Gut Microbiota and Phenotypic Changes Induced by Ablation of Liver- and Intestinal-Type Fatty Acid-Binding Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Body Weight and Food Intake
2.3. Intestinal Transit Time
2.4. Total Fecal Excretion
2.5. Gut Microbiota Analyses
2.6. Statistical Analysis
2.7. GC/MS Analysis of SCFAs in Fecal Samples
3. Results
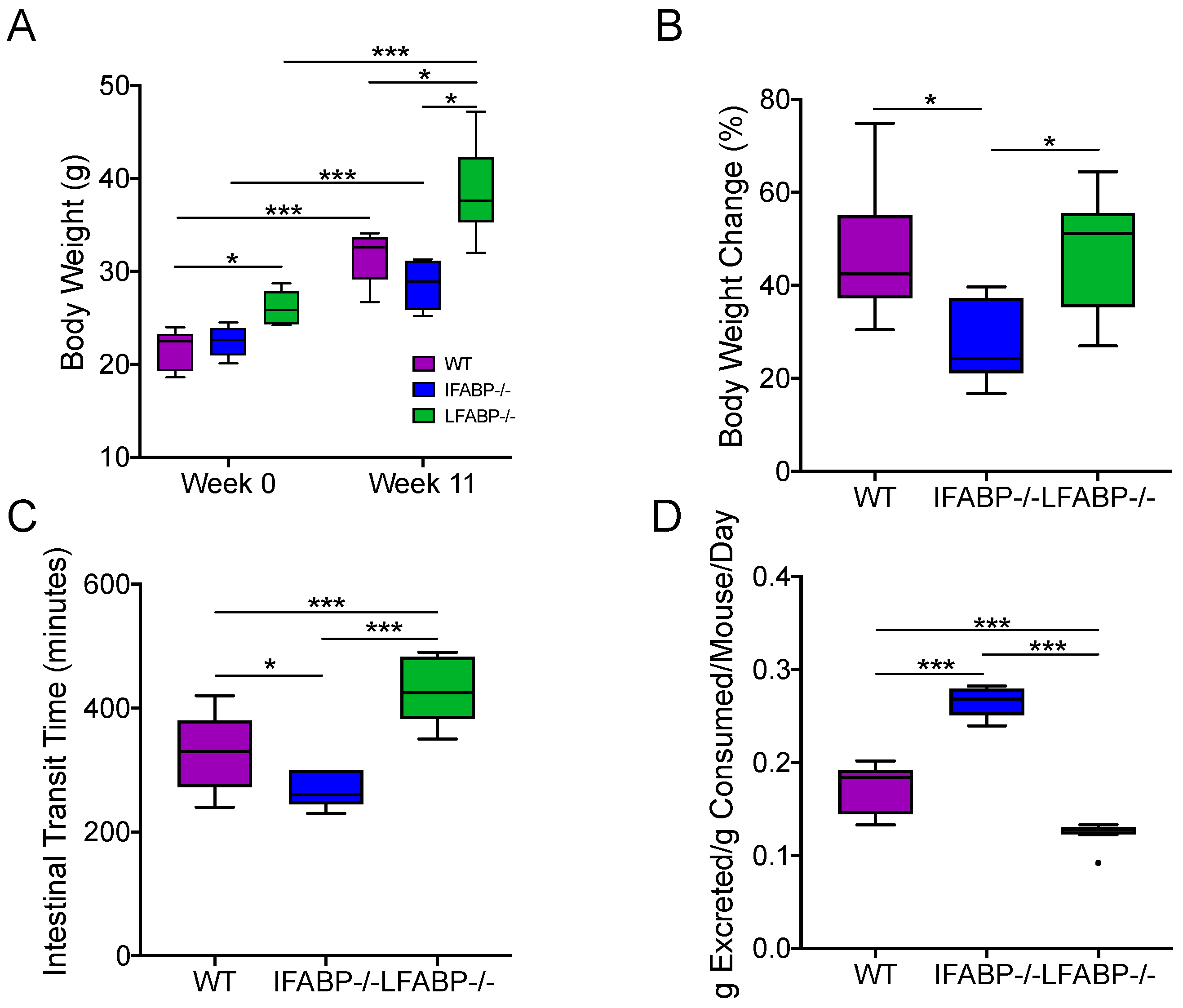
3.1. Body Weight Gain Differs in WT, IFABP−/−, and LFABP−/− Mice after Chronic HF Feeding
3.2. Intestinal Transit Time and Total Fecal Excretion Are Altered in Mice Lacking IFABP and LFABP
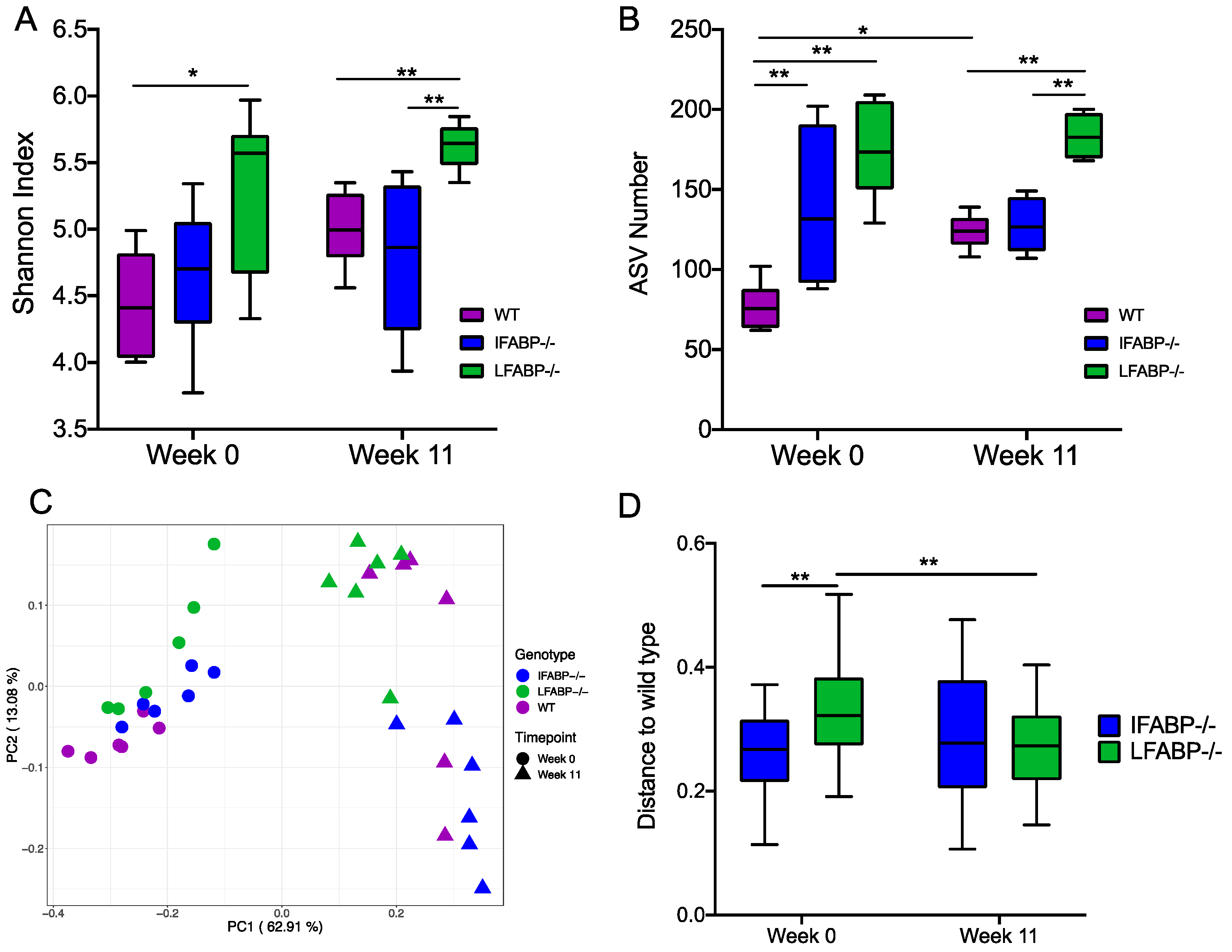
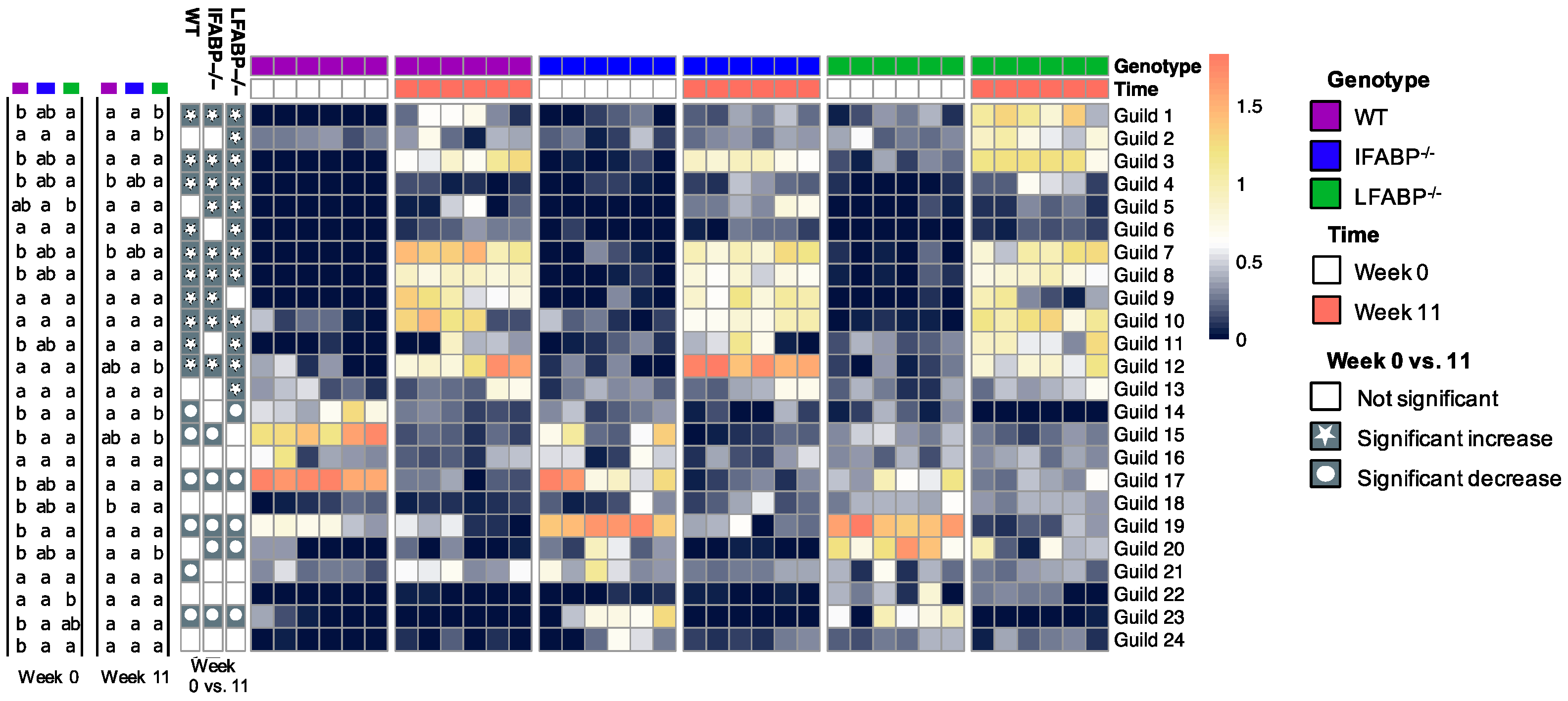
3.3. The Microbiota Composition Is Altered by IFABP and LFABP Ablation and Shows Different Responses to HFD
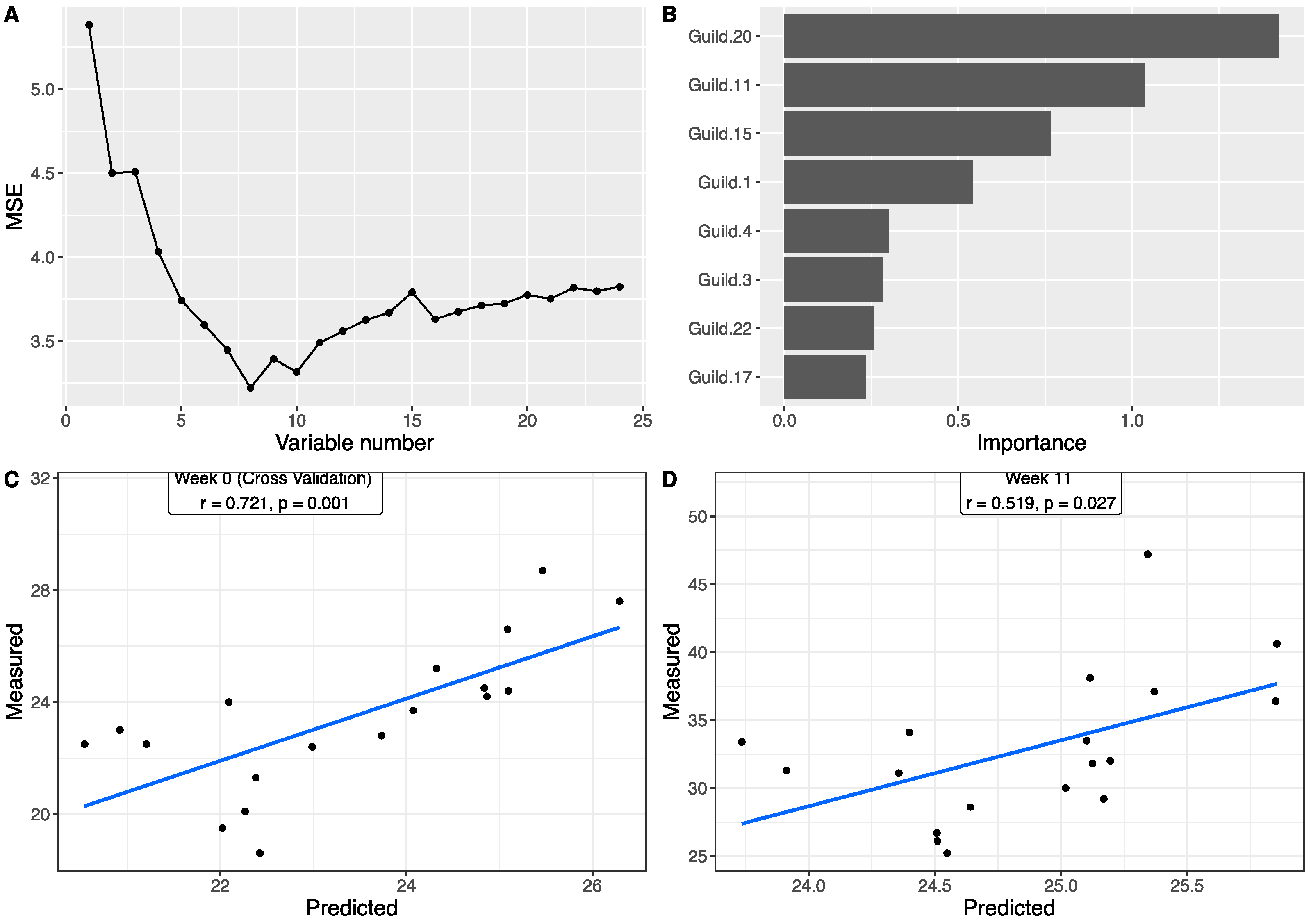
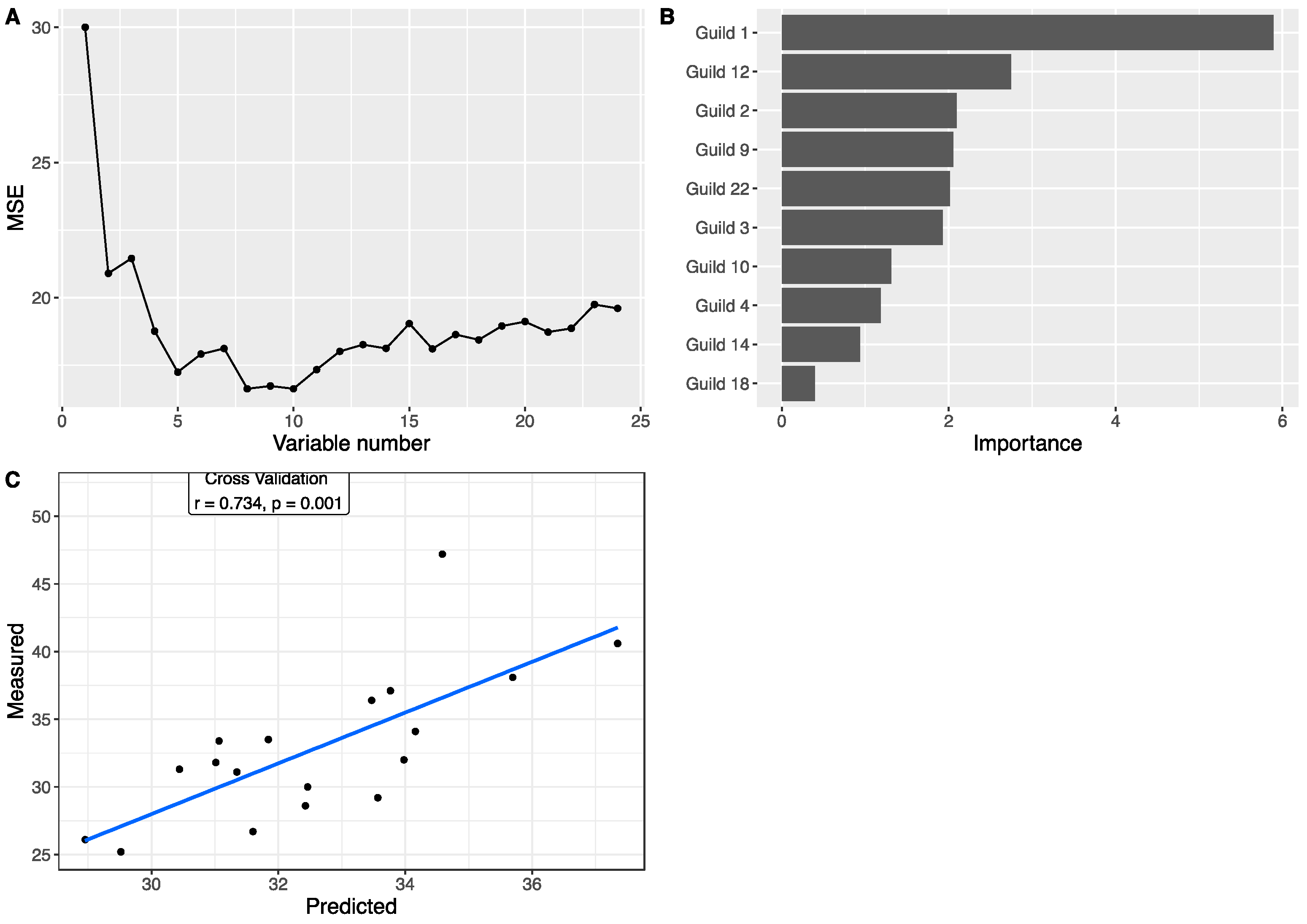
3.4. Associations between Gut Microbiota and Body Weight
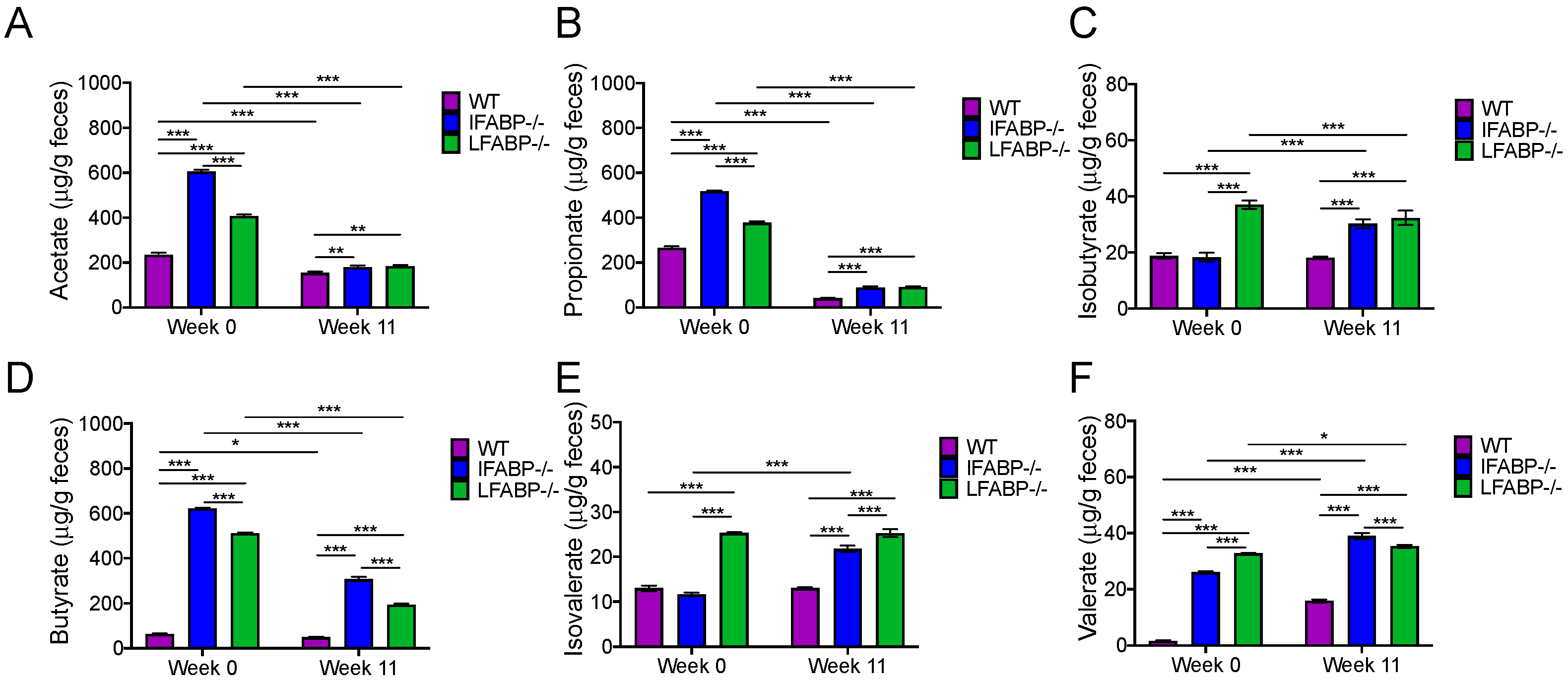
3.5. IFABP and LFABP Ablation and HFD Feeding Alter Fecal SCFA Levels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zimmerman, A.W.; Veerkamp, J.H. New insights into the structure and function of fatty acid-binding proteins. Cell. Mol. Life Sci. CMLS 2002, 59, 1096–1116. [Google Scholar] [CrossRef] [PubMed]
