Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer
Abstract
1. Introduction
2. Materials and Methods
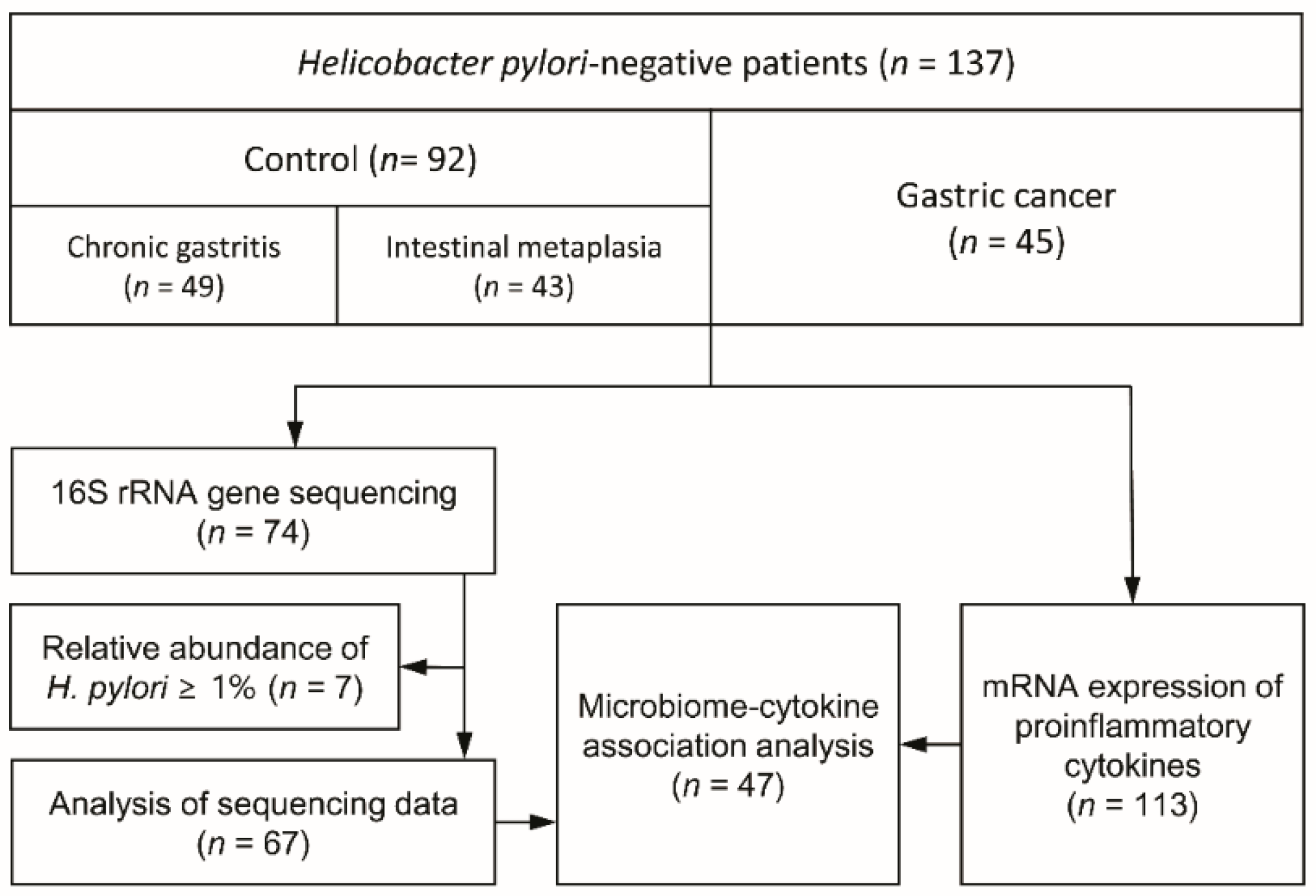
2.1. Study Participants and Sample Collection
2.2. DNA Extraction from Gastric Biopsy Samples and 16S rRNA Gene Sequencing
2.3. Sequence Data Processing
2.4. Microbial Profiling
2.5. RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-qPCR)
2.6. Correlation between the Altered Gastric Microbiome and Cytokine Response
3. Results
3.1. Study Demographics
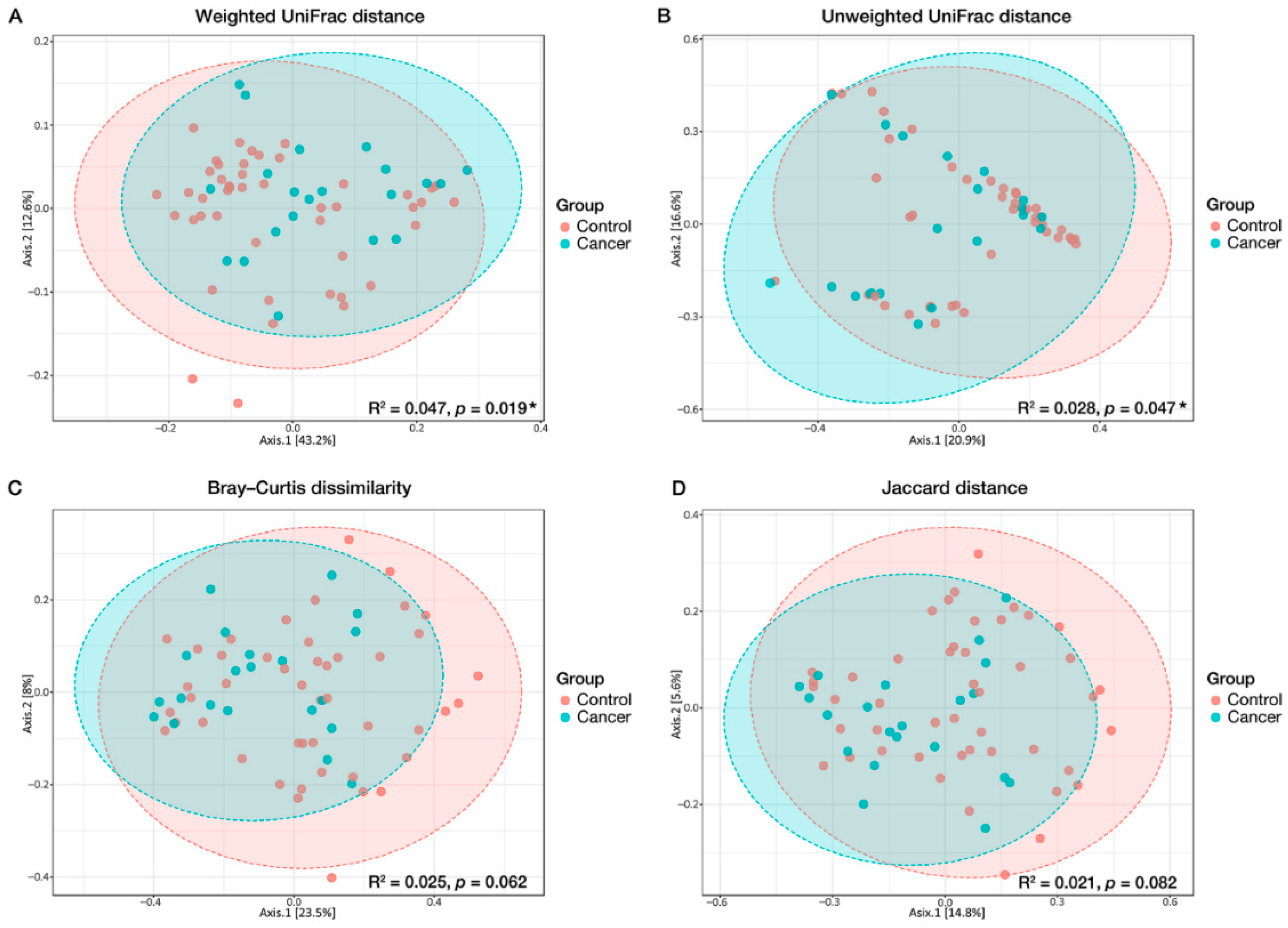
3.2. Microbial Diversity Associated with GC
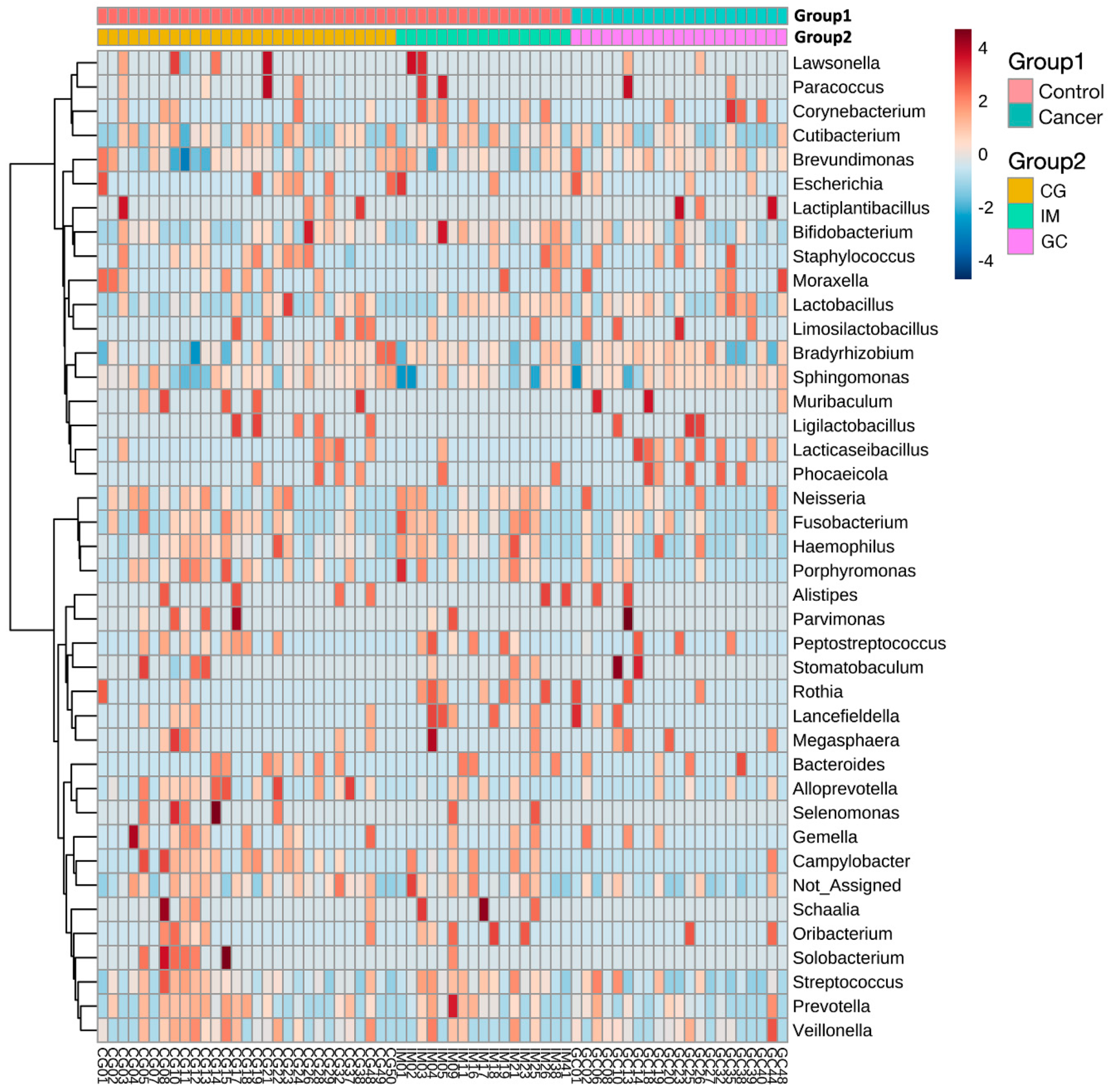
3.3. Taxonomic Alterations in GC
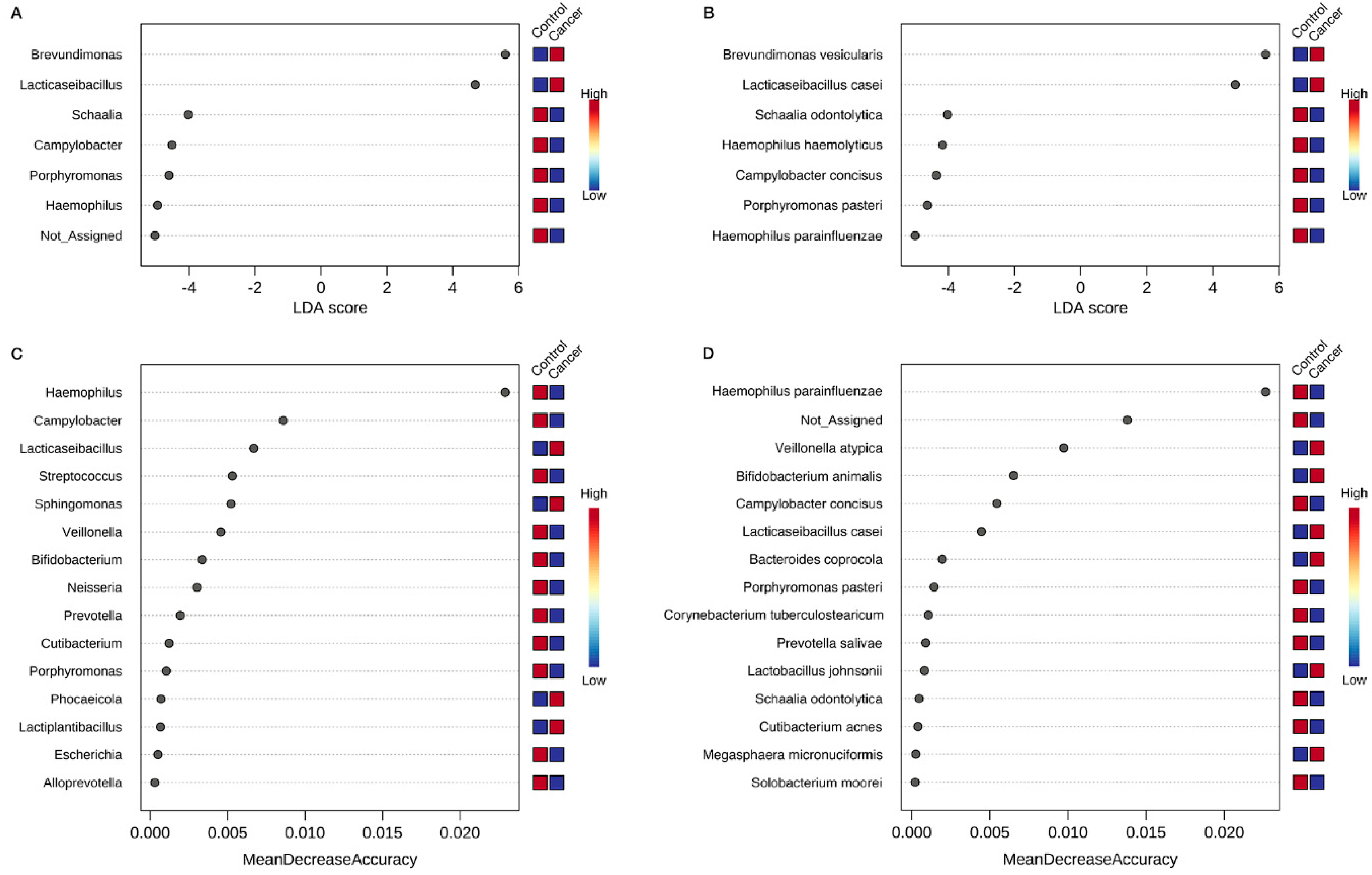
3.4. Identification of Key Gastric Microbiota Associated with GC via LEfSe and RF Analyses
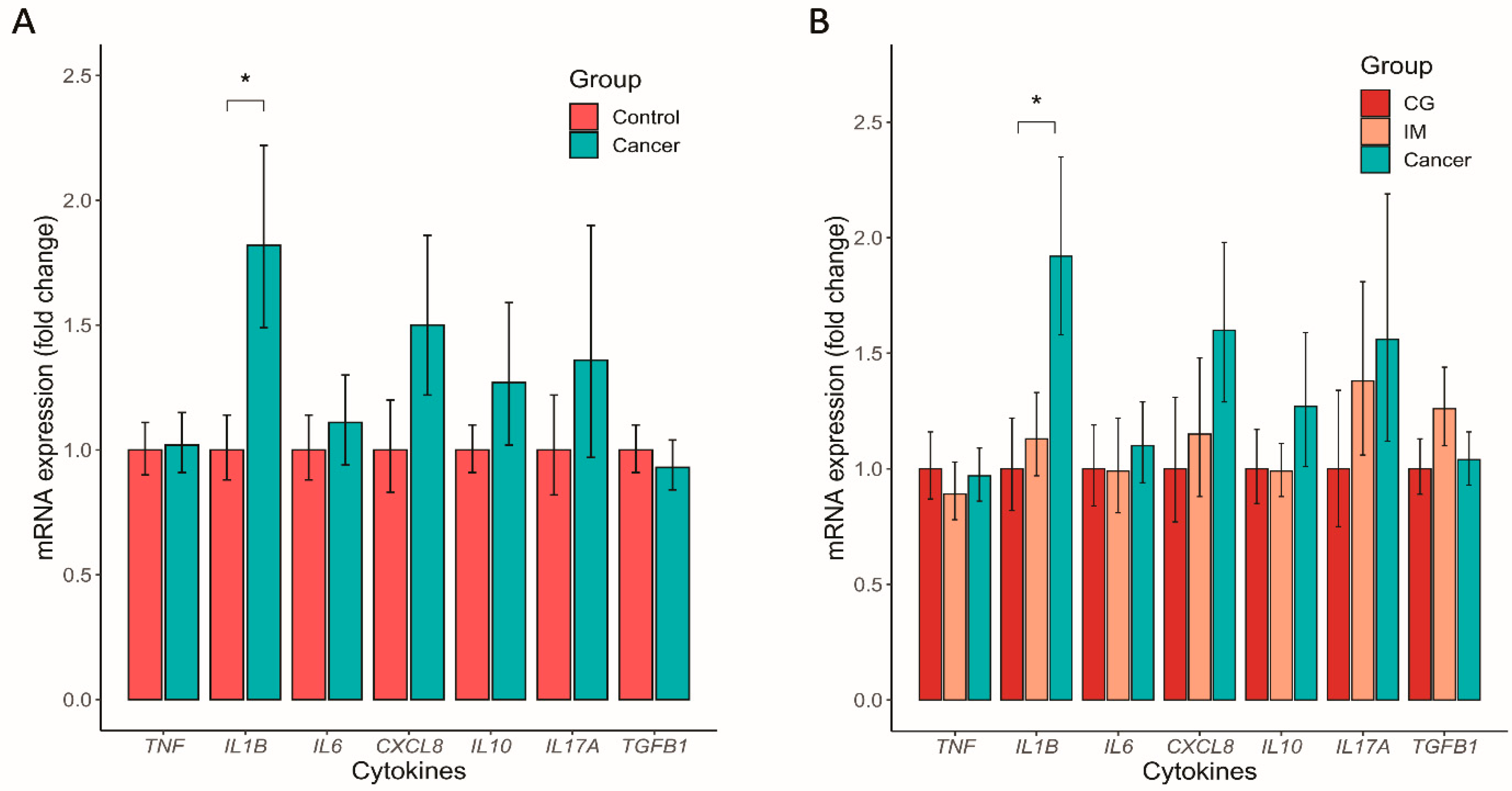
3.5. Altered Mucosal Inflammatory Cytokine Expression in GC
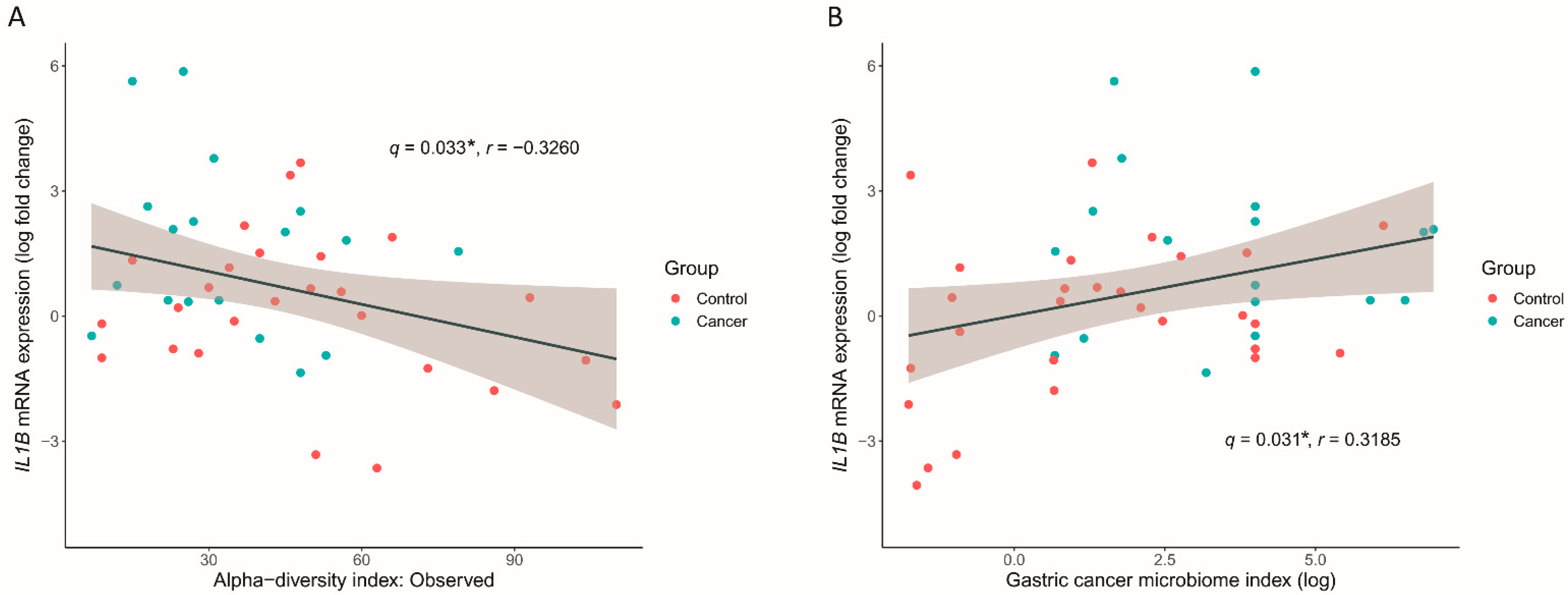
3.6. GC Microbiome Index Associated with the Inflammatory Cytokine Response
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Engstrand, L.; Graham, D.Y. Microbiome and Gastric Cancer. Dig. Dis. Sci. 2020, 65, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Correa, P. Human gastric carcinogenesis: A multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar] [PubMed]
