Anti-Adipogenic Activity of High-Phenolic Sorghum Brans in Pre-Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sorghum Processing
2.3. Total Phenolic Content (TPC) Assay
2.4. Vanillin Assay for Condensed Tannin Quantification
2.5. HPLC Analysis of Sorghum Extracts
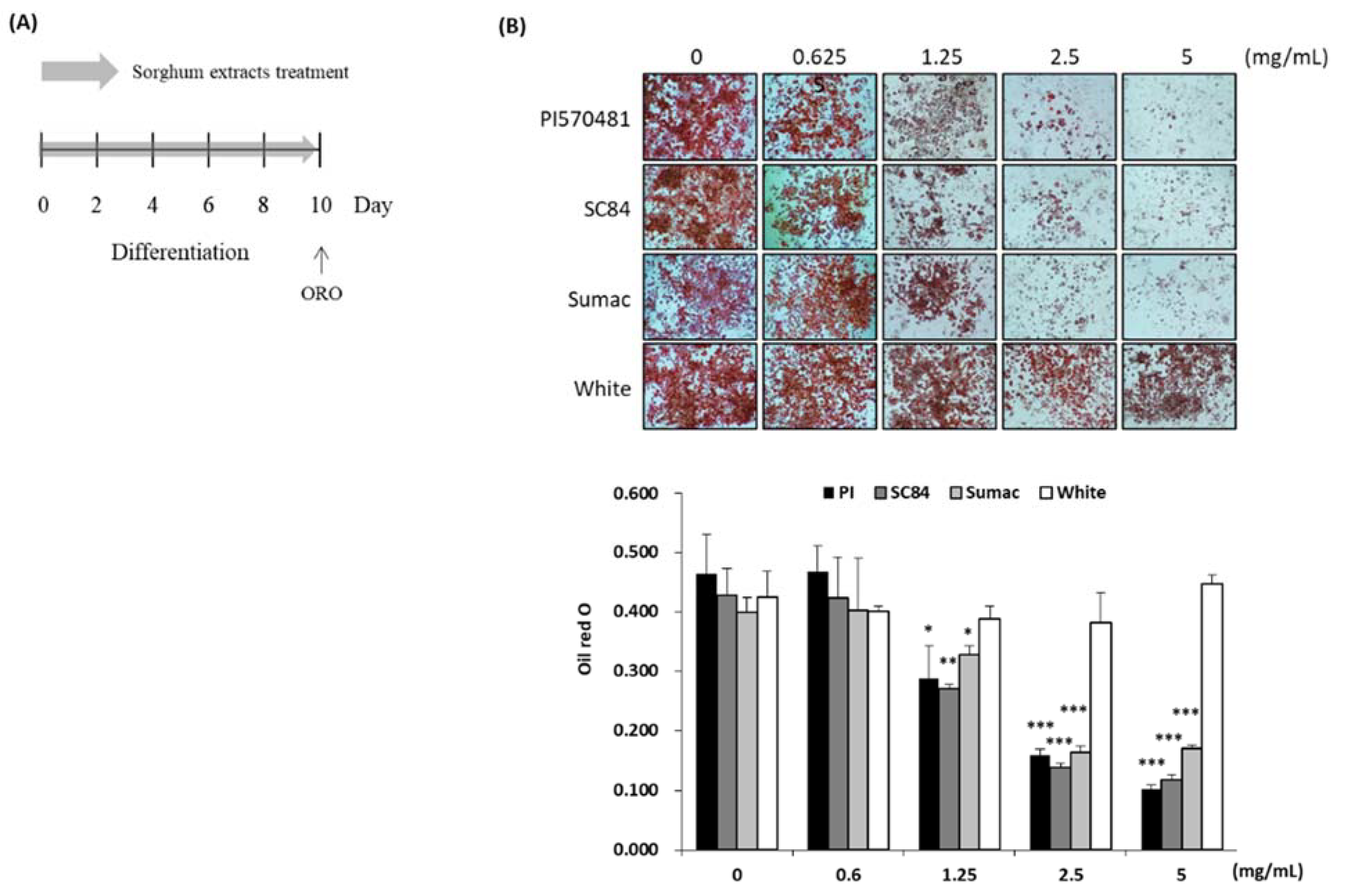
2.6. Cell Culture, Differentiation of 3T3-L1 and Treatment of Sorghum Bran Extracts
2.7. Oil Red O (ORO) Staining
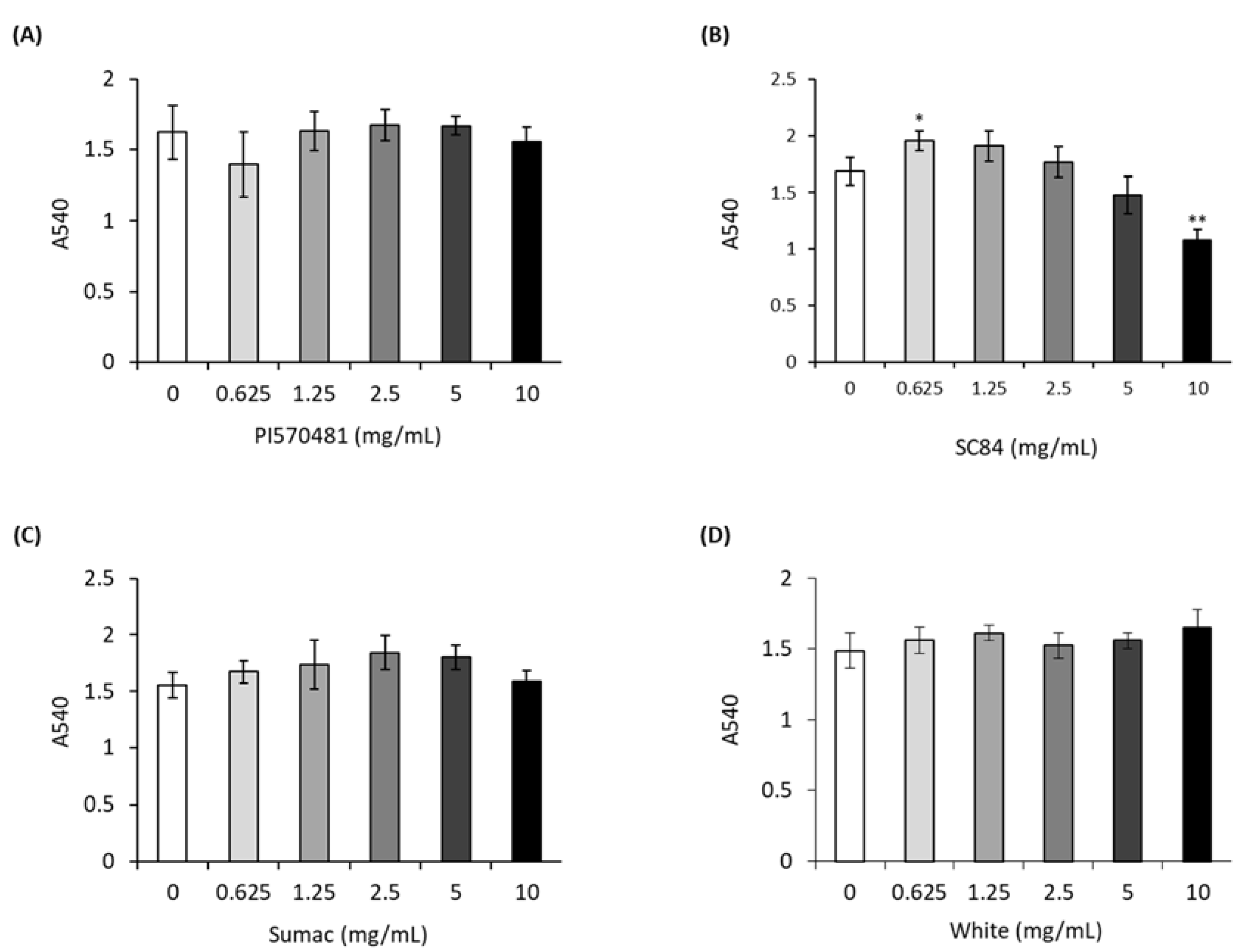
2.8. Cell Viability
2.9. Reactive Oxygen Species (ROS) Production
2.10. Glucose Uptake
2.11. Lipolysis
2.12. Western Blot
2.13. Statistical Analysis
3. Results
3.1. Chemical Analysis of Sorghum Bran Extracts
3.2. Sorghum Extracts Do Not Cause Cytotoxicity in 3T3-L1 Cells
3.3. Sorghum Bran Extracts Reduced Intracellular Lipid Accumulation in Differentiated 3T3-L1 Cells
3.4. Sorghum Bran Extracts Repressed Expression of Adipogenic and Lipogenic Proteins in Differentiated 3T3-L1 Cells
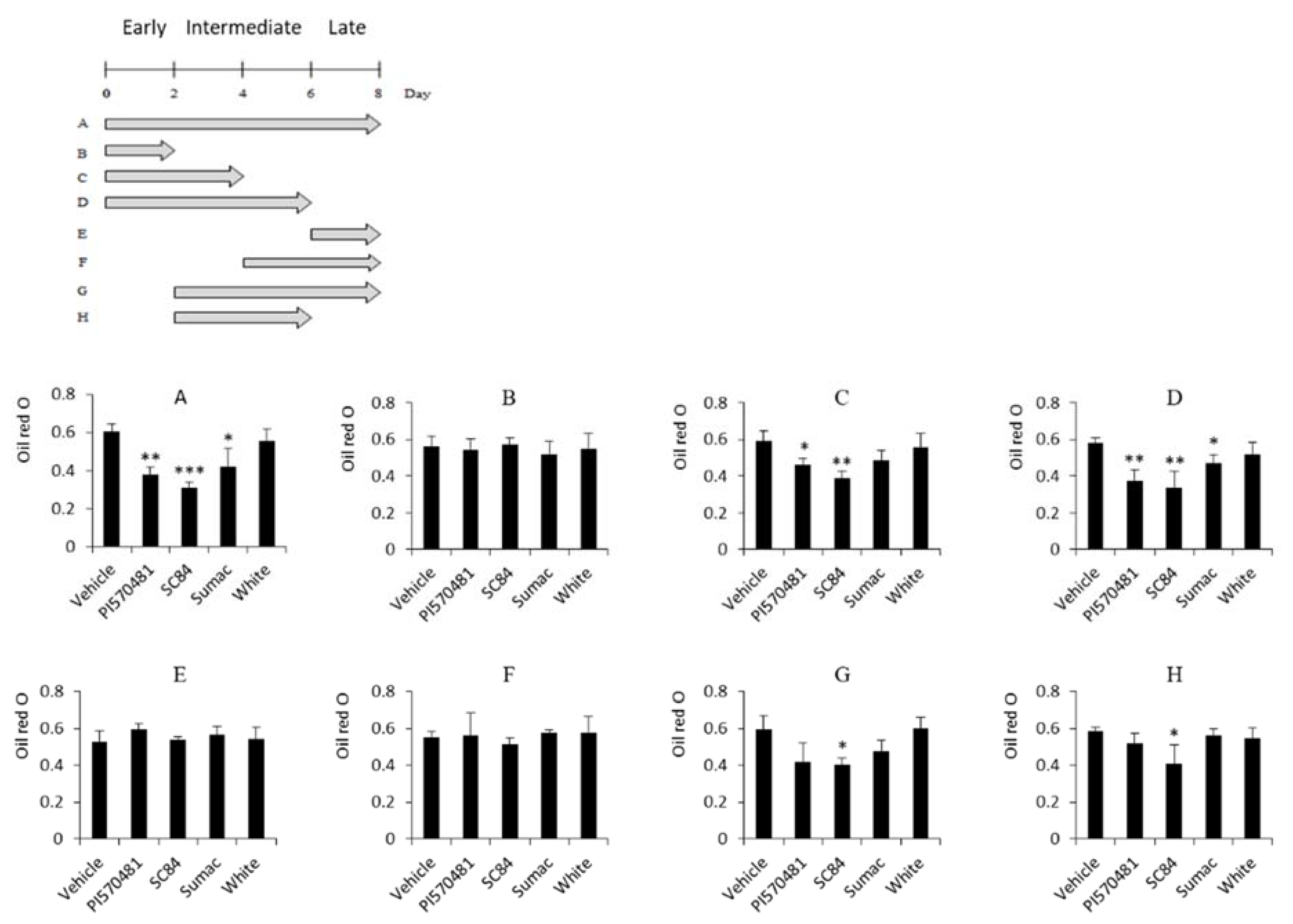
3.5. Anti-Adipogenic Activities of Sorghum Bran Extracts Depend on Early Phase of Adipogenesis
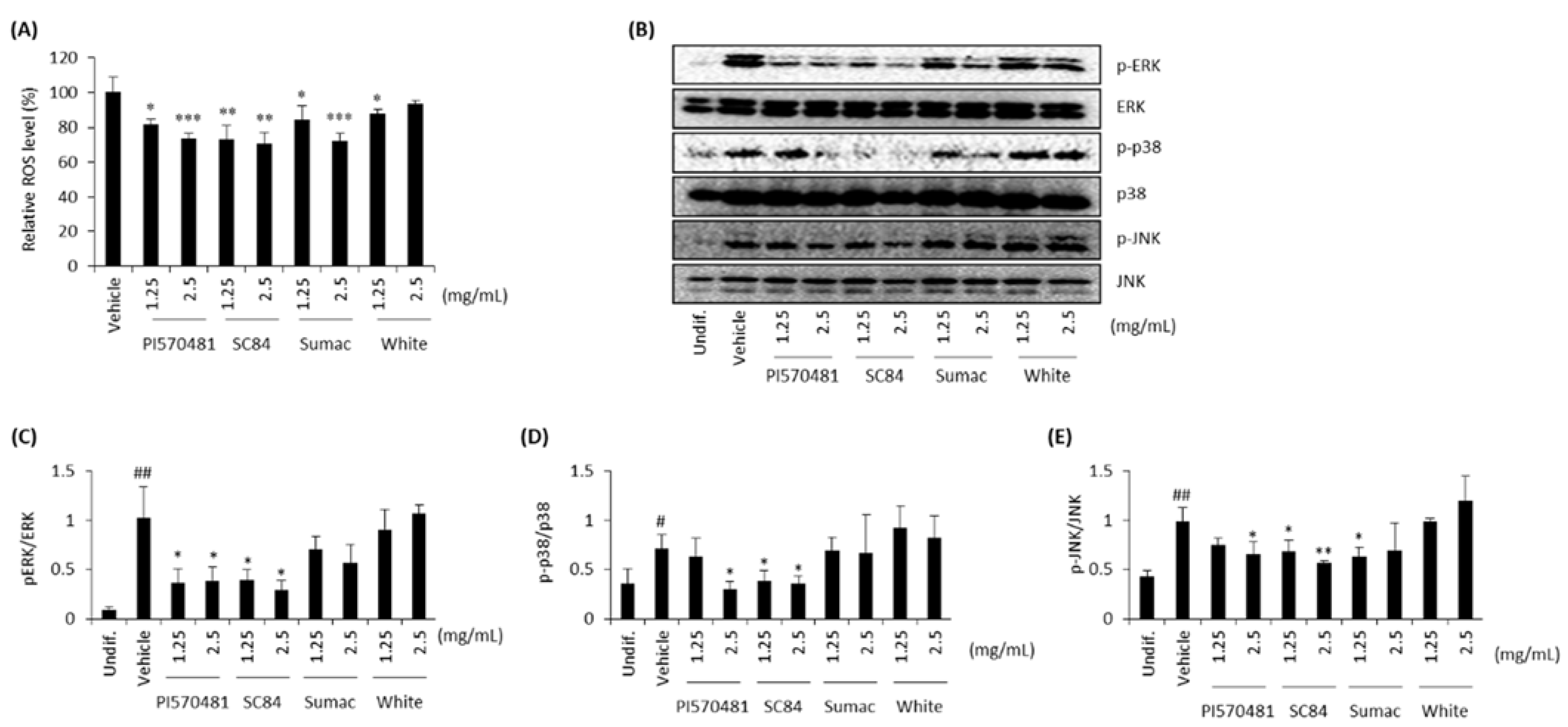
3.6. Sorghum Bran Extracts Repressed ROS Production and MAPK Signaling Pathways in Differentiated 3T3-L1 Cells
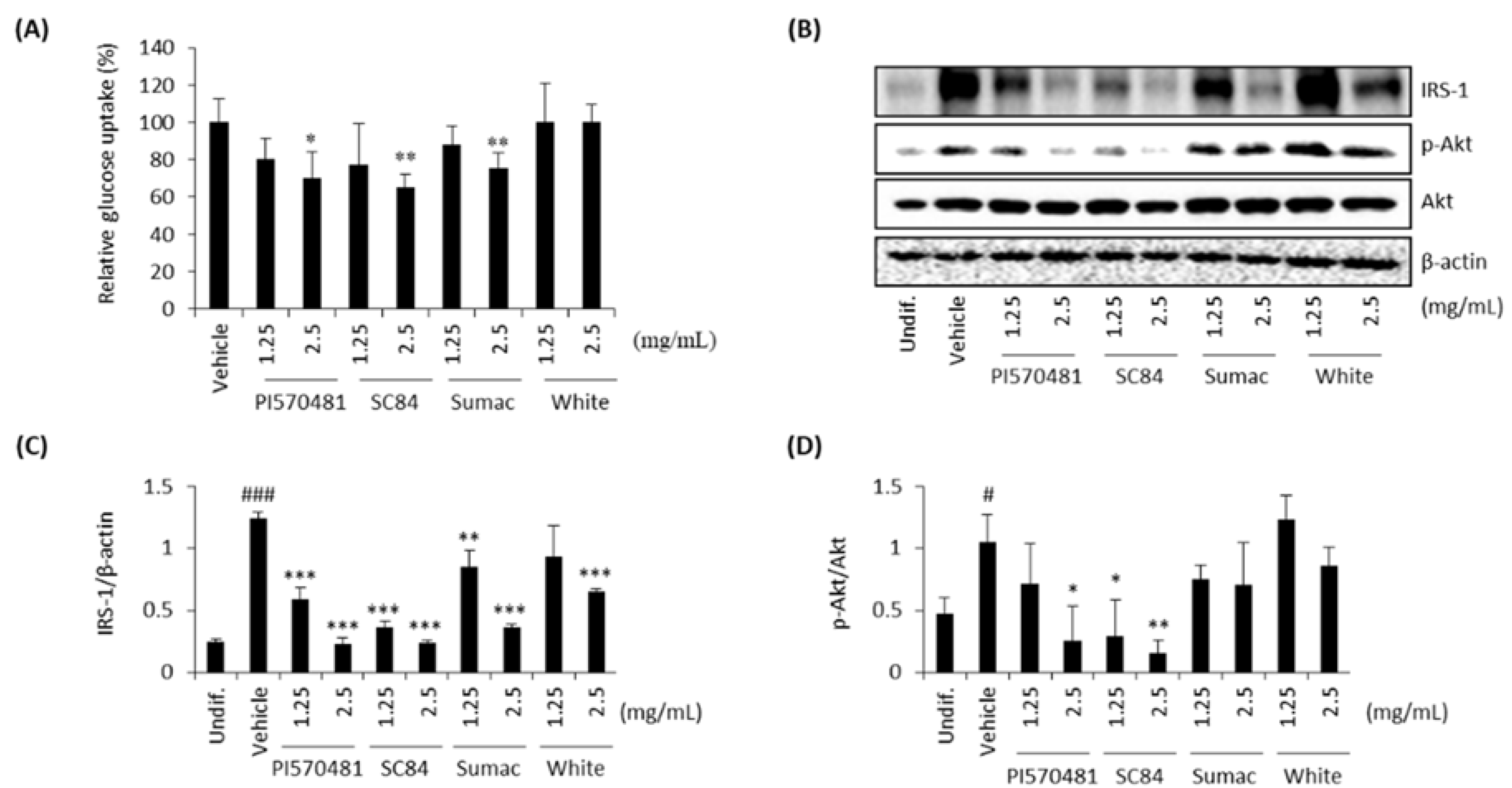
3.7. Sorghum Bran Extracts Repressed Insulin Signaling and Glucose Uptake in Differentiated 3T3-L1 Cells
3.8. Sorghum Bran Extracts Did Not Affect Lipolysis and Apoptosis of Differentiated in 3T3-L1 Cells
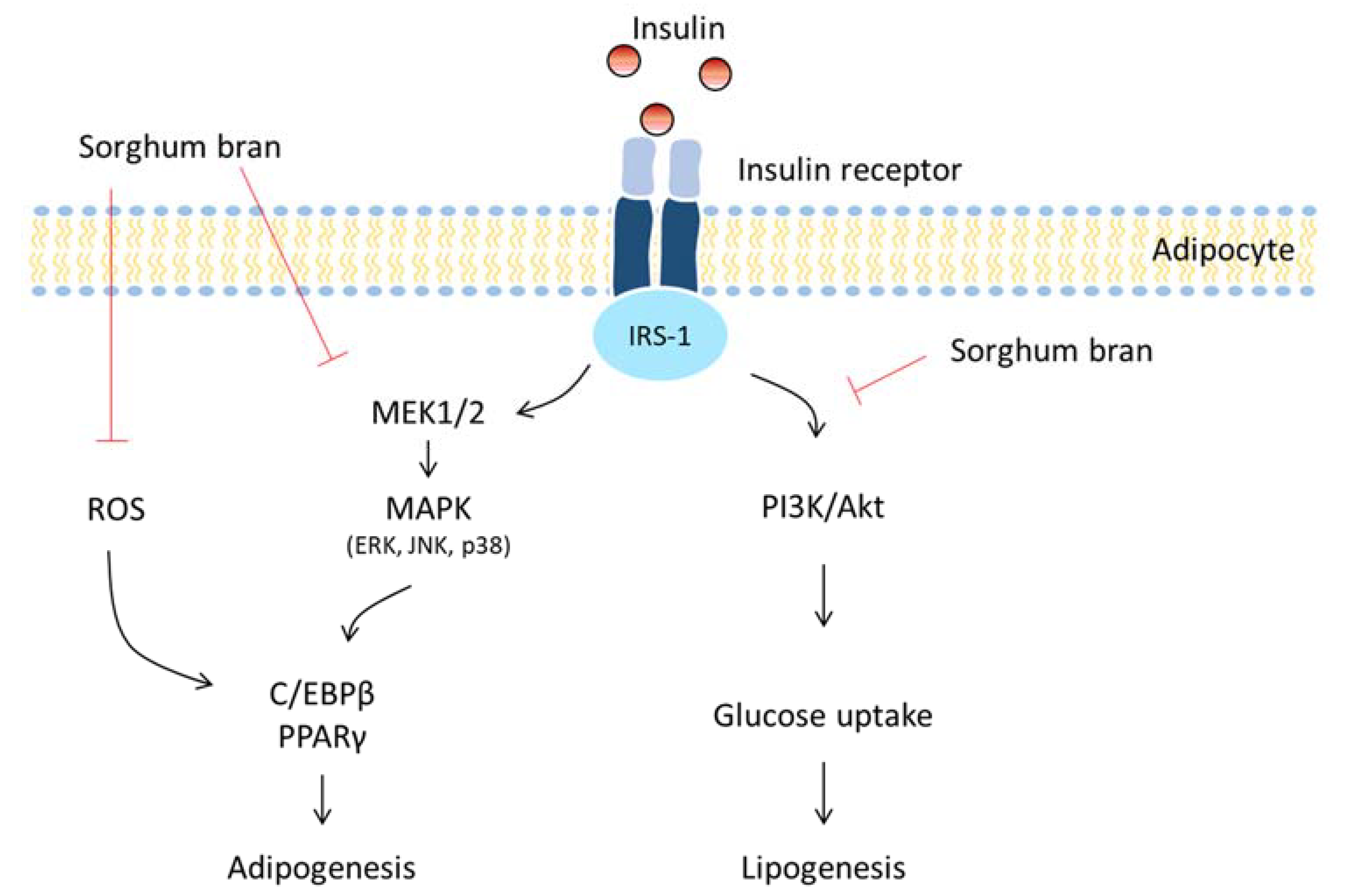
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Finkelstein, E.A.; Khavjou, O.A.; Thompson, H.; Trogdon, J.G.; Pan, L.; Sherry, B.; Dietz, W. Obesity and severe obesity forecasts through 2030. Am. J. Prev. Med. 2012, 42, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Maki, K.C.; Palacios, O.M.; Koecher, K.; Sawicki, C.M.; Livingston, K.A.; Bell, M.; Nelson Cortes, H.; McKeown, N.M. The Relationship between