- Storch, J.; Thumser, A.E. Tissue-specific functions in the fatty acid-binding protein family. J. Biol. Chem. 2010, 285, 32679–32683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishkin, S.; Stein, L.; Gatmaitan, Z.; Arias, I.M. The binding of fatty acids to cytoplasmic proteins: Binding to Z protein in liver and other tissues of the rat. Biochem. Biophys. Res. Commun. 1972, 47, 997–1003. [Google Scholar] [CrossRef]
- Ockner, R.K.; Manning, J.A.; Poppenhausen, R.B.; Ho, W.K. A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science 1972, 177, 56–58. [Google Scholar] [CrossRef]
- Gordon, J.I.; Elshourbagy, N.; Lowe, J.B.; Liao, W.S.; Alpers, D.H.; Taylor, J.M. Tissue specific expression and developmental regulation of two genes coding for rat fatty acid binding proteins. J. Biol. Chem. 1985, 260, 1995–1998. [Google Scholar] [CrossRef]
- Richieri, G.V.; Ogata, R.T.; Zimmerman, A.W.; Veerkamp, J.H.; Kleinfeld, A.M. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry 2000, 39, 7197–7204. [Google Scholar] [CrossRef]
- He, Y.; Yang, X.; Wang, H.; Estephan, R.; Francis, F.; Kodukula, S.; Storch, J.; Stark, R.E. Solution-state molecular structure of apo and oleate-liganded liver fatty acid-binding protein. Biochemistry 2007, 46, 12543–12556. [Google Scholar] [CrossRef]
- Lagakos, W.S.; Gajda, A.M.; Agellon, L.; Binas, B.; Choi, V.; Mandap, B.; Russnak, T.; Zhou, Y.X.; Storch, J. Different functions of intestinal and liver-type fatty acid-binding proteins in intestine and in whole body energy homeostasis. Am. J. Physiol.-Gastrointest. Liver Physiol. 2011, 300, G803–G814. [Google Scholar] [CrossRef] [Green Version]