- Ushijima, T.; Hattori, N. Molecular pathways: Involvement of Helicobacter pylori-triggered inflammation in the formation of an epigenetic field defect, and its usefulness as cancer risk and exposure markers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.C.; Yuan, Y.; Moayyedi, P. Helicobacter pylori eradication therapy to prevent gastric cancer: Systematic review and meta-analysis. Gut 2020, 69, 2113–2121. [Google Scholar] [CrossRef]
- Li, W.Q.; Zhang, J.Y.; Ma, J.L.; Li, Z.X.; Zhang, L.; Zhang, Y.; Guo, Y.; Zhou, T.; Li, J.Y.; Shen, L.; et al. Effects of Helicobacter pylori treatment and vitamin and garlic supplementation on gastric cancer incidence and mortality: Follow-up of a randomized intervention trial. BMJ 2019, 366, l5016. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.J.; Kook, M.C.; Kim, Y.I.; Cho, S.J.; Lee, J.Y.; Kim, C.G.; Park, B.; Nam, B.H. Helicobacter pylori Therapy for the Prevention of Metachronous Gastric Cancer. N. Engl. J. Med. 2018, 378, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, X.; Zeng, R.; Wu, Q.; Sun, H.; Wu, W.; Zhang, X.; Sun, G.; Yan, B.; Wu, L.; et al. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front. Microbiol. 2020, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Gantuya, B.; El Serag, H.B.; Matsumoto, T.; Ajami, N.J.; Uchida, T.; Oyuntsetseg, K.; Bolor, D.; Yamaoka, Y. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment. Pharmacol. Ther. 2020, 51, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Lee, A.R.; Lee, Y.R.; Eun, C.S.; Lee, S.K.; Han, D.S. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter 2019, 24, e12547. [Google Scholar] [CrossRef] [PubMed]