Whole Grain Intake and Body Weight: Results of Meta-Analyses of Observational Studies and Randomized Controlled Trials. Nutrients 2019, 11, 1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadebe, S.T.; Modi, A.T.; Mabhaudhi, T. Drought Tolerance and Water Use of Cereal Crops: A Focus on Sorghum as a Food Security Crop in Sub-Saharan Africa. J. Agron. Crop Sci. 2017, 203, 177–191. [Google Scholar] [CrossRef]
- Awika, J.M.; Rooney, L.W. Sorghum phytochemicals and their potential impact on human health. Phytochemistry 2004, 65, 1199–1221. [Google Scholar] [CrossRef]
- Dykes, L. Sorghum Phytochemicals and Their Potential Impact on Human Health. Methods Mol. Biol. 2019, 1931, 121–140. [Google Scholar]
- Amarakoon, D.; Lou, Z.; Lee, W.J.; Smolensky, D.; Lee, S.H. A mechanistic review: Potential chronic disease-preventive properties of sorghum. J. Sci. Food Agric. 2020, 101, 2641–2649. [Google Scholar] [CrossRef]
- Al-Mamary, M.; Molham, A.H.; Abdulwali, A.A.; Al-Obeidi, A. In vivo effects of dietary sorghum tannins on rabbit digestive enzymes and mineral absorption. Nutr. Res. 2001, 21, 1393–1401. [Google Scholar] [CrossRef]
- Chung, I.M.; Yeo, M.A.; Kim, S.J.; Kim, M.J.; Park, D.S.; Moon, H.I. Antilipidemic activity of organic solvent extract from Sorghum bicolor on rats with diet-induced obesity. Hum. Exp. Toxicol. 2011, 30, 1865–1868. [Google Scholar] [CrossRef]
- de Sousa, A.R.; de Castro Moreira, M.E.; Toledo, R.C.L.; dos Anjos Benjamin, L.; Queiroz, V.A.V.; Veloso, M.P.; de Souza Reis, K.; Martino, H.S.D. Extruded sorghum (Sorghum bicolor L.) reduces metabolic risk of hepatic steatosis in obese rats consuming a high fat diet. Food Res. Int. 2018, 112, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Lee, S.H.; Chung, I.M.; Park, Y. Sorghum extract exerts an anti-diabetic effect by improving insulin sensitivity via PPAR-γ in mice fed a high-fat diet. Nutr. Res. Pract. 2012, 6, 322–327. [Google Scholar] [CrossRef] [Green Version]
