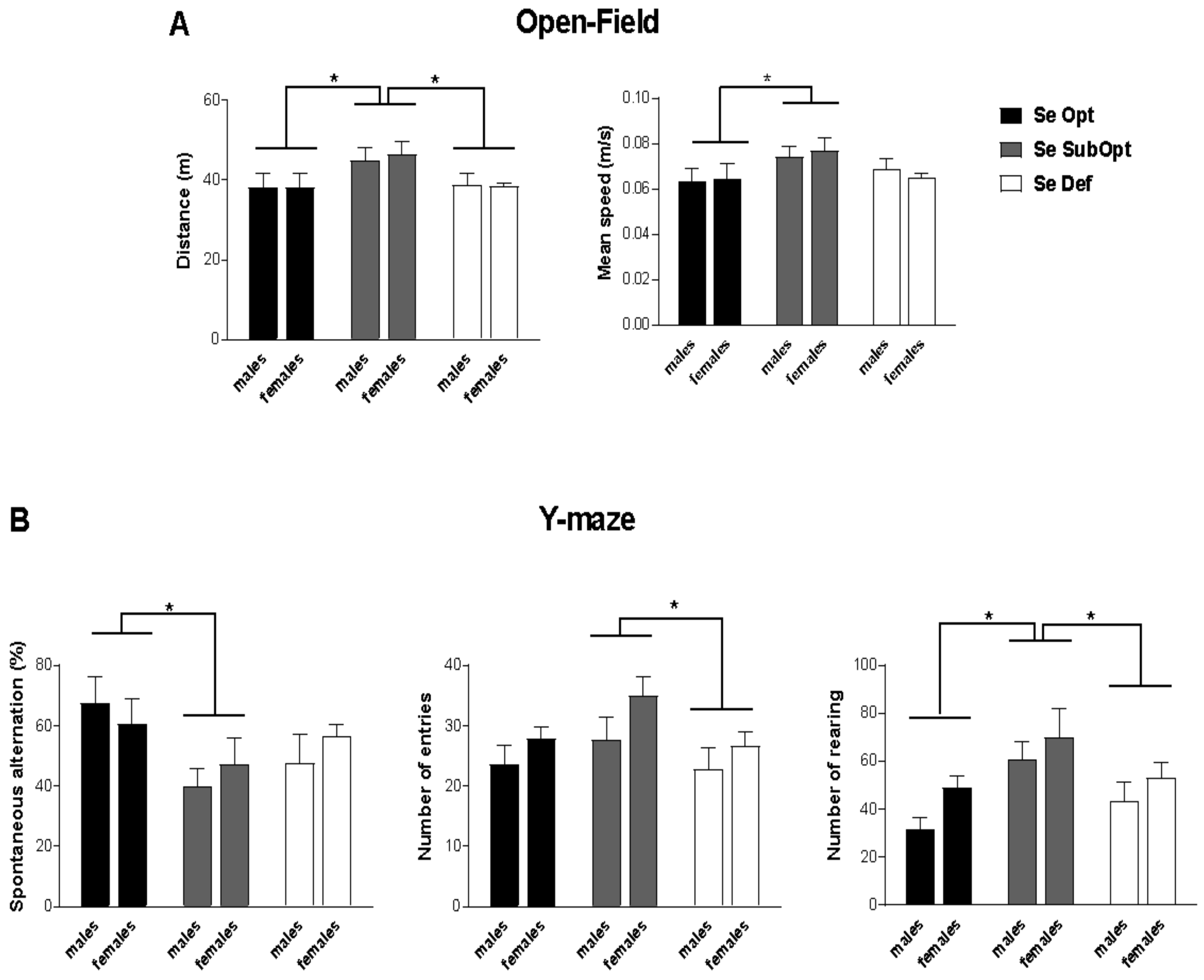
2.6. Behavioral Testing
2.6.1. Open-Field (OF)
2.6.2. Y-Maze
2.6.3. Light/Dark Test
2.6.4. Rotarod Test
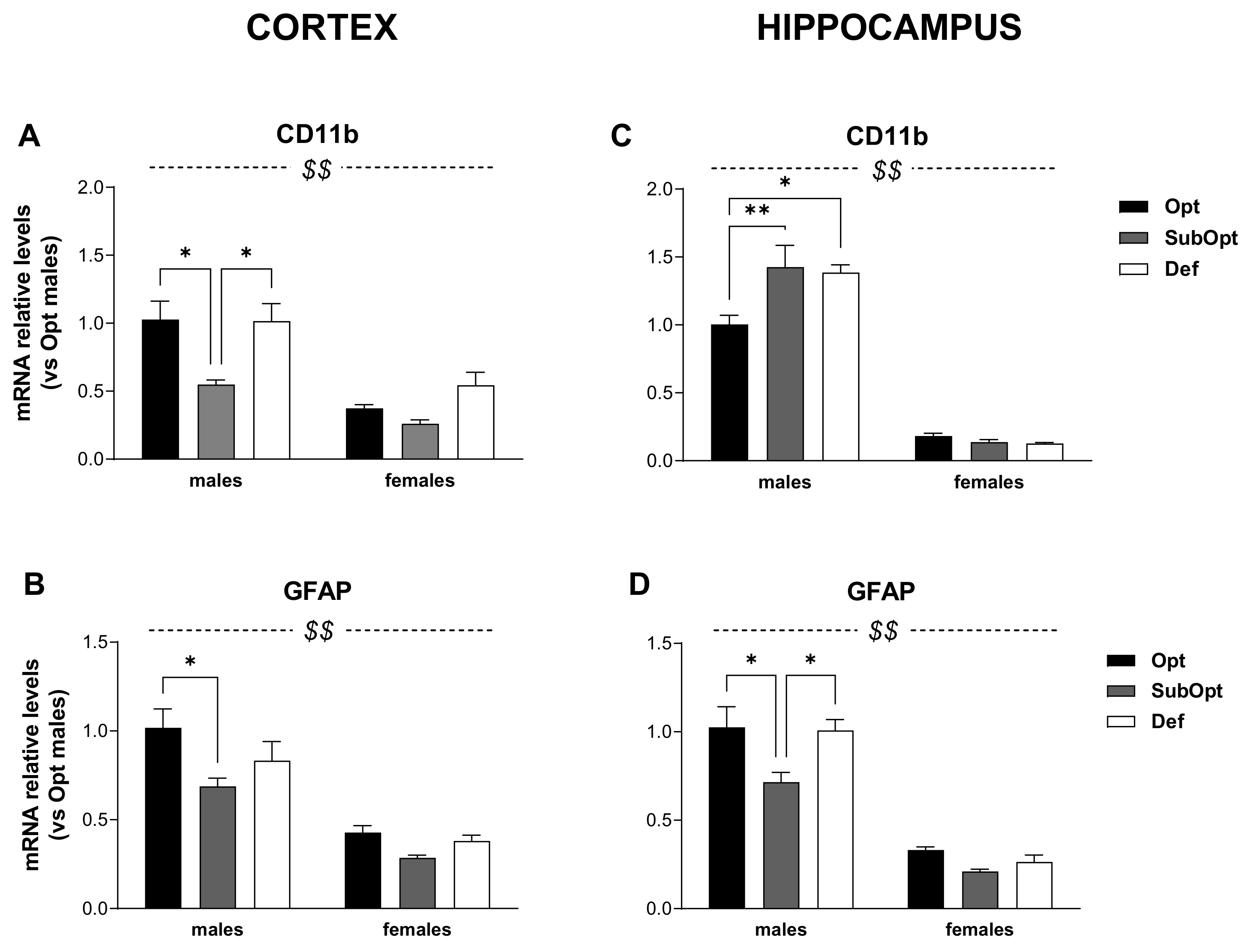
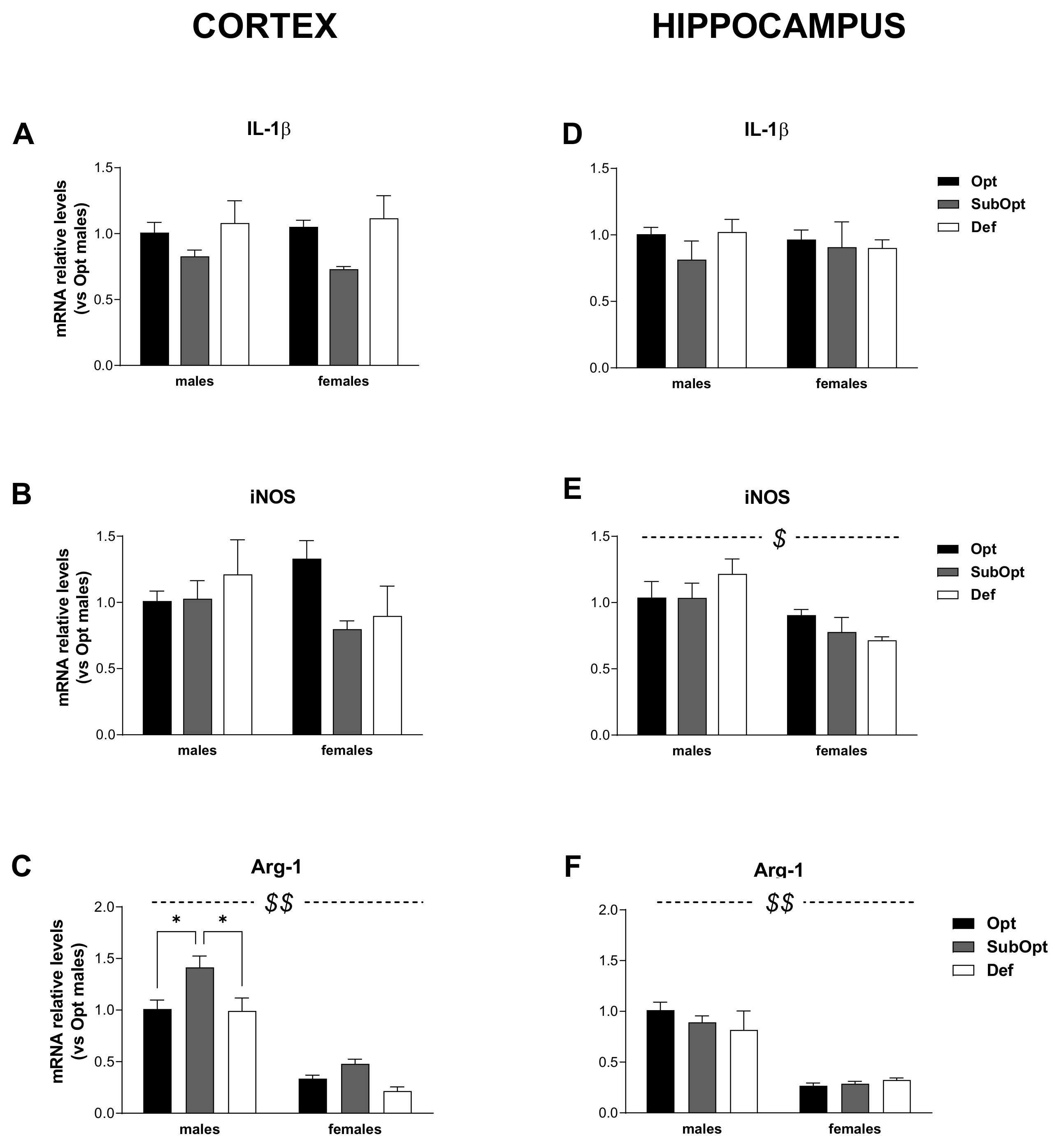
2.7. Real-Time Quantitative Polymerase Chain Reaction (RT-PCR)
2.8. Glutathione Peroxidase Enzymatic Activity
2.9. Statistical Analyses
3. Results
3.1. Selenium Levels in Dams and Offspring
3.2. Effect of Diet on Reproductive Performance and Pup Somatic Growth
3.3. Effects of Maternal Se Intake on Behavioral Responses in Juvenile Offspring
3.4. Effects of Maternal Se Intake on Brain Immune Profile in Juvenile Rats
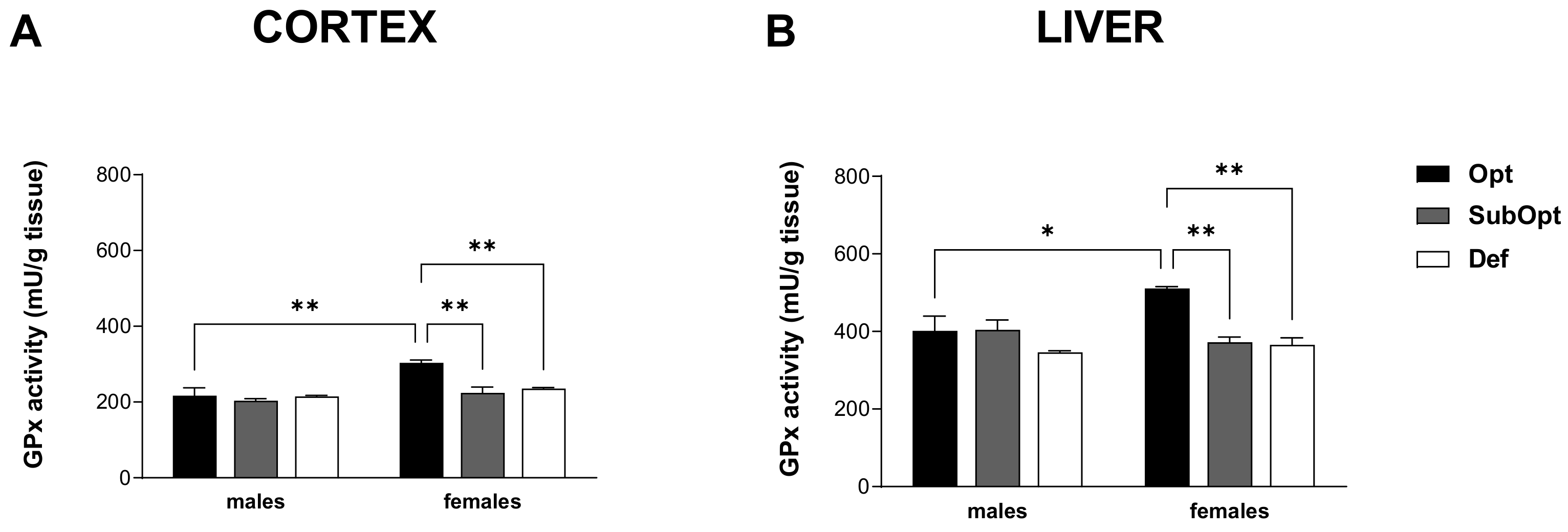
3.5. Effects of Maternal Se Intake on GPx Activity in Brain Cortex and Liver of Juvenile Rats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barker, D.J.; Fall, C.H. Fetal and infant origins of cardiovascular disease. Arch. Dis. Child. 1993, 68, 797–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, D.J. Intrauterine programming of adult disease. Mol. Med. Today 1995, 1, 418–423. [Google Scholar] [CrossRef]
- Hibbeln, J.R.; Davis, J.M.; Steer, C.; Emmett, P.; Rogers, I.; Williams, C.; Golding, J. Maternal seafood consumption in pregnancy and neurodevelopmental outcomes in childhood (ALSPAC study): An observational cohort study. Lancet 2007, 369, 578–585. [Google Scholar] [CrossRef]
- Cusick, S.E.; Georgieff, M.K. The Role of Nutrition in Brain Development: The Golden Opportunity of the “First 1000 Days”. J. Pediatr. 2016, 175, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Krebs, N.F.; Lozoff, B.; Georgieff, M.K. Neurodevelopment: The Impact of Nutrition and Inflammation during Infancy in Low-Resource Settings. Pediatrics 2017, 139, S50–S58. [Google Scholar] [CrossRef] [Green Version]
- Georgieff, M.K. Nutrition and the developing brain: Nutrient priorities and measurement. Am. J. Clin. Nutr. 2007, 85, 614S–620S. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [Green Version]
- Solovyev, N.D. Importance of selenium and selenoprotein for brain function: From antioxidant protection to neuronal signaling. J. Inorg. Biochem. 2015, 153, 1–12. [Google Scholar] [CrossRef]