- Li, T.H.; Qin, Y.; Sham, P.C.; Lau, K.S.; Chu, K.M.; Leung, W.K. Alterations in Gastric Microbiota After H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis. Sci. Rep. 2017, 7, 44935. [Google Scholar] [CrossRef] [PubMed]
- Aviles-Jimenez, F.; Vazquez-Jimenez, F.; Medrano-Guzman, R.; Mantilla, A.; Torres, J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014, 4, 4202. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Ono, S.; Ichikura, T.; Matsumoto, Y.; Yamamoto, J.; Hase, K. Roles of inflammatory cytokines in the progression of gastric cancer: Friends or foes? Gastric Cancer Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2010, 13, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Dreskin, B.W.; Luu, K.; Dong, T.S.; Benhammou, J.; Lagishetty, V.; Vu, J.; Sanford, D.; Durazo, F.; Agopian, V.G.; Jacobs, J.P.; et al. Specimen Collection and Analysis of the Duodenal Microbiome. J. Vis. Exp. 2021, 167, 33522511. [Google Scholar] [CrossRef] [PubMed]
- Son, B.K.; Kim, B.W.; Kim, W.H.; Myung, D.S.; Cho, Y.S.; Jang, B.I.; Disinfection Management and Conscious Sedation Committee of Korean Society of Gastrointestinal Endoscopy. Korean Society of Gastrointestinal Endoscopy Guidelines for Endoscope Reprocessing. Clin. Endosc. 2017, 50, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Cao, Q.; Sun, X.; Rajesh, K.; Chalasani, N.; Gelow, K.; Katz, B.; Shah, V.H.; Sanyal, A.J.; Smirnova, E. Effects of Rare Microbiome Taxa Filtering on Statistical Analysis. Front. Microbiol. 2020, 11, 607325. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.H. HuangLin/ANCOM: Third Release of ANCOM (V2.1); Zenodo: Genève, Switzerland, 2019. [Google Scholar] [CrossRef]
