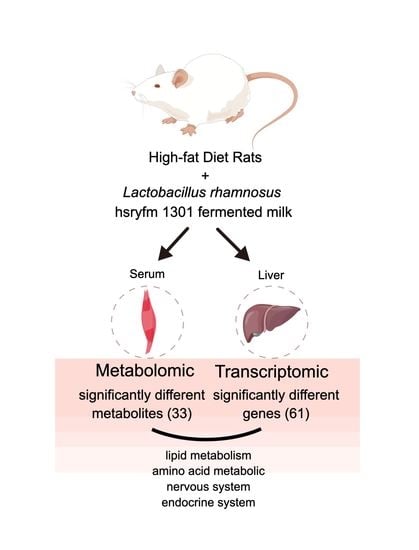
Effect of Lactobacillus rhamnosus hsryfm 1301 Fermented Milk on Lipid Metabolism Disorders in High-Fat-Diet Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Probiotic Bacteria and Fermented Milk Preparation
2.2. Animals
2.3. Blood Lipid Levels and FFAs in Serum
2.4. Metabolomics
2.4.1. Sample Preparation
2.4.2. Mass Spectrometry
2.4.3. HPLC
2.4.4. MS/MS Analysis
2.5. Transcriptomic
2.5.1. RNA Quantification and Qualification
2.5.2. Library Preparation for Transcriptome Sequencing
2.5.3. Clustering and Sequencing
2.6. Statistical Analysis
2.6.1. Biochemical Data Analysis
2.6.2. Differential Metabolite Analysis
2.6.3. Differential Expression Analysis and GO Enrichment Analysis
2.6.4. Bioinformatic Joint Analysis
3. Results
3.1. Lipid Metabolism Disorders
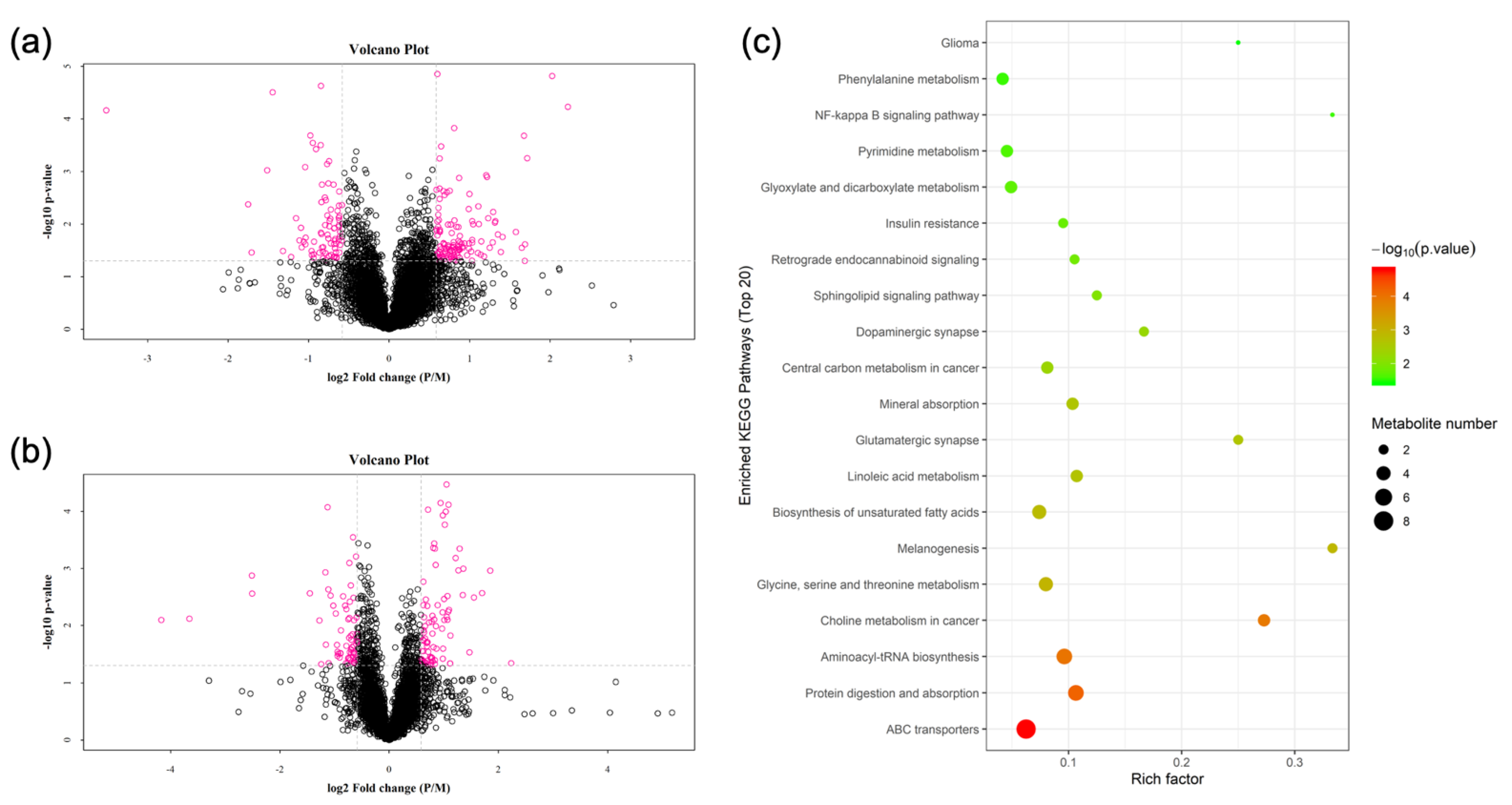
3.2. Metabolomics
3.3. Transcriptomics
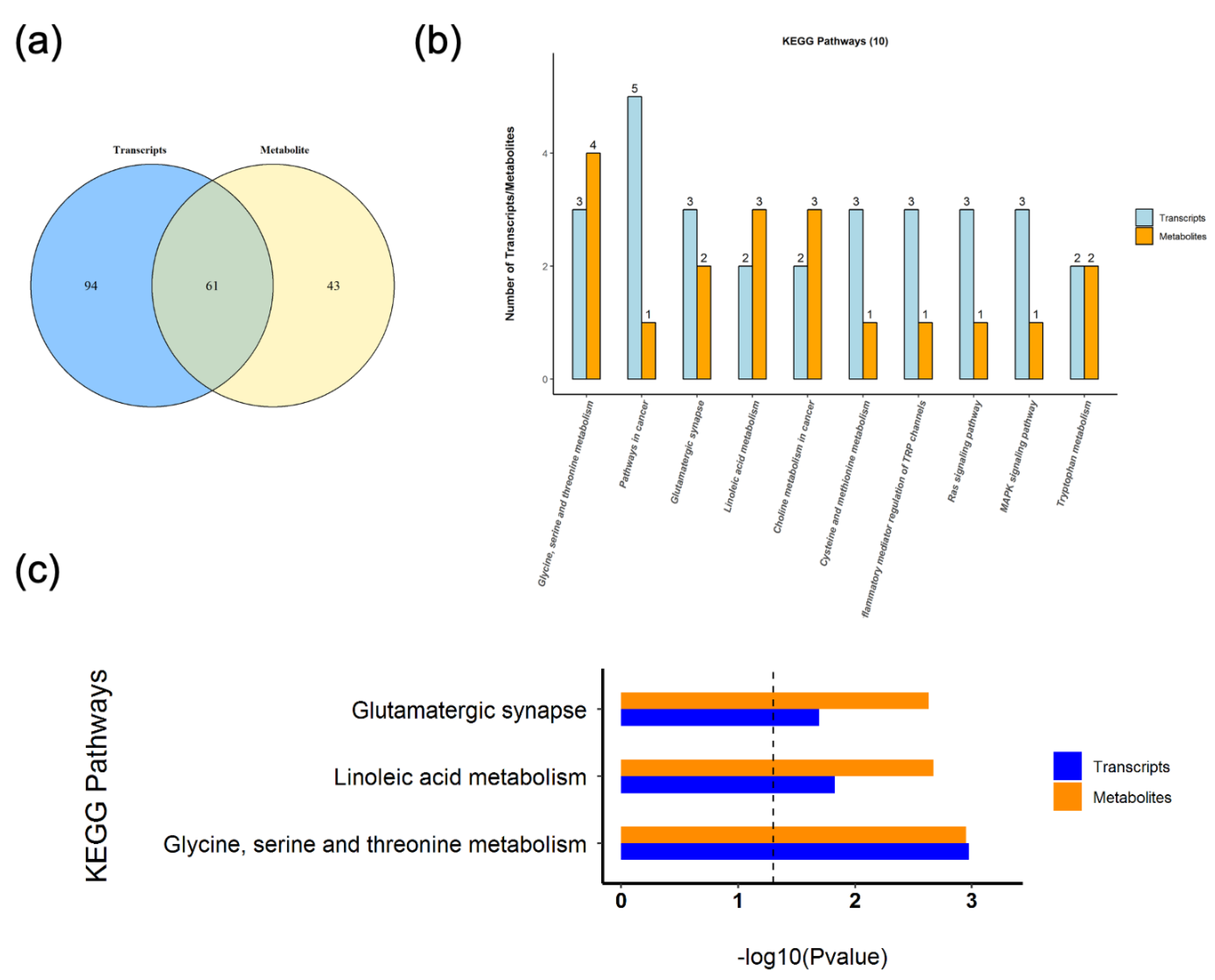
3.4. Correlation
3.5. KEGG Pathway Joint Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shattat, G.F. A Review Article on Hyperlipidemia: Types, Treatments and New Drug Targets. Biomed. Pharmacol. J. 2015, 7, 399–409. [Google Scholar] [CrossRef]
