Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life
Abstract
:1. Introduction
2. What Are the ‘Infant-Type’ Bifidobacterium Species and the Effect of Cross-Feeding?
3. Establishment and Evolution of Infant-Type Bifidobacteria
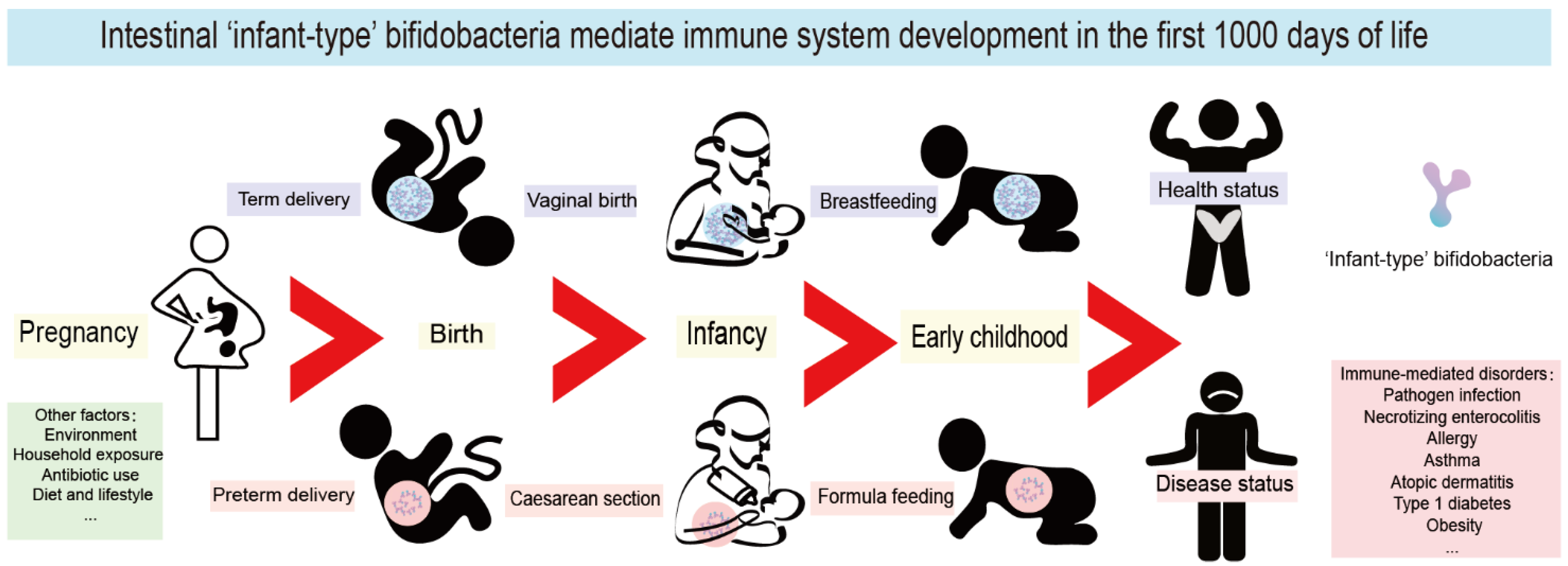
4. The First 1000 Days of Life Are a Key Window of Opportunity for Immune System Maturation
5. Infant-Type Bifidobacteria Affect the Establishment of Immunity in Early Life
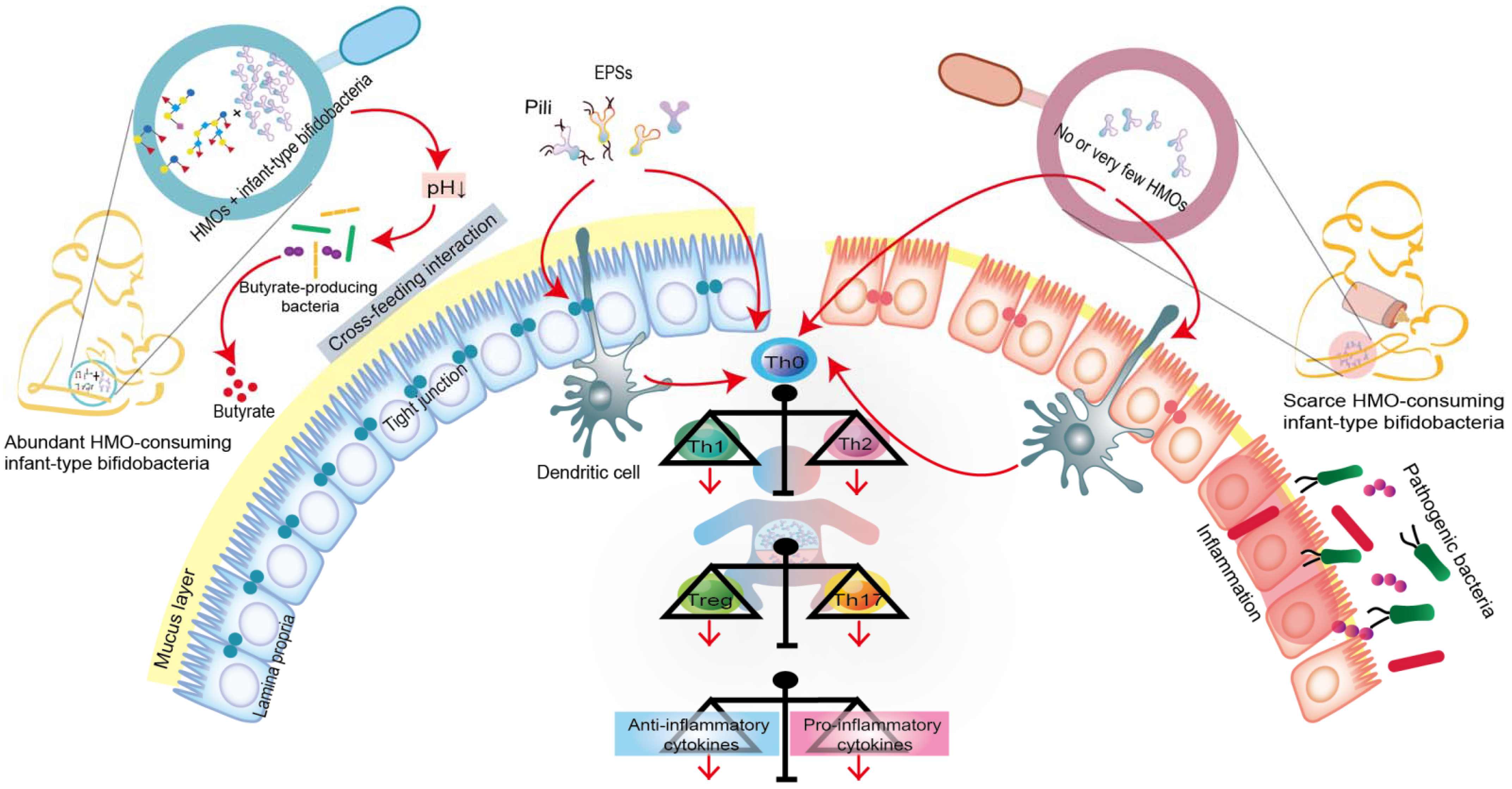
5.1. Infant-Type Bifidobacteria Occupy Intestinal Ecological Sites
5.2. Infant-Type Bifidobacteria Facilitate Breast-Milk Metabolism
5.3. Infant-Type Bifidobacteria Promote Immune Development and Prime the Anti-Inflammatory Gene Pool
5.4. The Potential Role of Infant-Type Bifidobacteria in Early Neuroimmune Development
6. Infant-Type Bifidobacteria Supplementation Is a Promising Strategy for Immune-Mediated Disorders
6.1. Effects of Infant-Type Bifidobacteria on Pathogen Infection and NEC
6.2. Effects of Infant-Type Bifidobacteria on Allergic Diseases
6.3. Effects of Infant-Type Bifidobacteria on T1D and Obesity
7. Conclusions
8. Outlook: How Do We Accelerate the Colonization of Infant-Type Bifidobacteria in the Intestinal Flora of Infants during Early Life?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, K.M.; Gerlach, M.J.; Adam, T.; Heimesaat, M.M.; Rossi, L.; Surette, M.G.; Sloboda, D.M.; Braun, T. Fetal Meconium Does Not Have a Detectable Microbiota before Birth. Nat. Microbiol. 2021, 6, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Perez-Munoz, M.E.; Arrieta, M.-C.; Ramer-Tait, A.E.; Walter, J. A Critical Assessment of the “Sterile Womb” and “in Utero Colonization” Hypotheses: Implications for Research on the Pioneer Infant Microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How Colonization by Microbiota in Early Life Shapes the Immune System. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of Bifidobacteria within the Infant Gut Microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turroni, F.; van Sinderen, D.; Ventura, M. Genomics and Ecological Overview of the Genus Bifidobacterium. Int. J. Food Microbiol. 2011, 149, 37–44. [Google Scholar] [CrossRef]
- Asakuma, S.; Hatakeyama, E.; Urashima, T.; Yoshida, E.; Katayama, T.; Yamamoto, K.; Kumagai, H.; Ashida, H.; Hirose, J.; Kitaoka, M. Physiology of Consumption of Human Milk Oligosaccharides by Infant Gut-Associated Bifidobacteria. J. Biol. Chem. 2011, 286, 34583–34592. [Google Scholar] [CrossRef] [Green Version]
- Katayama, T. Host-Derived Glycans Serve as Selected Nutrients for the Gut Microbe: Human Milk Oligosaccharides and Bifidobacteria. Biosci. Biotechnol. Biochem. 2016, 80, 621–632. [Google Scholar] [CrossRef] [Green Version]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-Mediated Immune System Imprinting Early in Life. Cell 2021, 184, 3884–3898. [Google Scholar] [CrossRef]
- Frost, G.; Sleeth, M.L.; Sahuri-Arisoylu, M.; Lizarbe, B.; Cerdan, S.; Brody, L.; Anastasovska, J.; Ghourab, S.; Hankir, M.; Zhang, S.; et al. The Short-Chain Fatty Acid Acetate Reduces Appetite via a Central Homeostatic Mechanism. Nat. Commun. 2014, 5, 3611. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, J.T.; Roesch, L.F.W.; Ordberg, M.; Ilonen, J.; Atkinson, M.A.; Schatz, D.A.; Triplett, E.W.; Ludvigsson, J. Genetic Risk for Autoimmunity Is Associated with Distinct Changes in the Human Gut Microbiome. Nat. Commun. 2019, 10, 3621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hamalainen, A.-M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted Microbiota and Opportunistic Pathogen Colonization in Caesarean-Section Birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Wang, S.; Ryan, C.A.; Boyaval, P.