- Kim, J.; Kim, N.; Jo, H.J.; Park, J.H.; Nam, R.H.; Seok, Y.J.; Kim, Y.R.; Kim, J.S.; Kim, J.M.; Kim, J.M.; et al. An Appropriate Cutoff Value for Determining the Colonization of Helicobacter pylori by the Pyrosequencing Method: Comparison with Conventional Methods. Helicobacter 2015, 20, 370–380. [Google Scholar] [CrossRef]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef]
- Castano-Rodriguez, N.; Goh, K.L.; Fock, K.M.; Mitchell, H.M.; Kaakoush, N.O. Dysbiosis of the microbiome in gastric carcinogenesis. Sci. Rep. 2017, 7, 15957. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Seo, H.; Kang, C.S.; Shin, T.S.; Kim, J.W.; Park, J.M.; Kim, J.G.; Kim, Y.K. Dysbiotic change in gastric microbiome and its functional implication in gastric carcinogenesis. Sci. Rep. 2022, 12, 4285. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.S.; Kim, B.K.; Han, D.S.; Kim, S.Y.; Kim, K.M.; Choi, B.Y.; Song, K.S.; Kim, Y.S.; Kim, J.F. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014, 19, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Stewart, O.A.; Wu, F.; Chen, Y. The role of gastric microbiota in gastric cancer. Gut Microbes 2020, 11, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.J.Y.; Coker, O.O.; Chu, E.; Szeto, C.H.; Luk, S.T.Y.; Lau, H.C.H.; Yu, J. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut 2020, 69, 1572–1580. [Google Scholar] [CrossRef]