- Arbex, P.M.; de Castro Moreira, M.E.; Toledo, R.C.L.; de Morais Cardoso, L.; Pinheiro-Sant’ana, H.M.; dos Anjos Benjamin, L.; Licursi, L.; Carvalho, C.W.P.; Queiroz, V.A.V.; Martino, H.S.D. Extruded sorghum flour (Sorghum bicolor L.) modulate adiposity and inflammation in high fat diet-induced obese rats. J. Funct. Foods 2018, 42, 346–355. [Google Scholar] [CrossRef]
- Anunciação, P.C.; Cardoso, L.D.M.; Queiroz, V.A.V.; de Menezes, C.B.; de Carvalho, C.W.P.; Pinheiro-Sant’Ana, H.M.; Alfenas, R.D.C.G. Consumption of a drink containing extruded sorghum reduces glycaemic response of the subsequent meal. Eur. J. Nutr. 2018, 57, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Stefoska-Needham, A.; Beck, E.J.; Johnson, S.K.; Chu, J.; Tapsell, L.C. Flaked sorghum biscuits increase postprandial GLP-1 and GIP levels and extend subjective satiety in healthy subjects. Mol. Nutr. Food Res. 2016, 60, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Anunciação, P.C.; de Morais Cardoso, L.; Alfenas, R.D.C.G.; Queiroz, V.A.V.; Carvalho, C.W.P.; Martino, H.S.D.; Pinheiro-Sant’Ana, H.M. Extruded sorghum consumption associated with a caloric restricted diet reduces body fat in overweight men: A randomized controlled trial. Food Res. Int. 2019, 119, 693–700. [Google Scholar] [CrossRef] [Green Version]
- Uchimiya, M. Proton-Coupled Electron Transfers of Defense Phytochemicals in Sorghum (Sorghum bicolor (L.) Moench). J. Agric. Food Chem. 2020, 68, 12978–12983. [Google Scholar] [CrossRef]
- Smolensky, D.; Rhodes, D.; McVey, D.S.; Fawver, Z.; Perumal, R.; Herald, T.; Noronha, L. High-Polyphenol Sorghum Bran Extract Inhibits Cancer Cell Growth Through ROS Induction, Cell Cycle Arrest, and Apoptosis. J. Med. Food 2018, 21, 990–998. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.; Herald, T.; Cox, S.; Noronha, L.; Perumal, R.; Lee, H.S.; Smolensky, D. Anti-cancer activity of a novel high phenolic sorghum bran in human colon cancer cells. Oxid. Med. Cell. Longev. 2020, 2020, 2890536. [Google Scholar] [CrossRef]