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Maternal Vertical Transmission Affecting Early-Life Microbiota Development. Trends Microbiol. 2020, 28, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Arriola, J.; Gerber, C.W.; Kaveti, A.; Kalanetra, K.M.; Kananurak, A.; Bevins, C.L.; Mills, D.A.; Dvorak, B. Bifidobacterium Longum Subsp Infantis in Experimental Necrotizing Enterocolitis: Alterations in Inflammation, Innate Immune Response, and the Microbiota. Pediatr. Res. 2014, 76, 326–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frese, S.A.; Hutton, A.A.; Contreras, L.N.; Shaw, C.A.; Palumbo, M.C.; Casaburi, G.; Xu, G.; Davis, J.C.C.; Lebrilla, C.B.; Henrick, B.M.; et al. Persistence of Supplemented Bifidobacterium Longum Subsp. Infantis EVC001 in Breastfed Infants. Msphere 2017, 2, e00501-17. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, A.-G.; Arnolds, S.; Kolln, A.; Achenbach, P.; Berner, R.; Bonifacio, E.; Casteels, K.; Elding Larsson, H.; Gundert, M.; Hasford, J.; et al. Supplementation with Bifidobacterium Longum Subspecies Infantis EVC001 for Mitigation of Type 1 Diabetes Autoimmunity: The GPPAD-SINT1A Randomised Controlled Trial Protocol. BMJ Open 2021, 11, e052449. [Google Scholar] [CrossRef]
- Seppo, A.E.; Bu, K.; Jumabaeva, M.; Thakar, J.; Choudhury, R.A.; Yonemitsu, C.; Bode, L.; Martina, C.A.; Allen, M.; Tamburini, S.; et al. Infant Gut Microbiome Is Enriched with Bifidobacterium Longum Ssp. Infantis in Old Order Mennonites with Traditional Farming Lifestyle. Allergy 2021, 76, 3489–3503. [Google Scholar] [CrossRef]
- Wong, C.B.; Sugahara, H.; Odamaki, T.; Xiao, J.Z. Different Physiological Properties of Human-Residential and Non-Human-Residential Bifidobacteria in Human Health. Benef. Microbes 2018, 9, 111–122. [Google Scholar] [CrossRef]
- Odamaki, T.; Bottacini, F.; Kato, K.; Mitsuyama, E.; Yoshida, K.; Horigome, A.; Xiao, J.; van Sinderen, D. Genomic Diversity and Distribution of Bifidobacterium Longum Subsp Longum across the Human Lifespan. Sci. Rep. 2018, 8, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Li, L.-Q.; Liu, F.; Wu, J.-Y. Human Milk Oligosaccharides and Infant Gut Microbiota: Molecular Structures, Utilization Strategies and Immune Function. Carbohydr. Polym. 2022, 276, 118738. [Google Scholar] [CrossRef]
- Bode, L. The Functional Biology of Human Milk Oligosaccharides. Early Hum. Dev. 2015, 91, 619–622. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, W.; Mu, W. Human Milk Oligosaccharides: The New Gold Standard for Premium Infant Formula. J. Agric. Food Chem. 2022, 70, 2061–2063. [Google Scholar] [CrossRef] [PubMed]
- Puccio, G.; Alliet, P.; Cajozzo, C.; Janssens, E.; Corsello, G.; Sprenger, N.; Wernimont, S.; Egli, D.; Gosoniu, L.; Steenhout, P. Effects of Infant Formula with Human Milk Oligosaccharides on Growth and Morbidity: A Randomized Multicenter Trial. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 624–631. [Google Scholar] [CrossRef] [Green Version]
- Akkerman, R.; Faas, M.M.; de Vos, P. Non-Digestible Carbohydrates in Infant Formula as Substitution for Human Milk Oligosaccharide Functions: Effects on Microbiota and Gut Maturation. Crit. Rev. Food Sci. Nutr. 2019, 59, 1486–1497. [Google Scholar] [CrossRef]
- Asnicar, F.; Manara, S.; Zolfo, M.; Truong, D.T.; Scholz, M.; Armanini, F.; Ferretti, P.; Gorfer, V.; Pedrotti, A.; Tett, A.; et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. Msystems 2017, 2, e00164-16. [Google Scholar] [CrossRef] [Green Version]