- Vinasco, K.; Mitchell, H.M.; Kaakoush, N.O.; Castano-Rodriguez, N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188309. [Google Scholar] [CrossRef]
- Hwang, J.W.; Baek, Y.M.; Yang, K.E.; Yoo, H.S.; Cho, C.K.; Lee, Y.W.; Park, J.; Eom, C.Y.; Lee, Z.W.; Choi, J.S.; et al. Lactobacillus casei extract induces apoptosis in gastric cancer by inhibiting NF-kappaB and mTOR-mediated signaling. Integr. Cancer Ther. 2013, 12, 165–173. [Google Scholar] [CrossRef]
- Li, Z.P.; Liu, J.X.; Lu, L.L.; Wang, L.L.; Xu, L.; Guo, Z.H.; Dong, Q.J. Overgrowth of Lactobacillus in gastric cancer. World J. Gastrointest. Oncol. 2021, 13, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.M.; Mercante, J.W.; Neish, A.S. Reactive oxygen production induced by the gut microbiota: Pharmacotherapeutic implications. Curr. Med. Chem. 2012, 19, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Lertpiriyapong, K.; Whary, M.T.; Muthupalani, S.; Lofgren, J.L.; Gamazon, E.R.; Feng, Y.; Ge, Z.; Wang, T.C.; Fox, J.G. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014, 63, 54–63. [Google Scholar] [CrossRef]
- Huang, K.; Gao, X.; Wu, L.; Yan, B.; Wang, Z.; Zhang, X.; Peng, L.; Yu, J.; Sun, G.; Yang, Y. Salivary Microbiota for Gastric Cancer Prediction: An Exploratory Study. Front. Cell. Infect. Microbiol. 2021, 11, 640309. [Google Scholar] [CrossRef]