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef] [Green Version]
- Pitts, M.W.; Byrns, C.N.; Ogawa-Wong, A.N.; Kremer, P.; Berry, M.J. Selenoproteins in nervous system development and function. Biol. Trace Elem. Res. 2014, 161, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Burk, R.F.; Hill, K.E.; Motley, A.K.; Winfrey, V.P.; Kurokawa, S.; Mitchell, S.L.; Zhang, W. Selenoprotein P and apolipoprotein E receptor-2 interact at the blood-brain barrier and also within the brain to maintain an essential selenium pool that protects against neurodegeneration. FASEB J. 2014, 28, 3579–3588. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, B.R.; Roberts, B.R.; Bush, A.I.; Hare, D.J. Selenium, selenoproteins and neurodegenerative diseases. Metallomics 2015, 7, 1213–1228. [Google Scholar] [CrossRef] [Green Version]
- Seiler, A.; Schneider, M.; Forster, H.; Roth, S.; Wirth, E.K.; Culmsee, C.; Plesnila, N.; Kremmer, E.; Radmark, O.; Wurst, W.; et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008, 8, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Hill, K.E.; Zhou, J.; McMahan, W.J.; Motley, A.K.; Burk, R.F. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J. Nutr. 2004, 134, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Soerensen, J.; Jakupoglu, C.; Beck, H.; Forster, H.; Schmidt, J.; Schmahl, W.; Schweizer, U.; Conrad, M.; Brielmeier, M. The role of thioredoxin reductases in brain development. PLoS ONE 2008, 3, e1813. [Google Scholar] [CrossRef] [Green Version]
- Wirth, E.K.; Conrad, M.; Winterer, J.; Wozny, C.; Carlson, B.A.; Roth, S.; Schmitz, D.; Bornkamm, G.W.; Coppola, V.; Tessarollo, L.; et al. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 2010, 24, 844–852. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on Dietary Reference Values for selenium. EFSA J. 2014, 12, 3846. [Google Scholar] [CrossRef]
- Fairweather-Tait, S.J.; Bao, Y.; Broadley, M.R.; Collings, R.; Ford, D.; Hesketh, J.E.; Hurst, R. Selenium in human health and disease. Antioxid. Redox Signal. 2011, 14, 1337–1383. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Ojeda, M.L.; Nogales, F.; Romero-Herrera, I.; Carreras, O. Fetal Programming Is Deeply Related to Maternal Selenium Status and Oxidative Balance; Experimental Offspring Health Repercussions. Nutrients 2021, 13, 2085. [Google Scholar] [CrossRef]