- Myers, J.; Kokkinos, P.; Nyelin, E. Physical Activity, Cardiorespiratory Fitness, and the Metabolic Syndrome. Nutrients 2019, 11, 1652. [Google Scholar] [CrossRef] [PubMed]
- Lehrke, M.; Lazar, M.A. The many faces of PPARgamma. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef]
- Yoon, I.C.; Eun, J.R. Pharmacologic therapy for nonalcoholic steatohepatitis focusing on pathophysiology. Yeungnam Univ. J. Med. 2019, 36, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Martín-Mateos, R.; Albillos, A. The Role of the Gut-Liver Axis in Metabolic Dysfunction-Associated Fatty Liver Disease. Front. Immunol. 2021, 12, 660179. [Google Scholar] [CrossRef] [PubMed]
- Shida, K.; Nomoto, K. Probiotics as efficient immunopotentiators: Translational role in cancer prevention. Indian J. Med. Res. 2013, 138, 808–814. [Google Scholar] [PubMed]
- Hu, X.; Wang, T.; Li, W.; Jin, F.; Wang, L. Effects of NS lactobacillus strains on lipid metabolism of rats fed a high-cholesterol diet. Lipids Health Dis. 2013, 12, 1–12. [Google Scholar] [CrossRef]
- Xin, J.; Zheng, D.; Wang, H.; Sun, N.; Zhao, Y.; Dan, Y.; Pan, K.; Jing, B.; Ni, X. Live Probiotic Lactobacillus johnsonii BS15 Promotes Growth Performance and Lowers Fat Deposition by Improving Lipid Metabolism, Intestinal Development, and Gut Microflora in Broilers. Front. Microbiol. 2017, 8, 1073. [Google Scholar]