- Akutko, K.; Matusiewicz, K. Campylobacter concisus as the etiologic agent of gastrointestinal diseases. Adv. Clin. Exp. Med. 2017, 26, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Raubenheimer, K.; Bondonno, C.; Blekkenhorst, L.; Wagner, K.H.; Peake, J.M.; Neubauer, O. Effects of dietary nitrate on inflammation and immune function, and implications for cardiovascular health. Nutr. Rev. 2019, 77, 584–599. [Google Scholar] [CrossRef]
- Liu, F.; Ma, R.; Wang, Y.; Zhang, L. The Clinical Importance of Campylobacter concisus and Other Human Hosted Campylobacter Species. Front. Cell. Infect. Microbiol. 2018, 8, 243. [Google Scholar] [CrossRef]
- Blackett, K.L.; Siddhi, S.S.; Cleary, S.; Steed, H.; Miller, M.H.; Macfarlane, S.; Macfarlane, G.T.; Dillon, J.F. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: Association or causality? Aliment. Pharmacol. Ther. 2013, 37, 1084–1092. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Ter Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016, 167, 1125–1136.e8. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Chen, C.C.; Chiang, H.L.; Liou, J.M.; Chang, C.M.; Lu, T.P.; Chuang, E.Y.; Tai, Y.C.; Cheng, C.; Lin, H.Y.; et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflamm. 2019, 16, 129. [Google Scholar] [CrossRef]
- Outlioua, A.; Badre, W.; Desterke, C.; Echarki, Z.; El Hammani, N.; Rabhi, M.; Riyad, M.; Karkouri, M.; Arnoult, D.; Khalil, A.; et al. Gastric IL-1beta, IL-8, and IL-17A expression in Moroccan patients infected with Helicobacter pylori may be a predictive signature of severe pathological stages. Cytokine 2020, 126, 154893. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, N.; Park, J.H.; Choi, S.; Shin, C.M.; Lee, O.J. Helicobacter pylori Eradication Induced Constant Decrease in Interleukin- 1B Expression over More Than 5 Years in Patients with Gastric Cancer and Dysplasia. Gut Liver 2020, 14, 735–745. [Google Scholar] [CrossRef]

| Gastric Cancer (n = 45) | Control (n = 92) | p Value | |
|---|---|---|---|
| Age, years, mean ± SD | 62.9 ± 10.2 | 50.7 ± 13.6 | <0.001 |
| Sex, n (%) | 0.048 | ||
| Female | 14 (31.1) | 45 (48.9) | |
| Male | 31 (68.9) | 47 (51.1) | |
| Body mass index, n (%) | 0.016 | ||
| <25 kg/m2 | 24 (53.3) | 67 (73.6) | |
| ≥25 kg/m2 | 21 (46.7) | 24 (26.4) | |
| Gastric mucosal atrophy, n (%) | <0.001 | ||
| Absent | 2 (4.4) | 44 (47.8) | |
| Present | 43 (95.6) | 48 (52.2) | |
| Intestinal metaplasia, n (%) | <0.001 | ||
| Absent | 4 (8.9) | 49 (53.3) | |
| Present | 41 (91.1) | 43 (46.7) |
| Taxonomic Assignment a | Relative Abundance (Mean%) | ANCOM2 b (Unadjusted) | ANCOM2 b (Adjusted for Age, Sex, and BMI) | MaAsLin2 (Unadjusted) | MaAsLin2 (Adjusted for Age, Sex, and BMI) | |||
|---|---|---|---|---|---|---|---|---|
| Control | Cancer | Detection level | Detection level | log2FC c | pd | log2FC c | pd | |
| p_Proteobacteria; | ||||||||
| o_Gammaproteobacteria; | ||||||||
| c_Pasteurellales; | 6.29% | 4.45% | 0.8 | 0.8 | −2.283 | 0.006 | −1.862 | 0.052 |