- Skroder, H.M.; Hamadani, J.D.; Tofail, F.; Persson, L.A.; Vahter, M.E.; Kippler, M.J. Selenium status in pregnancy influences children’s cognitive function at 1.5 years of age. Clin. Nutr. 2015, 34, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Polanska, K.; Krol, A.; Sobala, W.; Gromadzinska, J.; Brodzka, R.; Calamandrei, G.; Chiarotti, F.; Wasowicz, W.; Hanke, W. Selenium status during pregnancy and child psychomotor development-Polish Mother and Child Cohort study. Pediatr. Res. 2016, 79, 863–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, L.; Nogales, F.; Murillo, L.; Carreras, O. The role of folic acid and selenium against oxidative damage from ethanol in early life programming: A review. Biochem. Cell Biol. 2018, 96, 178–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappas, A.C.; Zoidis, E.; Chadio, S.E. Maternal Selenium and Developmental Programming. Antioxidants 2019, 8, 145. [Google Scholar] [CrossRef] [Green Version]
- Sunde, R.A.; Evenson, J.K.; Thompson, K.M.; Sachdev, S.W. Dietary selenium requirements based on glutathione peroxidase-1 activity and mRNA levels and other Se-dependent parameters are not increased by pregnancy and lactation in rats. J. Nutr. 2005, 135, 2144–2150. [Google Scholar] [CrossRef] [Green Version]
- Tartaglione, A.M.; Serafini, M.M.; Raggi, A.; Iacoponi, F.; Zianni, E.; Scalfari, A.; Minghetti, L.; Ricceri, L.; Cubadda, F.; Calamandrei, G.; et al. Sex-Dependent Effects of Developmental Lead Exposure in Wistar Rats: Evidence from Behavioral and Molecular Correlates. Int. J. Mol. Sci. 2020, 21, 2664. [Google Scholar] [CrossRef] [Green Version]
- Gasparin, F.R.S.; Carreno, F.O.; Mewes, J.M.; Gilglioni, E.H.; Pagadigorria, C.L.S.; Natali, M.R.M.; Utsunomiya, K.S.; Constantin, R.P.; Ouchida, A.T.; Curti, C.; et al. Sex differences in the development of hepatic steatosis in cafeteria diet-induced obesity in young mice. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2018, 1864, 2495–2509. [Google Scholar] [CrossRef]