- Lawson, M.A.E.; O’Neill, I.J.; Kujawska, M.; Javavdi, S.G.; Wijeyesekera, A.; Flegg, Z.; Chalklen, L.; Hall, L.J. Breast Milk-Derived Human Milk Oligosaccharides Promote Bifidobacterium Interactions within a Single Ecosystem. Isme J. 2020, 14, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Tannock, G.W.; Lawley, B.; Munro, K.; Pathmanathan, S.G.; Zhou, S.J.; Makrides, M.; Gibson, R.A.; Sullivan, T.; Prosser, C.G.; Lowry, D.; et al. Comparison of the Compositions of the Stool Microbiotas of Infants Fed Goat Milk Formula, Cow Milk-Based Formula, or Breast Milk. Appl. Environ. Microbiol. 2013, 79, 3040–3048. [Google Scholar] [CrossRef] [Green Version]
- Centanni, M.; Ferguson, S.A.; Sims, I.M.; Biswas, A.; Tannock, G.W. Bifidobacterium Bifidum ATCC 15696 and Bifidobacterium Breve 24b Metabolic Interaction Based on 2 ’-O-Fucosyl-Lactose Studied in Steady-State Cultures in a Freter-Style Chemostat. Appl. Environ. Microbiol. 2019, 85, e02783-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turroni, F.; Milani, C.; Duranti, S.; Mahony, J.; van Sinderen, D.; Ventura, M. Glycan Utilization and Cross-Feeding Activities by Bifidobacteria. Trends Microbiol. 2018, 26, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Moyano, S.; Totten, S.M.; Garrido, D.A.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Variation in Consumption of Human Milk Oligosaccharides by Infant Gut-Associated Strains of Bifidobacterium Breve. Appl. Environ. Microbiol. 2013, 79, 6040–6049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Cai, X.; Ye, Y.; Wang, F.; Chen, F.; Zheng, C. The Role of Microbiota in Infant Health: From Early Life to Adulthood. Front. Immunol. 2021, 12, 708472. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Impact of Human Milk Bacteria and Oligosaccharides on Neonatal Gut Microbiota Establishment and Gut Health. Nutr. Rev. 2015, 73, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal Microbiota Composition of Breast-Fed Infants Is Correlated With Human Milk Oligosaccharides Consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Blaser, M.J.; Devkota, S.; McCoy, K.D.; Relman, D.A.; Yassour, M.; Young, V.B. Lessons Learned from the Prenatal Microbiome Controversy. Microbiome 2021, 9, 8. [Google Scholar] [CrossRef]
- Brugman, S.; Perdijk, O.; van Neerven, R.J.J.; Savelkoul, H.F.J. Mucosal Immune Development in Early Life: Setting the Stage. Arch. Immunol. Ther. Ex. 2015, 63, 251–268. [Google Scholar] [CrossRef] [Green Version]
- Carolyn, A.T.; Kathy, D.M. The Role of Mom’s Microbes during Pregnancy. Scientist 2021. Available online: https://www.the-scientist.com/features/the-role-of-mom-s-microbes-during-pregnancy-69009 (accessed on 1 August 2021).
- Gopalakrishna, K.P.; Macadangdang, B.R.; Rogers, M.B.; Tometich, J.T.; Firek, B.A.; Baker, R.; Ji, J.; Burr, A.H.P.; Ma, C.; Good, M.; et al. Maternal IgA Protects against the Development of Necrotizing Enterocolitis in Preterm Infants. Nat. Med. 2019, 25, 1110–1115. [Google Scholar] [CrossRef]
- Neu, J. The Microbiome during Pregnancy and Early Postnatal Life. Semin. Fetal. Neonatal. Med. 2016, 21, 373–379. [Google Scholar] [CrossRef]
- Hobbs, A.J.; Mannion, C.A.; McDonald, S.W.; Brockway, M.; Tough, S.C. The Impact of Caesarean Section on Breastfeeding Initiation, Duration and Difficulties in the First Four Months Postpartum. BMC Pregnancy Childbirth 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matamoros, S.; Gras-Leguen, C.; Le Vacon, F.; Potel, G.; de La Cochetiere, M.-F. Development of Intestinal Microbiota in Infants and Its Impact on Health. Trends Microbiol. 