- Thumser, A.E.; Wilton, D.C. The binding of cholesterol and bile salts to recombinant rat liver fatty acid-binding protein. Biochem. J. 1996, 320 Pt 3, 729–733. [Google Scholar] [CrossRef] [Green Version]
- Thumser, A.E.; Voysey, J.E.; Wilton, D.C. The binding of lysophospholipids to rat liver fatty acid-binding protein and albumin. Biochem. J. 1994, 301 Pt 3, 801–806. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; McIntosh, A.L.; Martin, G.G.; Landrock, D.; Chung, S.; Landrock, K.K.; Dangott, L.J.; Li, S.; Kier, A.B.; Schroeder, F. FABP1: A Novel Hepatic Endocannabinoid and Cannabinoid Binding Protein. Biochemistry 2016, 55, 5243–5255. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.P.; Katz, F.S.; Bernard, C.; Storch, J.; Stark, R.E. Two fatty acid-binding proteins expressed in the intestine interact differently with endocannabinoids. Protein Sci. A Publ. Protein Soc. 2020, 29, 1606–1617. [Google Scholar] [CrossRef]
- Vassileva, G.; Huwyler, L.; Poirier, K.; Agellon, L.B.; Toth, M.J. The intestinal fatty acid binding protein is not essential for dietary fat absorption in mice. FASEB J. 2000, 14, 2040–2046. [Google Scholar] [CrossRef]
- Lowe, J.B.; Sacchettini, J.C.; Laposata, M.; McQuillan, J.J.; Gordon, J.I. Expression of rat intestinal fatty acid-binding protein in Escherichia coli. Purification and comparison of ligand binding characteristics with that of Escherichia coli-derived rat liver fatty acid-binding protein. J. Biol. Chem. 1987, 262, 5931–5937. [Google Scholar] [CrossRef]