- Rhodes, D.; Gadgil, P.; Perumal, R.; Tesso, T.; Herald, T.J. Natural Variation and Genome-Wide Association Study of Antioxidants in a Diverse Sorghum Collection. Cereal Chem. 2017, 94, 190–198. [Google Scholar] [CrossRef]
- Cox, S.; Noronha, L.; Herald, T.; Bean, S.; Lee, S.H.; Perumal, R.; Wang, W.; Smolensky, D. Evaluation of ethanol-based extraction conditions of sorghum bran bioactive compounds with downstream anti-proliferative properties in human cancer cells. Heliyon 2019, 5, e01589. [Google Scholar] [CrossRef] [Green Version]
- Singleton, V.L.; Rossi, J.A.J. Colorimetry of total phenolics with phosphomolybdic–phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Herald, T.J.; Gadgil, P.; Tilley, M. High-throughput micro plate assays for screening flavonoid content and DPPH-scavenging activity in sorghum bran and flour. J. Sci. Food Agric. 2012, 92, 2326–2331. [Google Scholar] [CrossRef] [PubMed]
- Herald, T.J.; Gadgil, P.; Perumal, R.; Bean, S.R.; Wilson, J.D. High-throughput micro-plate HCl–vanillin assay for screening tannin content in sorghum grain. J. Sci. Food Agric. 2014, 94, 2133–2136. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Santhakumar, A.B.; Chinkwo, K.A.; Wu, G.; Johnson, S.K.; Blanchard, C.L. Characterization of phenolic compounds and antioxidant activity in sorghum grains. J. Cereal Sci. 2018, 84, 103–111. [Google Scholar] [CrossRef]
- Rhodes, D.H.; Hoffmann, L., Jr.; Rooney, W.L.; Ramu, P.; Morris, G.P.; Kresovich, S. Genome-Wide Association Study of Grain Polyphenol Concentrations in Global Sorghum [Sorghum bicolor (L.) Moench] Germplasm. J. Agric. Food Chem. 2014, 62, 10916–10927. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.A.; Njobeh, P.B.; Kayitesi, E. Fermentation by Lactobacillus fermentum strains (singly and in combination) enhances the properties of ting from two whole grain sorghum types. J. Cereal Sci. 2018, 82, 49–56. [Google Scholar] [CrossRef]