2013, 21, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Martin, R.; Ishikawa, E.; Gawad, A.; Kubota, H.; Sakai, T.; Oishi, K.; Tanaka, R.; Ben-Amor, K.; Knol, J.; et al. Multilocus Sequence Typing of Bifidobacterial Strains from Infant’s Faeces and Human Milk: Are Bifidobacteria Being Sustainably Shared during Breastfeeding? Benef. Microbes 2015, 6, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Hesla, H.M.; Stenius, F.; Jaderlund, L.; Nelson, R.; Engstrand, L.; Alm, J.; Dicksved, J. Impact of Lifestyle on the Gut Microbiota of Healthy Infants and Their Mothers-The ALADDIN Birth Cohort. Fems Microbiol. Ecol. 2014, 90, 791–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogra, S.; Sakwinska, O.; Soh, S.-E.; Ngom-Bru, C.; Brueck, W.M.; Berger, B.; Bruessow, H.; Lee, Y.S.; Yap, F.; Chong, Y.-S.; et al. Dynamics of Infant Gut Microbiota Are Influenced by Delivery Mode and Gestational Duration and Are Associated with Subsequent Adiposity. Mbio 2015, 6, e02419-14. [Google Scholar] [CrossRef] [Green Version]
- Boudry, G.; Charton, E.; Le Huerou-Luron, I.; Ferret-Bernard, S.; Le Gall, S.; Even, S.; Blat, S. The Relationship Between Breast Milk Components and the Infant Gut Microbiota. Front. Nutr. 2021, 8, 629740. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Berger, B.; Carnielli, V.P.; Ksiazyk, J.; Lagstrom, H.; Sanchez Luna, M.; Migacheva, N.; Mosselmans, J.-M.; Picaud, J.-C.; Possner, M.; et al. Human Milk Oligosaccharides: 2-Fucosyllactose (2-FL) and Lacto-N-Neotetraose (LNnT) in Infant Formula. Nutrients 2018, 10, 1161. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Prince, A.L.; Bader, D.; Hu, M.; Ganu, R.; Baquero, K.; Blundell, P.; Harris, R.A.; Frias, A.E.; Grove, K.L.; et al. High-Fat Maternal Diet during Pregnancy Persistently Alters the Offspring Microbiome in a Primate Model. Nat. Commun. 2014, 5, 3889. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Ferrario, C.; Lugli, G.A.; Mancabelli, L.; van Sinderen, D.; Ventura, M. Bifidobacteria and the Infant Gut: An Example of Co-Evolution and Natural Selection. Cell. Mol. Life Sci. 2018, 75, 103–118. [Google Scholar] [CrossRef]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The Microbiome in Early Life: Implications for Health Outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef] [Green Version]
- De Muinck, E.J.; Trosvik, P. Individuality and Convergence of the Infant Gut Microbiota during the First Year of Life. Nat. Commun. 2018, 9, 2233. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Victora, C.G.; Adair, L.; Fall, C.; Hallal, P.C.; Martorell, R.; Richter, L.; Sachdev, H.S. Maternal and Child Undernutrition 2-Maternal and Child Undernutrition: Consequences for Adult Health and Human Capital. Lancet 2008, 371, 340–357. [Google Scholar] [CrossRef] [Green Version]
- Balbus, J.M.; Barouki, R.; Birnbaum, L.S.; Etzel, R.A.; Gluckman, P.D.; Grandjean, P.; Hancock, C.; Hanson, M.A.; Heindel, J.J.; Hoffman, K.; et al. Early-Life Prevention of Non-Communicable Diseases. Lancet 2013, 381, 3–4. [Google Scholar] [CrossRef] [Green Version]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated Fecal PH Indicates a Profound Change in the Breastfed Infant Gut Microbiome Due to Reduction of Bifidobacterium over the Past Century. Msphere 2018, 3, e00041-18. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, C.E.; Meier, A.K.; Cernioglo, K.; Mitchell, R.D.; Casaburi, G.; Frese, S.A.; Henrick, B.M.; Underwood, M.A.; Smilowitz, J.T. Early Probiotic Supplementation with B. Infantis in Breastfed Infants Leads to Persistent Colonization at 1 Year. Pediatr. Res. 2022, 91, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Kalanetra, K.M.; Bokulich, N.A.; Lewis, Z.T.; Mirmiran, M.; Tancredi, D.J.; Mills, D.A. A Comparison of Two Probiotic Strains of Bifidobacteria in Premature Infants. J. Pediatr. 2013, 163, 1585–1591. [Google Scholar] [CrossRef] [Green Version]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Morbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Molgaard, C.; et al. Bifidobacterium Species Associated with Breastfeeding Produce Aromatic Lactic Acids in the Infant Gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Renz, H.; Skevaki, C. Early Life Microbial Exposures and Allergy Risks: Opportunities for Prevention. Nat. Rev. Immunol. 2021, 21, 177–191. [Google Scholar] [CrossRef]