- Yanguas-Casas, N. Physiological sex differences in microglia and their relevance in neurological disorders. Neuroimmunol. Neuroinflamm. 2020, 7, 13–22. [Google Scholar] [CrossRef]
- Saunders, A.; Macosko, E.Z.; Wysoker, A.; Goldman, M.; Krienen, F.M.; de Rivera, H.; Bien, E.; Baum, M.; Bortolin, L.; Wang, S.; et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 2018, 174, 1015–1030.e16. [Google Scholar] [CrossRef] [Green Version]
- Santos-Galindo, M.; Acaz-Fonseca, E.; Bellini, M.J.; Garcia-Segura, L.M. Sex differences in the inflammatory response of primary astrocytes to lipopolysaccharide. Biol. Sex Differ. 2011, 2, 7. [Google Scholar] [CrossRef] [Green Version]
- Villa, A.; Della Torre, S.; Maggi, A. Sexual differentiation of microglia. Front. Neuroendocrinol. 2019, 52, 156–164. [Google Scholar] [CrossRef]
- Smith, C.J.; Bilbo, S.D. Microglia Sculpt Sex Differences in Social Behavior. Neuron 2019, 102, 275–277. [Google Scholar] [CrossRef] [Green Version]
- Lenz, K.M.; Nelson, L.H. Microglia and Beyond: Innate Immune Cells as Regulators of Brain Development and Behavioral Function. Front. Immunol. 2018, 9, 698. [Google Scholar] [CrossRef] [Green Version]
- Guneykaya, D.; Ivanov, A.; Hernandez, D.P.; Haage, V.; Wojtas, B.; Meyer, N.; Maricos, M.; Jordan, P.; Buonfiglioli, A.; Gielniewski, B.; et al. Transcriptional and Translational Differences of Microglia from Male and Female Brains. Cell Rep. 2018, 24, 2773–2783.e6. [Google Scholar] [CrossRef] [Green Version]
- Bordt, E.A.; Ceasrine, A.M.; Bilbo, S.D. Microglia and sexual differentiation of the developing brain: A focus on ontogeny and intrinsic factors. Glia 2020, 68, 1085–1099. [Google Scholar] [CrossRef]
- Lu, C.W.; Chang, H.H.; Yang, K.C.; Chiang, C.H.; Yao, C.A.; Huang, K.C. Gender Differences with Dose–Response Relationship between Serum Selenium Levels and Metabolic Syndrome—A Case-Control Study. Nutrients 2019, 11, 477. [Google Scholar] [CrossRef] [Green Version]
- Schomburg, L.; Schweizer, U. Hierarchical regulation of selenoprotein expression and sex-specific effects of selenium. Biochim. Biophys. Acta BBA Gen. Subj. 2009, 1790, 1453–1462. [Google Scholar] [CrossRef]