- Richieri, G.V.; Ogata, R.T.; Kleinfeld, A.M. Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J. Biol. Chem. 1994, 269, 23918–23930. [Google Scholar] [CrossRef]
- Gajda, A.M.; Storch, J. Enterocyte fatty acid-binding proteins (FABPs): Different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot. Essent. Fat. Acids 2015, 93, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Makowski, L.; Hotamisligil, G.S. Fatty acid binding proteins--the evolutionary crossroads of inflammatory and metabolic responses. J. Nutr. 2004, 134, 2464S–2468S. [Google Scholar] [CrossRef] [Green Version]
- Hsu, K.T.; Storch, J. Fatty acid transfer from liver and intestinal fatty acid-binding proteins to membranes occurs by different mechanisms. J. Biol. Chem. 1996, 271, 13317–13323. [Google Scholar] [CrossRef] [Green Version]
- Gajda, A.M.; Zhou, Y.X.; Agellon, L.B.; Fried, S.K.; Kodukula, S.; Fortson, W.; Patel, K.; Storch, J. Direct comparison of mice null for liver or intestinal fatty acid-binding proteins reveals highly divergent phenotypic responses to high fat feeding. J. Biol. Chem. 2013, 288, 30330–30344. [Google Scholar] [CrossRef] [Green Version]
- Atshaves, B.P.; McIntosh, A.L.; Storey, S.M.; Landrock, K.K.; Kier, A.B.; Schroeder, F. High dietary fat exacerbates weight gain and obesity in female liver fatty acid binding protein gene-ablated mice. Lipids 2010, 45, 97–110. [Google Scholar] [CrossRef] [Green Version]