- Chichlowski, M.; De Lartigue, G.; German, J.B.; Raybould, H.E.; Mills, D.A. Bifidobacteria Isolated From Infants and Cultured on Human Milk Oligosaccharides Affect Intestinal Epithelial Function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Sela, D.A.; Mills, D.A. Nursing Our Microbiota: Molecular Linkages between Bifidobacteria and Milk Oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios-Covian, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de los Reyes-Gavilan, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and Their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, S.H.; Holtrop, G.; Lobley, G.E.; Calder, A.G.; Stewart, C.S.; Flint, H.J. Contribution of Acetate to Butyrate Formation by Human Faecal Bacteria. Br. J. Nutr. 2004, 91, 915–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef]
- Alessandri, G.; Ossiprandi, M.C.; MacSharry, J.; van Sinderen, D.; Ventura, M. Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019, 10, 2348. [Google Scholar] [CrossRef] [Green Version]
- Saturio, S.; Nogacka, A.M.; Alvarado-Jasso, G.M.; Salazar, N.; de los Reyes-Gavilan, C.G.; Gueimonde, M.; Arboleya, S. Role of Bifidobacteria on Infant Health. Microorganisms 2021, 9, 2415. [Google Scholar] [CrossRef]
- Pungel, D.; Treveil, A.; Dalby, M.J.; Caim, S.; Colquhoun, I.J.; Booth, C.; Ketskemety, J.; Korcsmaros, T.; van Sinderen, D.; Lawson, M.A.E.; et al. Bifidobacterium Breve UCC2003 Exopolysaccharide Modulates the Early Life Microbiota by Acting as a Potential Dietary Substrate. Nutrients 2020, 12, 948. [Google Scholar] [CrossRef] [Green Version]
- Schiavi, E.; Gleinser, M.; Molloy, E.; Groeger, D.; Frei, R.; Ferstl, R.; Rodriguez-Perez, N.; Ziegler, M.; Grant, R.; Moriarty, T.F.; et al. The Surface-Associated Exopolysaccharide of Bifidobacterium Longum 35624 Plays an Essential Role in Dampening Host Proinflammatory Responses and Repressing Local T(H)17 Responses. Appl. Environ. Microbiol. 2016, 82, 7185–7196. [Google Scholar] [CrossRef] [Green Version]
- Bottacini, F.; van Sinderen, D.; Ventura, M. Omics of Bifidobacteria: Research and Insights into Their Health-Promoting Activities. Biochem. J. 2017, 474, 4137–4152. [Google Scholar] [CrossRef]
- Turroni, F.; Serafini, F.; Foroni, E.; Duranti, S.; Motherway, M.O.; Taverniti, V.; Mangifesta, M.; Milani, C.; Viappiani, A.; Roversi, T.; et al. Role of Sortase-Dependent Pili of Bifidobacterium Bifidum PRL2010 in Modulating Bacterium-Host Interactions. Proc. Natl. Acad. Sci. USA 2013, 110, 11151–11156. [Google Scholar] [CrossRef] [Green Version]
- de Roock, S.; Stoppelenburg, A.J.; Scholman, R.; Hoeks, S.B.E.A.; Meerding, J.; Prakken, B.J.; Boes, M. Defective T(H)17 Development in Human Neonatal T Cells Involves Reduced RORC2 MRNA Content. J. Allergy Clin. Immunol. 2013, 132, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.N.; Ahmad, S.M.; Alam, M.J.; Khanam, A.; Kalanetra, K.M.; Taft, D.H.; Raqib, R.