- Byrns, C.N.; Pitts, M.W.; Gilman, C.A.; Hashimoto, A.C.; Berry, M.J. Mice lacking selenoprotein P and selenocysteine lyase exhibit severe neurological dysfunction, neurodegeneration, and audiogenic seizures. J. Biol. Chem. 2014, 289, 9662–9674. [Google Scholar] [CrossRef] [Green Version]
- Kilonzo, V.W.; Sasuclark, A.R.; Torres, D.J.; Coyle, C.; Pilat, J.M.; Williams, C.S.; Pitts, M.W. Juvenile Selenium Deficiency Impairs Cognition, Sensorimotor Gating, and Energy Homeostasis in Mice. Front. Nutr. 2021, 8, 667587. [Google Scholar] [CrossRef]
- Hofstee, P.; McKeating, D.R.; Bartho, L.A.; Anderson, S.T.; Perkins, A.V.; Cuffe, J.S.M. Maternal Selenium Deficiency in Mice Alters Offspring Glucose Metabolism and Thyroid Status in a Sexually Dimorphic Manner. Nutrients 2020, 12, 267. [Google Scholar] [CrossRef] [Green Version]
- Pitts, M.W.; Kremer, P.M.; Hashimoto, A.C.; Torres, D.J.; Byrns, C.N.; Williams, C.S.; Berry, M.J. Competition between the Brain and Testes under Selenium-Compromised Conditions: Insight into Sex Differences in Selenium Metabolism and Risk of Neurodevelopmental Disease. J. Neurosci. 2015, 35, 15326–15338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seale, L.A.; Ogawa-Wong, A.N.; Berry, M.J. Sexual Dimorphism in Selenium Metabolism and Selenoproteins. Free Radic. Biol. Med. 2018, 127, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Hofstee, P.; Cuffe, J.S.M.; Perkins, A.V. Analysis of Selenoprotein Expression in Response to Dietary Selenium Deficiency during Pregnancy Indicates Tissue Specific Differential Expression in Mothers and Sex Specific Changes in the Fetus and Offspring. Int. J. Mol. Sci. 2020, 21, 2210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, A.; Hill, K.E.; Austin, L.M.; Motley, A.K.; Burk, R.F. All regions of mouse brain are dependent on selenoprotein P for maintenance of selenium. J. Nutr. 2007, 137, 690–693. [Google Scholar] [CrossRef]
- Mattei, D.; Notter, T. Basic Concept of Microglia Biology and Neuroinflammation in Relation to Psychiatry. Curr. Top. Behav. Neurosci. 2020, 44, 9–34. [Google Scholar] [CrossRef]
- Pillai, R.; Uyehara-Lock, J.H.; Bellinger, F.P. Selenium and selenoprotein function in brain disorders. IUBMB Life 2014, 66, 229–239. [Google Scholar] [CrossRef]