- Kumari, P.K.; Umakanth, A.V.; Narsaiah, T.B.; Uma, A. Exploring anthocyanins, antioxidant capacity and α-glucosidase inhibition in bran and flour extracts of selected sorghum genotypes. Food Biosci. 2021, 41, 100979. [Google Scholar] [CrossRef]
- Barros, F.; Dykes, L.; Awika, J.M.; Rooney, L.W. Accelerated solvent extraction of phenolic compounds from sorghum brans. J. Cereal Sci. 2013, 58, 305–312. [Google Scholar] [CrossRef]
- Shelembe, J.S.; Cromarty, D.; Bester, M.; Minnaar, A.; Duodu, K.G. Effect of acidic condition on phenolic composition and antioxidant potential of aqueous extracts from sorghum (Sorghum bicolor) bran. J. Food Biochem. 2014, 38, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Chiremba, C.; Taylor, J.R.; Rooney, L.W.; Beta, T. Phenolic acid content of sorghum and maize cultivars varying in hardness. Food Chem. 2012, 134, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Irakli, M.N.; Samanidou, V.F.; Biliaderis, C.G.; Papadoyannis, I.N. Simultaneous determination of phenolic acids and flavonoids in rice using solid-phase extraction and RP-HPLC with photodiode array detection. J. Sep. Sci. 2012, 35, 1603–1611. [Google Scholar] [CrossRef]
- Lee, J.; Yue, Y.; Park, Y.; Lee, S.H. 3,3’-Diindolylmethane Suppresses Adipogenesis Using AMPKalpha-Dependent Mechanism in 3T3-L1 Adipocytes and Caenorhabditis elegans. J. Med. Food 2017, 20, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Imm, J.Y.; Lee, S.H. Beta-Catenin Mediates Anti-adipogenic and Anticancer Effects of Arctigenin in Preadipocytes and Breast Cancer Cells. J. Agric. Food Chem. 2017, 65, 2513–2520. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.; Lee, J.; Lee, S.H. Synergistic anticancer activity of capsaicin and 3,3’-diindolylmethane in human colorectal cancer. J. Agric. Food Chem. 2015, 63, 4297–4304. [Google Scholar] [CrossRef] [PubMed]