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Bifidobacterium Abundance in Early Infancy and Vaccine Response at 2 Years of Age. Pediatrics 2019, 143, e20181489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Bai, Y.; Zhou, J.; Huang, W.; Yan, J.; Tao, J.; Fan, Q.; Liu, Y.; Mei, D.; Yan, Q.; et al. Core Fucosylation of Maternal Milk N-Glycan Evokes B Cell Activation by Selectively Promoting the L-Fucose Metabolism of Gut Bifidobacterium spp. and Lactobacillus spp. Mbio 2019, 10, e00128-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luck, B.; Engevik, M.A.; Ganesh, B.P.; Lackey, E.P.; Lin, T.; Balderas, M.; Major, A.; Runge, J.; Luna, R.A.; Sillitoe, R.; et al. Bifidobacteria Shape Host Neural Circuits during Postnatal Development by Promoting Synapse Formation and Microglial Function. Sci. Rep. 2020, 10, 7737. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Ventura, M.; van Sinderen, D. The Human Gut Microbiota during the Initial Stages of Life: Insights from Bifidobacteria. Curr. Opin. Biotechnol. 2022, 73, 81–87. [Google Scholar] [CrossRef]
- Kelsey, C.M.; Prescott, S.; McCulloch, J.A.; Trinchieri, G.; Valladares, T.L.; Dreisbach, C.; Alhusen, J.; Grossmann, T. Gut Microbiota Composition Is Associated with Newborn Functional Brain Connectivity and Behavioral Temperament. Brain. Behav. Immun. 2021, 91, 472–486. [Google Scholar] [CrossRef]
- Hoyos, A.B. Reduced Incidence of Necrotizing Enterocolitis Associated with Enteral Administration of Lactobacillus Acidophilus and Bifidobacterium Infantis to Neonates in an Intensive Care Unit. Int. J. Infect. Dis. 1999, 3, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Azagra-Boronat, I.; Massot-Cladera, M.; Knipping, K.; Garssen, J.; Ben Amor, K.; Knol, J.; Franch, A.; Castell, M.; Rodriguez-Lagunas, M.J.; Perez-Cano, F.J. Strain-Specific Probiotic Properties of Bifidobacteria and Lactobacilli for the Prevention of Diarrhea Caused by Rotavirus in a Preclinical Model. Nutrients 2020, 12, 498. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.P.; Duffy, L.C.; Griffiths, E.; Dryja, D.; Leavens, A.; Rossman, J.; Rich, G.; Riepenhoff-Talty, M.; Locniskar, M. Immune Responses in Rhesus Rotavirus-Challenged Balb/c Mice Treated with Bifidobacteria and Prebiotic Supplements. Pediatr. Res. 2002, 51, 750–755. [Google Scholar] [CrossRef]
- Kang, J.Y.; Lee, D.K.; Ha, N.J.; Shin, H.S. Antiviral Effects of Lactobacillus Ruminis SPM0211 and Bifidobacterium Longum SPM1205 and SPM1206 on Rotavirus-Infected Caco-2 Cells and a Neonatal Mouse Model. J. Microbiol. 2015, 53, 796–803. [Google Scholar] [CrossRef]
- Bergmann, K.R.; Liu, S.X.L.; Tian, R.; Kushnir, A.; Turner, J.R.; Li, H.-L.; Chou, P.M.; Weber, C.R.; De Plaen, I.G. Bifidobacteria Stabilize Claudins at Tight Junctions and Prevent Intestinal Barrier Dysfunction in Mouse Necrotizing Enterocolitis. Am. J. Pathol. 2013, 182, 1595–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, T.; Izumi, H.; Iwabuchi, N.; Odamaki, T.; Namba, K.; Abe, F.; Xiao, J.Z. Bifidobacterium Breve Prevents Necrotising Enterocolitis by Suppressing Inflammatory Responses in a Preterm Rat Model. Benef. Microbes 2016, 7, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Kananurak, A.; Coursodon, C.F.; Adkins-Reick, C.K.; Chu, H.; Bennett, S.H.; Wehkamp, J.; Castillo, P.A.; Leonard, B.C.; Tancredi, D.J.; et al. Bifidobacterium Bifidum in a Rat Model of Necrotizing Enterocolitis: Antimicrobial Peptide and Protein Responses. Pediatr. Res. 2012, 71, 546–551. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-F.; Chiu, H.-Y.; Chen, A.-C.; Lin, H.-Y.; Lin, H.-C.; Caplan, M. Efficacy of Different Probiotic Combinations on Death and Necrotizing Enterocolitis in a Premature Rat Model. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.L.; Preidis, G.A.; Kashyap, P.C.; Weizman, A.; Sadeghirad, B. Probiotics Reduce Mortality and Morbidity in Preterm, Low-Birth-Weight Infants: A Systematic Review and Network Meta-Analysis of Randomized Trials. Gastroenterology 2020, 159, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Niemarkt, H.J.; De Meij, T.G.; van Ganzewinkel, C.; de Boer, N.K.H.; Andriessen, P.; Hutten, M.C.; Kramer, B.W. Necrotizing Enterocolitis, Gut Microbiota, and Brain Development: Role of the Brain-Gut Axis. Neonatology 2019, 115, 423–431. [Google Scholar] [CrossRef]