- Newberry, E.P.; Xie, Y.; Kennedy, S.M.; Luo, J.; Davidson, N.O. Protection against Western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology 2006, 44, 1191–1205. [Google Scholar] [CrossRef]
- Xu, H.; Gajda, A.M.; Zhou, Y.X.; Panetta, C.; Sifnakis, Z.; Fatima, A.; Henderson, G.C.; Storch, J. Muscle metabolic reprogramming underlies the resistance of liver fatty acid-binding protein (LFABP)-null mice to high-fat feeding-induced decline in exercise capacity. J. Biol. Chem. 2019, 294, 15358–15372. [Google Scholar] [CrossRef]
- Lackey, A.I.; Chen, T.; Zhou, Y.X.; Bottasso Arias, N.M.; Doran, J.M.; Zacharisen, S.M.; Gajda, A.M.; Jonsson, W.O.; Corsico, B.; Anthony, T.G.; et al. Mechanisms underlying reduced weight gain in intestinal fatty acid-binding protein (IFABP) null mice. Am. J. Physiol.-Gastrointest. Liver Physiol. 2020, 318, G518–G530. [Google Scholar] [CrossRef]
- Ghazalpour, A.; Cespedes, I.; Bennett, B.J.; Allayee, H. Expanding role of gut microbiota in lipid metabolism. Curr. Opin. Lipidol. 2016, 27, 141–147. [Google Scholar] [CrossRef]
- Cani, P.D.; Van Hul, M.; Lefort, C.; Depommier, C.; Rastelli, M.; Everard, A. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar] [CrossRef]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef]
- Fei, N.; Bruneau, A.; Zhang, X.J.; Wang, R.R.; Wang, J.X.; Rabot, S.; Gerard, P.; Zhao, L.P. Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. Mbio 2020, 11, e03263-19. [Google Scholar] [CrossRef] [Green Version]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Backhed, F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef] [Green Version]