- Price, M.L.; Van Scoyoc, S.; Butler, L.G. A critical evaluation of the vanillin reaction as an assay for tannin in sorghum grain. J. Agric. Food Chem. 1978, 26, 1214–1218. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Puigserver, P.; Spiegelman, B.M. Transcriptional activation of adipogenesis. Curr. Opin. Cell Biol. 1999, 11, 689–694. [Google Scholar] [CrossRef]
- Chang, E.; Kim, C.Y. Natural Products and Obesity: A Focus on the Regulation of Mitotic Clonal Expansion during Adipogenesis. Molecules 2019, 24, 1157. [Google Scholar] [CrossRef] [Green Version]
- Villiers, D.D.; Potgieter, M.; Ambele, M.A.; Adam, L.; Durandt, C.; Pepper, M.S. The Role of Reactive Oxygen Species in Adipogenic Differentiation. Adv. Exp. Med. Biol. 2018, 1083, 125–144. [Google Scholar]
- Lee, H.; Lee, Y.J.; Choi, H.; Ko, E.H.; Kim, J.W. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J. Biol. Chem. 2009, 284, 10601–10609. [Google Scholar] [CrossRef] [Green Version]
- Bost, F.; Aouadi, M.; Caron, L.; Binétruy, B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 2005, 87, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Son, Y.; Cheong, Y.K.; Kim, N.H.; Chung, H.T.; Kang, D.G.; Pae, H.O. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J. Signal Transduct. 2011, 2011, 792639. [Google Scholar] [CrossRef] [PubMed]
- Cignarelli, A.; Genchi, V.A.; Perrini, S.; Natalicchio, A.; Laviola, L.; Giorgino, F. Insulin and Insulin Receptors in Adipose Tissue Development. Int. J. Mol. Sci. 2019, 20, 759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krycer, J.R.; Quek, L.E.; Francis, D.; Zadoorian, A.; Weiss, F.C.; Cooke, K.C.; Nelson, M.E.; Diaz-Vegas, A.; Humphrey, S.J.; Scalzo, R.; et al. Insulin signaling requires glucose to promote lipid anabolism in adipocytes. J. Biol. Chem. 2020, 295, 13250–13266. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.M.; Griesel, B.A.; Gurley, J.M.; Szweda, L.I.; Olson, A.L. Glucose availability controls adipogenesis in mouse 3T3-L1 adipocytes via up-regulation of nicotinamide metabolism. J. Biol. Chem. 2017, 292, 18556–18564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhao, H.; Jin, Q.; You, W.; Cheng, H.; Liu, Y.; Song, E.; Liu, G.; Tan, X.; Zhang, X.; et al. Resveratrol induces apoptosis and inhibits adipogenesis by stimulating the SIRT1-AMPKalpha-FOXO1 signalling pathway in bovine intramuscular adipocytes. Mol. Cell. Biochem. 2018, 439, 213–223. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, N.; Zhang, Z.; Li, W.; Zhu, W. Resveratrol induces cell apoptosis in adipocytes via AMPK activation. Biochem. Biophys. Res. Commun. 2015, 457, 608–613. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, H.S.; Lee, J.; Amarakoon, D.; Lou, Z.; Noronha, L.E.; Herald, T.J.; Perumal, R.; Smolensky, D. Polyphenol Containing Sorghum Brans Exhibit an Anti-Cancer Effect in Apc Min/+ Mice Treated with Dextran Sodium Sulfate. Int. J. Mol. Sci. 2021, 22, 8286. [Google Scholar] [CrossRef]
- Li, W.; Saud, S.M.; Young, M.R.; Chen, G.; Hua, B. Targeting AMPK for cancer prevention and treatment. Oncotarget 2015, 6, 7365–7378. [Google Scholar] [CrossRef] [Green Version]
- Dagon, Y.; Avraham, Y.; Berry, E.M. AMPK activation regulates apoptosis, adipogenesis, and lipolysis by eIF2alpha in adipocytes. Biochem. Biophys. Res. Commun. 2006, 340, 43–47. [Google Scholar] [CrossRef]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef] [Green Version]
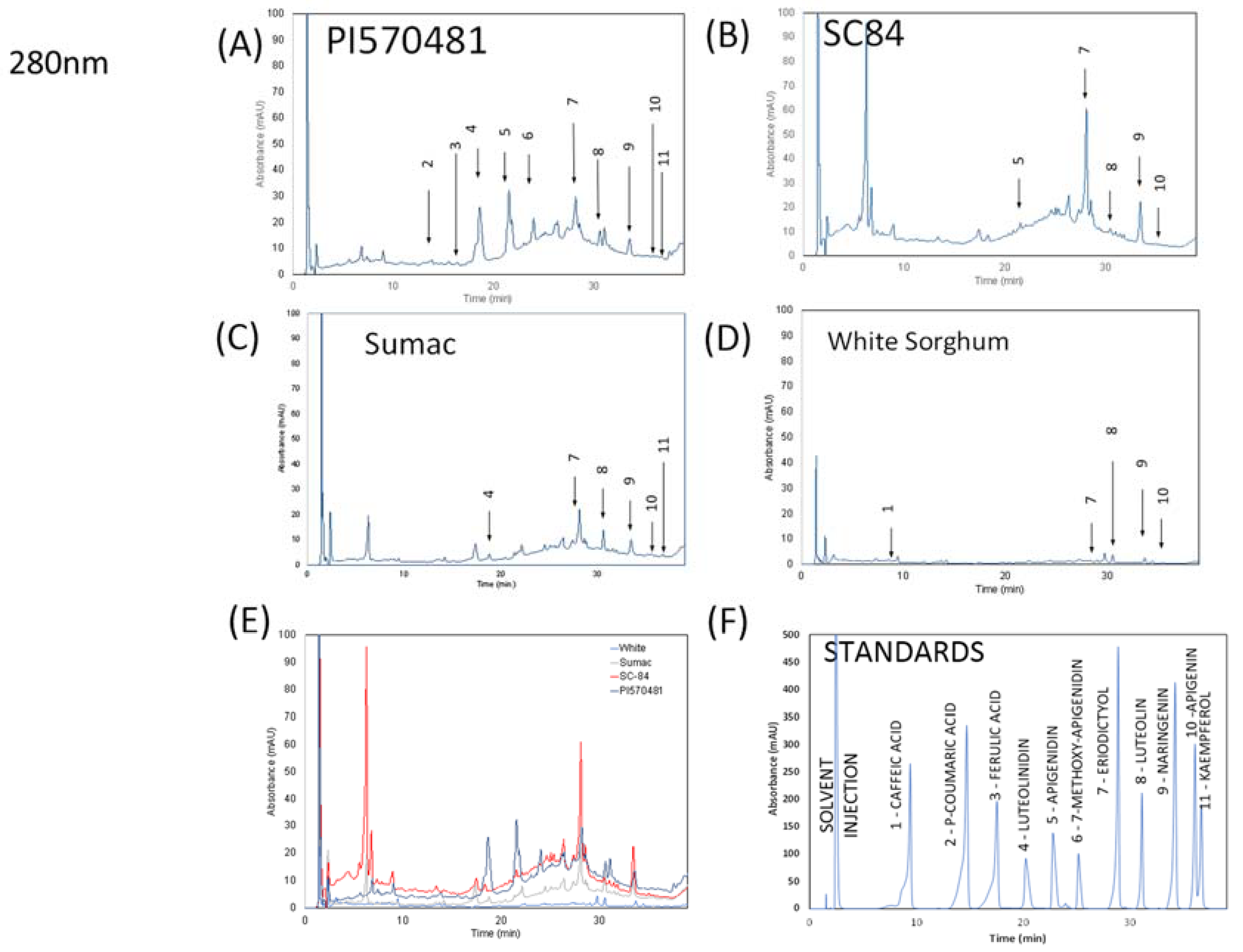
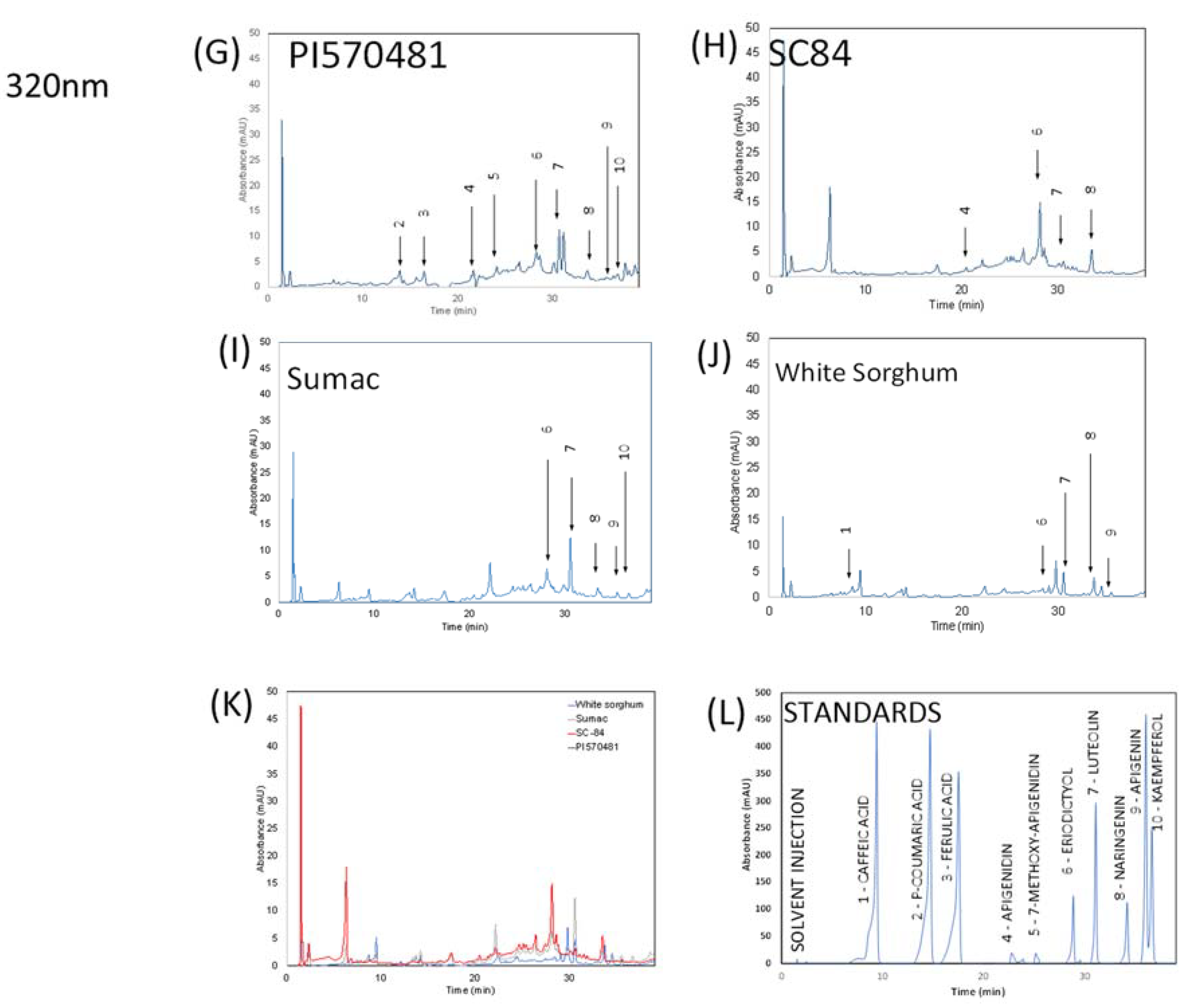

| Compound | Peak Number | Wavelength (nm) | Retention Time (min) | PI570481 | SC84 | Sumac | White |
|---|---|---|---|---|---|---|---|
| Caffeic acid | 1 | 320 | 8.64 ± 0.05 | ND† | ND† | ND† | 9.84 ± 0.04 |
| p-Coumaric acid | 2 | 320 | 13.43 ± 0.07 | 50.23 ± 0.33 | ND† | ND† | ND† |
| Ferulic acid | 3 | 320 | 16.06 ± 0.08 | 15.70 ± 1.71 | ND† | ND† | ND† |