- Claud, E.C.; Walker, W.A. Hypothesis: Inappropriate Colonization of the Premature Intestine Can Cause Neonatal Necrotizing Enterocolitis. Faseb J. 2001, 15, 1398–1403. [Google Scholar] [CrossRef] [Green Version]
- Groer, M.W.; Luciano, A.A.; Dishaw, L.J.; Ashmeade, T.L.; Miller, E.; Gilbert, J.A. Development of the Preterm Infant Gut Microbiome: A Research Priority. Microbiome 2014, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Raba, A.A.; O’Sullivan, A.; Miletin, J. Pathogenesis of Necrotising Enterocolitis: The Impact of the Altered Gut Microbiota and Antibiotic Exposure in Preterm Infants. Acta Paediatr. 2021, 110, 433–440. [Google Scholar] [CrossRef]
- Henderickx, J.G.E.; Zwittink, R.D.; van Lingen, R.A.; Knol, J.; Belzer, C. The Preterm Gut Microbiota: An Inconspicuous Challenge in Nutritional Neonatal Care. Front. Cell. Infect. Microbiol. 2019, 9, 85. [Google Scholar] [CrossRef]
- Elgin, T.G.; Kern, S.L.; McElroy, S.J. Development of the Neonatal Intestinal Microbiome and Its Association With Necrotizing Enterocolitis. Clin. Ther. 2016, 38, 706–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawankar, R. Allergic Diseases and Asthma: A Global Public Health Concern and a Call to Action. World Allergy Organ. J. 2014, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberg, E.G. The WAO White Book on Allergy 2011–2012. Curr. Allergy Clin. Immunol. 2011, 24, 156–157. [Google Scholar]
- Sonnenburg, E.D.; Sonnenburg, J.L. The Ancestral and Industrialized Gut Microbiota and Implications for Human Health. Nat. Rev. Microbiol. 2019, 17, 383–390. [Google Scholar] [CrossRef]
- Kalliomaki, M.; Kirjavainen, P.; Eerola, E.; Kero, P.; Salminen, S.; Isolauri, E. Distinct Patterns of Neonatal Gut Microflora in Infants in Whom Atopy Was and Was Not Developing. J. Allergy Clin. Immunol. 2001, 107, 129–134. [Google Scholar] [CrossRef]
- Zimmermann, P.; Messina, N.; Mohn, W.W.; Finlay, B.B.; Curtis, N. Association between the Intestinal Microbiota and Allergic Sensitization, Eczema, and Asthma: A Systematic Review. J. Allergy Clin. Immunol. 2019, 143, 467–485. [Google Scholar] [CrossRef]
- Cukrowska, B.; Bierla, J.B.; Zakrzewska, M.; Klukowski, M.; Maciorkowska, E. The Relationship between the Infant Gut Microbiota and Allergy. The Role of Bifidobacterium Breve and Prebiotic Oligosaccharides in the Activation of Anti-Allergic Mechanisms in Early Life. Nutrients 2020, 12, 946. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Li, Y.; Fang, J.; Liu, G.; Yin, J.; Li, T.; Yin, Y. Gut Microbiota and Type 1 Diabetes. Int. J. Mol. Sci. 2018, 19, 995. [Google Scholar] [CrossRef] [Green Version]
- Durazzo, M.; Ferro, A.; Gruden, G. Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art. J. Clin. Med. 2019, 8, 1843. [Google Scholar] [CrossRef] [Green Version]
- White, R.A.; Bjornholt, J.V.; Baird, D.D.; Midtvedt, T.; Harris, J.R.; Pagano, M.; Hide, W.; Rudi, K.; Moen, B.; Iszatt, N.; et al. Novel Developmental Analyses Identify Longitudinal Patterns of Early Gut Microbiota That Affect Infant Growth. PLoS Comput. Biol. 2013, 9, e1003042. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Yoo, J.Y.; Valeria Ozorio Dutra, S.; Morgan, K.H.; Groer, M. The Association between Early-Life Gut Microbiota and Long-Term Health and Diseases. J. Clin. Med. 2021, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Conterno, L.; Fava, F.; Viola, R.; Tuohy, K.M. Obesity and the Gut Microbiota: Does up-Regulating Colonic Fermentation Protect against Obesity and Metabolic Disease? Genes Nutr. 2011, 6, 241–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Type of Study | Immune-Mediated Disorders | Study Object | Study Design | Study Outcomes | Conclusions | References |