- Lam, Y.Y.; Ha, C.W.Y.; Hoffmann, J.M.A.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef]
- Devkota, S.; Wang, Y.W.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Org, E.; Parks, B.W.; Joo, J.W.; Emert, B.; Schwartzman, W.; Kang, E.Y.; Mehrabian, M.; Pan, C.; Knight, R.; Gunsalus, R.; et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015, 25, 1558–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, G.G.; Danneberg, H.; Kumar, L.S.; Atshaves, B.P.; Erol, E.; Bader, M.; Schroeder, F.; Binas, B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J. Biol. Chem. 2003, 278, 21429–21438. [Google Scholar] [CrossRef] [Green Version]
- Nagakura, Y.; Ito, H.; Kiso, T.; Naitoh, Y.; Miyata, K. The selective 5-hydroxytryptamine (5-HT)4-receptor agonist RS67506 enhances lower intestinal propulsion in mice. Jpn. J. Pharmacol. 1997, 74, 209–212. [Google Scholar] [CrossRef]
- Kelly, J.R.; Borre, Y.; O’Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aqaut. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, Q.; Wu, G.; Zhang, C.; Zhang, M.; Zhao, L. Minimizing spurious features in 16S rRNA gene amplicon sequencing. PeerJ Prepr. 2018, 6, 2167–9843. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps; R Package Version 1.0. 8. 2015. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 24 March 2022).