- Kipp, A.; Banning, A.; van Schothorst, E.M.; Meplan, C.; Schomburg, L.; Evelo, C.; Coort, S.; Gaj, S.; Keijer, J.; Hesketh, J.; et al. Four selenoproteins, protein biosynthesis, and Wnt signalling are particularly sensitive to limited selenium intake in mouse colon. Mol. Nutr. Food Res. 2009, 53, 1561–1572. [Google Scholar] [CrossRef]
- Kipp, A.P.; Banning, A.; van Schothorst, E.M.; Meplan, C.; Coort, S.L.; Evelo, C.T.; Keijer, J.; Hesketh, J.; Brigelius-Flohe, R. Marginal selenium deficiency down-regulates inflammation-related genes in splenic leukocytes of the mouse. J. Nutr. Biochem. 2012, 23, 1170–1177. [Google Scholar] [CrossRef]
- Meplan, C.; Johnson, I.T.; Polley, A.C.; Cockell, S.; Bradburn, D.M.; Commane, D.M.; Arasaradnam, R.P.; Mulholland, F.; Zupanic, A.; Mathers, J.C.; et al. Transcriptomics and proteomics show that selenium affects inflammation, cytoskeleton, and cancer pathways in human rectal biopsies. FASEB J. 2016, 30, 2812–2825. [Google Scholar] [CrossRef] [Green Version]
- Muller, M.; Banning, A.; Brigelius-Flohe, R.; Kipp, A. Nrf2 target genes are induced under marginal selenium-deficiency. Genes Nutr. 2010, 5, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Paolicelli, R.C.; Gross, C.T. Microglia in development: Linking brain wiring to brain environment. Neuron Glia Biol. 2011, 7, 77–83. [Google Scholar] [CrossRef]
- Presumey, J.; Bialas, A.R.; Carroll, M.C. Complement System in Neural Synapse Elimination in Development and Disease. Adv. Immunol. 2017, 135, 53–79. [Google Scholar] [CrossRef]
- Wilhelmsson, U.; Pozo-Rodrigalvarez, A.; Kalm, M.; de Pablo, Y.; Widestrand, A.; Pekna, M.; Pekny, M. The role of GFAP and vimentin in learning and memory. Biol. Chem. 2019, 400, 1147–1156. [Google Scholar] [CrossRef]
- Dziegielewska, K.M.; Moller, J.E.; Potter, A.M.; Ek, J.; Lane, M.A.; Saunders, N.R. Acute-phase cytokines IL-1β and TNF-α in brain development. Cell Tissue Res. 2000, 299, 335–345. [Google Scholar] [CrossRef]
- Pozzi, D.; Menna, E.; Canzi, A.; Desiato, G.; Mantovani, C.; Matteoli, M. The Communication Between the Immune and Nervous Systems: The Role of IL-1β in Synaptopathies. Front. Mol. Neurosci. 2018, 11, 111. [Google Scholar] [CrossRef]