| f_Pasteurellaceae; | 6.29% | 4.45% | 0.8 | 0.8 | −2.283 | 0.006 | −1.862 | 0.052 |
| g_Haemophilus; | 6.29% | 4.45% | 0.8 | 0.8 | −2.283 | 0.006 | −1.862 | 0.052 |
| s_parainfluenzae; | 4.99% | 2.92% | 0.9 | 0.9 | −1.876 | 0.005 | −1.488 | 0.052 |
| p_Proteobacteria; | ||||||||
| o_Epsilonproteobacteria; | ||||||||
| c_Campylobacterales; | ||||||||
| f_Campylobacteraceae; | ||||||||
| g_Campylobacter | 1.04% | 0.38% | 0.7 | 0.6 | −2.457 | 0.002 | −1.784 | 0.047 |
| p_Firmicutes; | ||||||||
| o_Bacilli; | ||||||||
| c_Lactobacillales; | ||||||||
| f_Lactobacillaceae; | ||||||||
| g_Lacticaseibacillus; | 0.29% | 1.24% | 0.6 | 0.7 | 1.017 | 0.021 | 1.405 | 0.006 |
| s_casei | 0.29% | 1.24% | 0.7 | 0.8 | 1.017 | 0.021 | 1.405 | 0.006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-N.; Kim, M.-J.; Jacobs, J.P.; Yang, H.-J. Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer. Nutrients 2022, 14, 4981. https://doi.org/10.3390/nu14234981
Kim H-N, Kim M-J, Jacobs JP, Yang H-J. Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer. Nutrients. 2022; 14(23):4981. https://doi.org/10.3390/nu14234981
Chicago/Turabian StyleKim, Han-Na, Min-Jeong Kim, Jonathan P. Jacobs, and Hyo-Joon Yang. 2022. "Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer" Nutrients 14, no. 23: 4981. https://doi.org/10.3390/nu14234981
APA StyleKim, H.-N., Kim, M.-J., Jacobs, J. P., & Yang, H.-J. (2022). Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer. Nutrients, 14(23), 4981. https://doi.org/10.3390/nu14234981