| Luteolinidin | 4 | 280 | 18.40 ± 0.50 | 607.61 ± 145.21 | ND† | 22.15 ± 3.20 | ND† |
| Apigenidin | 5 | 280 | 21.32 ± 0.40 | 103.45 ± 19 | 82.96 ± 8.37 | ND† | ND† |
| 7-methoxy apigenidin | 6 | 320 | 24.03 ± 0.29 | 226.74 ± 20.72 | ND† | ND† | ND† |
| Eriodictyol | 7 | 280 | 28.23 ± 0.26 | 148.42 ± 95 | 483.77 ± 32.24 | 39.98 ± 2.40 | NQ‡ |
| Luteolin | 8 | 280 | 30.69 ± 0.30 | 128.06 ± 4.35 | 68.78 ± 1.05 | 57.01 ± 0.48 | 16.15 ± 0.34 |
| Naringenin | 9 | 280 | 33.48 ± 0.05 | 32.74 ± 0.75 | 98.78 ± 2.48 | 15.32 ± 0.48 | 1.94 ± 0.13 |
| Apigenin | 10 | 320 | 35.55 ± 0.06 | NQ‡ | NQ‡ | 33.52 ± 0.05 | NQ‡ |
| Kaempferol | 11 | 320 | 36.12 ± 0.13 | 35.36 ± 0.06 | ND† | 12.52 ± 0.03 | ND† |
| Total polyphenols mg GAE/g * | 38.11 ± 1.38 | 55.57 ± 4.78 | 12.4 ± 0.67 | 0.67 ± 0.88 | |||
| Condensed Tannins mg CE/g # | 179.01 ± 20.99 | 415.83 ± 89.61 | 13.03 ± 0.27 | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-S.; Santana, Á.L.; Peterson, J.; Yucel, U.; Perumal, R.; De Leon, J.; Lee, S.-H.; Smolensky, D. Anti-Adipogenic Activity of High-Phenolic Sorghum Brans in Pre-Adipocytes. Nutrients 2022, 14, 1493. https://doi.org/10.3390/nu14071493
Lee H-S, Santana ÁL, Peterson J, Yucel U, Perumal R, De Leon J, Lee S-H, Smolensky D. Anti-Adipogenic Activity of High-Phenolic Sorghum Brans in Pre-Adipocytes. Nutrients. 2022; 14(7):1493. https://doi.org/10.3390/nu14071493
Chicago/Turabian StyleLee, Hee-Seop, Ádina L. Santana, Jaymi Peterson, Umut Yucel, Ramasamy Perumal, Joaquin De Leon, Seong-Ho Lee, and Dmitriy Smolensky. 2022. "Anti-Adipogenic Activity of High-Phenolic Sorghum Brans in Pre-Adipocytes" Nutrients 14, no. 7: 1493. https://doi.org/10.3390/nu14071493
APA StyleLee, H.-S., Santana, Á. L., Peterson, J., Yucel, U., Perumal, R., De Leon, J., Lee, S.-H., & Smolensky, D. (2022). Anti-Adipogenic Activity of High-Phenolic Sorghum Brans in Pre-Adipocytes. Nutrients, 14(7), 1493. https://doi.org/10.3390/nu14071493