- Del Rey, A.; Balschun, D.; Wetzel, W.; Randolf, A.; Besedovsky, H.O. A cytokine network involving brain-borne IL-1β, IL-1ra, IL-18, IL-6, and TNFα operates during long-term potentiation and learning. Brain Behav. Immun. 2013, 33, 15–23. [Google Scholar] [CrossRef]
- Liu, X.; Quan, N. Microglia and CNS Interleukin-1: Beyond Immunological Concepts. Front. Neurol. 2018, 9, 8. [Google Scholar] [CrossRef] [Green Version]
- Krystofova, J.; Pathipati, P.; Russ, J.; Sheldon, A.; Ferriero, D. The Arginase Pathway in Neonatal Brain Hypoxia-Ischemia. Dev. Neurosci. 2018, 40, 437–450. [Google Scholar] [CrossRef]
- Barkley, R.A. Behavioral inhibition, sustained attention, and executive functions: Constructing a unifying theory of ADHD. Psychol. Bull. 1997, 121, 65–94. [Google Scholar] [CrossRef] [Green Version]
- Van der Kooij, M.A.; Glennon, J.C. Animal models concerning the role of dopamine in attention-deficit hyperactivity disorder. Neurosci. Biobehav. Rev. 2007, 31, 597–618. [Google Scholar] [CrossRef]
- Castano, A.; Ayala, A.; Rodriguez-Gomez, J.A.; Herrera, A.J.; Cano, J.; Machado, A. Low selenium diet increases the dopamine turnover in prefrontal cortex of the rat. Neurochem. Int. 1997, 30, 549–555. [Google Scholar] [CrossRef]
- Watanabe, C.; Satoh, H. Brain selenium status and behavioral development in selenium-deficient preweanling mice. Physiol. Behav. 1994, 56, 927–932. [Google Scholar] [CrossRef]
- Amoros, R.; Murcia, M.; Gonzalez, L.; Rebagliato, M.; Iniguez, C.; Lopez-Espinosa, M.J.; Vioque, J.; Broberg, K.; Ballester, F.; Llop, S. Maternal selenium status and neuropsychological development in Spanish preschool children. Environ. Res. 2018, 166, 215–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EFSA Scientific Committee. Statistical Significance and Biological Relevance. EFSA J. 2011, 9, 2372. [Google Scholar] [CrossRef]
| Plasma Se in Pre-Mating Females (mg/L) | Se in Early Milk (mg/L) | ||||
|---|---|---|---|---|---|
| GROUP | n | Median (IQR) a | n | Median (IQR) a | |
| Opt | (0.15 mg Se/kg) | 5 | 0.27 (0.02) | 6 | 0.16 (0.05) |