|---|---|---|---|---|---|---|
| Clinical trial | Pathogen infection | Healthy infants at 6 to 15 weeks of age | The association of Bifidobacterium abundance in the stool with T-cell and antibody responses |
| The abundance of bifidobacteria in early infants might improve the protective effect of the vaccine by enhancing immune memory. | Huda, M.N. et al. [73] |
| NEC | 1237 newborns (both inpatients and transfer patients) |
|
| Infant-type bifidobacteria showed therapeutic effects on NEC. | Hoyos, A.B. et al. [78] | |
| Allergic diseases | A cohort of 65 Old Order Mennonite (OOM) and 39 Rochester mother-infant pairs | The gut microbiome and metabolome composition of atopic diseases in rural OOM infants and urban/suburban Rochester infants. |
| A high rate of B. longum subsp. infantis colonization was found in the OOM infants at low risk of atopic diseases. | Seppo, A.E. et al. [19] | |
| Pre-clinical study | Pathogen infection (rotavirus (simian SA-11)) | Lewis pups |
|
| B. breve M-16V seemed to be a very effective probiotic strain in ameliorating and preventing RV-induced diarrhea in children. | Azagra-Boronat, I. et al. [79] |
| Pathogen infection (rhesus rotavirus) | Balb/c pups |
|
| Infant-type bifidobacteria might act as an adjuvant to alleviate the severity of diarrhea caused by rotavirus by regulating early mucous membrane and strong humoral rotavirus-specific immune response. | Qiao, H.P. et al. [80] | |
| Pathogen infection (Wa rotavirus) | 7 day-old Balb/c pups |
|
| B. longum SPM1205 and SPM1206 effectively inhibited rotavirus replication by promoting type I IFNs to regulate the immune response. | Kang, J.Y. et al. [81] | |
| NEC | Cesarean-section SD rats |
|
| NEC-related inflammation could be alleviated by supplementing B. longum subsp. infantis. | Underwood, M.A. et al. [16] | |
| NEC | Naturally delivered C57BL/6 newborn mice |
|
| Administration of B. infantis reduced NEC incidence, at least in part due to its barrier-preserving properties. | Bergmann, K.R. et al. [82] | |
| NEC | Cesarean-section SD rats |
|
| B. breve M-16V prevented the development of NEC by regulating the expression of TLR and inhibiting the inflammatory response. | Satoh, T. et al. [83] | |
| NEC | Cesarean-section SD rats |
| The expression of lysozyme, secretory phospholipase A2, pancreatic-associated proteins 1 and 3 mRNA was elevated. | Oral administration of B. bifidum OLB6378 could avert both NEC and the associated increase in expression of antimicrobial peptides. | Underwood, M.A. et al. [84] | |
| NEC | Premature SD rats |
|
| Administration of a mixture of B. bifidum and B. longum was most effective in preventing death and NEC. | Wu, S.-F. et al. [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Lin, Y.; Zhang, H.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients 2022, 14, 1498. https://doi.org/10.3390/nu14071498
Lin C, Lin Y, Zhang H, Wang G, Zhao J, Zhang H, Chen W. Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients. 2022; 14(7):1498. https://doi.org/10.3390/nu14071498
Chicago/Turabian StyleLin, Chunxiu, Yugui Lin, Heng Zhang, Gang Wang, Jianxin Zhao, Hao Zhang, and Wei Chen. 2022. "Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life" Nutrients 14, no. 7: 1498. https://doi.org/10.3390/nu14071498
APA StyleLin, C., Lin, Y., Zhang, H., Wang, G., Zhao, J., Zhang, H., & Chen, W. (2022). Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients, 14(7), 1498. https://doi.org/10.3390/nu14071498