- Bland, J.M.; Altman, D.G. Calculating correlation coefficients with repeated observations: Part 1—Correlation within subjects. BMJ 1995, 310, 446. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Zhao, N.; Zhang, C.; Lam, Y.Y.; Zhao, L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 2021, 13, 22. [Google Scholar] [CrossRef]
- García-Villalba, R.; Giménez-Bastida, J.A.; García-Conesa, M.T.; Tomás-Barberán, F.A.; Carlos Espín, J.; Larrosa, M. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J. Sep. Sci. 2012, 35, 1906–1913. [Google Scholar] [CrossRef]
- Muller, M.; Hermes, G.D.A.; Canfora, E.E.; Smidt, H.; Masclee, A.A.M.; Zoetendal, E.G.; Blaak, E.E. Distal colonic transit is linked to gut microbiota diversity and microbial fermentation in humans with slow colonic transit. Am. J. Physiol.-Gastrointest. Liver Physiol. 2020, 318, G361–G369. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Simberloff, D.; Dayan, T. The guild concept and the structure of ecological communities. Annu. Rev. Ecol. Syst. 1991, 22, 115–143. [Google Scholar] [CrossRef]
- Kaczocha, M.; Glaser, S.T.; Deutsch, D.G. Identification of intracellular carriers for the endocannabinoid anandamide. Proc. Natl. Acad. Sci. USA 2009, 106, 6375–6380. [Google Scholar] [CrossRef] [Green Version]
- Lagakos, W.S.; Guan, X.; Ho, S.Y.; Sawicki, L.R.; Corsico, B.; Kodukula, S.; Murota, K.; Stark, R.E.; Storch, J. Liver fatty acid-binding protein binds monoacylglycerol in vitro and in mouse liver cytosol. J. Biol. Chem. 2013, 288, 19805–19815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partosoedarso, E.R.; Abrahams, T.P.; Scullion, R.T.; Moerschbaecher, J.M.; Hornby, P.J. Cannabinoid1 receptor in the dorsal vagal complex modulates lower oesophageal sphincter relaxation in ferrets. J. Physiol. 2003, 550, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Vianna, C.R.; Donato, J., Jr.; Rossi, J.; Scott, M.; Economides, K.; Gautron, L.; Pierpont, S.; Elias, C.F.; Elmquist, J.K. Cannabinoid receptor 1 in the vagus nerve is dispensable for body weight homeostasis but required for normal gastrointestinal motility. J. Neurosci. 2012, 32, 10331–10337. [Google Scholar] [CrossRef] [Green Version]
- DiPatrizio, N.V. Endocannabinoids in the Gut. Cannabis Cannabinoid Res. 2016, 1, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Cristino, L.; Becker, T.; Di Marzo, V. Endocannabinoids and energy homeostasis: An update. BioFactors 2014, 40, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ueda, N. Biology of endocannabinoid synthesis system. Prostaglandins Other Lipid Mediat. 2009, 89, 112–119. [Google Scholar] [CrossRef]
- Koch, M. Cannabinoid Receptor Signaling in Central Regulation of Feeding Behavior: A Mini-Review. Front. Neurosci. 2017, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Hansen, L.B.S.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gobel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, N.; Tan, H.Y.; Li, S.; Zhang, C.; Feng, Y.B. Function of Akkermansia muciniphila in Obesity: Interactions With Lipid Metabolism, Immune Response and Gut Systems. Front. Microbiol. 2020, 11, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeney, D.D.; Gareau, M.G.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Li, D.; Ke, W.; Liang, D.; Hu, X.; Chen, F. Resveratrol-induced gut microbiota reduces obesity in high-fat diet-fed mice. Int. J. Obes. 2020, 44, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ni, Z.X.; Yu, J.; Cheng, W.; Cai, Z.L.; Yu, C.Q. Correlation Between Fecal Metabolomics and Gut Microbiota in Obesity and Polycystic Ovary Syndrome. Front. Endocrinol. 2020, 11, 628. [Google Scholar] [CrossRef]
- Chen, L.W.; Xu, J.; Soh, S.E.; Aris, I.M.; Tint, M.T.; Gluckman, P.D.; Tan, K.H.; Shek, L.P.C.; Chong, Y.S.; Yap, F.; et al. Implication of gut microbiota in the association between infant antibiotic exposure and childhood obesity and adiposity accumulation. Int. J. Obes. 2020, 44, 1508–1520. [Google Scholar] [CrossRef]
- Won, S.M.; Chen, S.; Lee, S.Y.; Lee, K.E.; Park, K.W.; Yoon, J.H. Lactobacillus sakei ADM14 Induces Anti-Obesity Effects and Changes in Gut Microbiome in High-Fat Diet-Induced Obese Mice. Nutrients 2020, 12, 3703. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef]
- Naudin, C.R.; Maner-Smith, K.; Owens, J.A.; Wynn, G.M.; Robinson, B.S.; Matthews, J.D.; Reedy, A.R.; Luo, L.; Wolfarth, A.A.; Darby, T.M.; et al. Lactococcus lactis Subspecies cremoris Elicits Protection Against Metabolic Changes Induced by a Western-Style Diet. Gastroenterology 2020, 159, 639–651.e5. [Google Scholar] [CrossRef] [PubMed]