| SubOpt | (0.04 mg Se/kg) | 5 | 0.24 (0.04) | 5 | 0.05 (0.03) * |
| Def | (0.02 mg Se/kg) | 5 | 0.24 (0.08) | 11 | 0.05 (0.02) ** |
| GROUP | n | M/F a | Median M/F (mg/kg) | Median M + F (IQR) b (mg/kg) | |
|---|---|---|---|---|---|
| Opt | (0.15 mg Se/kg) | 8 | 5/3 | 3.69/3.57 | 3.63 (0.52) |
| SubOpt | (0.04 mg Se/kg) | 7 | 4/3 | 0.70/0.69 | 0.69 (0.11) |
| Def | (0.02 mg Se/kg) | 8 | 5/3 | 0.33/0.33 | 0.33 (0.06) ** |
| GROUP | n | Median (IQR) a (mg/L) | |
|---|---|---|---|
| Opt | (0.15 mg Se/kg) | 5 | 0.22 (0.04) |
| SubOpt | (0.04 mg Se/kg) | 5 | 0.09 (0.03) |
| Def | (0.02 mg Se/kg) | 5 | 0.04 (0.02) ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajmone-Cat, M.A.; De Simone, R.; Tartaglione, A.M.; Di Biase, A.; Di Benedetto, R.; D’Archivio, M.; Varì, R.; Ricceri, L.; Aureli, F.; Iacoponi, F.; et al. Critical Role of Maternal Selenium Nutrition in Neurodevelopment: Effects on Offspring Behavior and Neuroinflammatory Profile. Nutrients 2022, 14, 1850. https://doi.org/10.3390/nu14091850
Ajmone-Cat MA, De Simone R, Tartaglione AM, Di Biase A, Di Benedetto R, D’Archivio M, Varì R, Ricceri L, Aureli F, Iacoponi F, et al. Critical Role of Maternal Selenium Nutrition in Neurodevelopment: Effects on Offspring Behavior and Neuroinflammatory Profile. Nutrients. 2022; 14(9):1850. https://doi.org/10.3390/nu14091850
Chicago/Turabian StyleAjmone-Cat, Maria Antonietta, Roberta De Simone, Anna Maria Tartaglione, Antonella Di Biase, Rita Di Benedetto, Massimo D’Archivio, Rosaria Varì, Laura Ricceri, Federica Aureli, Francesca Iacoponi, and et al. 2022. "Critical Role of Maternal Selenium Nutrition in Neurodevelopment: Effects on Offspring Behavior and Neuroinflammatory Profile" Nutrients 14, no. 9: 1850. https://doi.org/10.3390/nu14091850
APA StyleAjmone-Cat, M. A., De Simone, R., Tartaglione, A. M., Di Biase, A., Di Benedetto, R., D’Archivio, M., Varì, R., Ricceri, L., Aureli, F., Iacoponi, F., Raggi, A., Cubadda, F., Fairweather-Tait, S. J., Calamandrei, G., & Minghetti, L. (2022). Critical Role of Maternal Selenium Nutrition in Neurodevelopment: Effects on Offspring Behavior and Neuroinflammatory Profile. Nutrients, 14(9), 1850. https://doi.org/10.3390/nu14091850