- Loubinoux, J.; Bronowicki, J.P.; Pereira, I.A.C.; Mougenel, J.L.; Le Faou, A.E. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. Fems Microbiol. Ecol. 2002, 40, 107–112. [Google Scholar] [CrossRef]
- Zhang, C.H.; Zhang, M.H.; Wang, S.Y.; Han, R.J.; Cao, Y.F.; Hua, W.Y.; Mao, Y.J.; Zhang, X.J.; Pang, X.Y.; Wei, C.C.; et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. Isme J. 2010, 4, 312–313. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Pang, B.; Shao, D.; Jiang, C.; Hu, X.; Shi, J. Artemisia sphaerocephala Krasch polysaccharide mediates lipid metabolism and metabolic endotoxaemia in associated with the modulation of gut microbiota in diet-induced obese mice. Int. J. Biol. Macromol. 2020, 147, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med. Sci. Sports Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Ticinesi, A.; Lauretani, F.; Milani, C.; Nouvenne, A.; Tana, C.; Del Rio, D.; Maggio, M.; Ventura, M.; Meschi, T. Aging Gut Microbiota at the Cross-Road between Nutrition, Physical Frailty, and Sarcopenia: Is There a Gut-Muscle Axis? Nutrients 2017, 9, 1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosicki, G.J.; Fielding, R.A.; Lustgarten, M.S. Gut Microbiota Contribute to Age-Related Changes in Skeletal Muscle Size, Composition, and Function: Biological Basis for a Gut-Muscle Axis. Calcif. Tissue Int. 2018, 102, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat. Metab. 2020, 2, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Morino, K.; Ugi, S.; Nakagawa, F.; Lemecha, M.; Ida, S.; Ohashi, N.; Sato, D.; Fujita, Y.; Maegawa, H. Microbiome potentiates endurance exercise through intestinal acetate production. Am. J. Physiology. Endocrinol. Metab. 2019, 316, E956–E966. [Google Scholar] [CrossRef]
- Morita, H.; Kano, C.; Ishii, C.; Kagata, N.; Ishikawa, T.; Uchiyama, Y.; Hara, S.; Nakamura, T.; Fukuda, S.J.b. Bacteroides uniformis enhances endurance exercise performance through gluconeogenesis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPARγ-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Fan, C.; Li, P.; Lu, Y.; Chang, X.; Qi, K. Short Chain Fatty Acids Prevent High-fat-diet-induced Obesity in Mice by Regulating G Protein-coupled Receptors and Gut Microbiota. Sci. Rep. 2016, 6, 37589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Tottey, W.; Feria-Gervasio, D.; Gaci, N.; Laillet, B.; Pujos, E.; Martin, J.F.; Sebedio, J.L.; Sion, B.; Jarrige, J.F.; Alric, M.; et al. Colonic Transit Time Is a Driven Force of the Gut Microbiota Composition and Metabolism: In Vitro Evidence. J. Neurogastroenterol. 2017, 23, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Smathers, R.L.; Petersen, D.R. The human fatty acid-binding protein family: Evolutionary divergences and functions. Hum. Genom. 2011, 5, 170–191. [Google Scholar] [CrossRef]
| HFS Grams/4057 kcal | |
|---|---|
| C16 | 49.9 |
| C16:1 | 0.4 |
| C18 | 64.3 |
| C18:1 | 65.2 |
| C18:2 | 10.7 |
| C18:3 | 1.0 |
| % | |
| Saturated fatty acids | 60.0 |
| Monounsaturated fatty acids | 33.9 |
| Polyunsaturated fatty acids | 6.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.; Tawfeeq, H.R.; Lackey, A.I.; Zhou, Y.; Sifnakis, Z.; Zacharisen, S.M.; Xu, H.; Doran, J.M.; Sampath, H.; Zhao, L.; et al. Gut Microbiota and Phenotypic Changes Induced by Ablation of Liver- and Intestinal-Type Fatty Acid-Binding Proteins. Nutrients 2022, 14, 1762. https://doi.org/10.3390/nu14091762
Wu G, Tawfeeq HR, Lackey AI, Zhou Y, Sifnakis Z, Zacharisen SM, Xu H, Doran JM, Sampath H, Zhao L, et al. Gut Microbiota and Phenotypic Changes Induced by Ablation of Liver- and Intestinal-Type Fatty Acid-Binding Proteins. Nutrients. 2022; 14(9):1762. https://doi.org/10.3390/nu14091762
Chicago/Turabian StyleWu, Guojun, Hiba R. Tawfeeq, Atreju I. Lackey, Yinxiu Zhou, Zoe Sifnakis, Sophia M. Zacharisen, Heli Xu, Justine M. Doran, Harini Sampath, Liping Zhao, and et al. 2022. "Gut Microbiota and Phenotypic Changes Induced by Ablation of Liver- and Intestinal-Type Fatty Acid-Binding Proteins" Nutrients 14, no. 9: 1762. https://doi.org/10.3390/nu14091762
APA StyleWu, G., Tawfeeq, H. R., Lackey, A. I., Zhou, Y., Sifnakis, Z., Zacharisen, S. M., Xu, H., Doran, J. M., Sampath, H., Zhao, L., Lam, Y. Y., & Storch, J. (2022). Gut Microbiota and Phenotypic Changes Induced by Ablation of Liver- and Intestinal-Type Fatty Acid-Binding Proteins. Nutrients, 14(9), 1762. https://doi.org/10.3390/nu14091762