- Zhong, Z.; Zhang, W.; Du, R.; Meng, H.; Zhang, H. Lactobacillus casei Zhang stimulates lipid metabolism in hypercholesterolemic rats by affecting gene expression in the liver. Eur. J. Lipid Sci. Technol. 2012, 114, 224–252. [Google Scholar] [CrossRef]
- Chen, D.; Yang, Z.; Chen, X.; Huang, Y.; Yin, B.; Guo, F.; Zhao, H.; Zhao, T.; Zhenquan, Y.; Huang, J.; et al. The effect of Lactobacillus rhamnosus hsryfm 1301 on the intestinal microbiota of a hyperlipidemic rat model. BMC Complement. Altern. Med. 2014, 14, 1–9. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Santoso, A.; Heriansyah, T.; Rohman, M.S. Phospholipase A2 is an Inflammatory Predictor in Cardiovascular Diseases: Is there any Spacious Room to Prove the Causation? Curr. Cardiol. Rev. 2020, 16, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Blackie, J.A.; Bloomer, J.C.; Brown, M.J.; Cheng, H.-Y.; Hammond, B.; Hickey, D.M.; Ife, R.J.; Leach, C.A.; Lewis, V.; Macphee, C.H.; et al. The identification of clinical candidate SB-480848: A potent inhibitor of lipoprotein-associated phospholipase A2. Bioorganic Med. Chem. Lett. 2003, 13, 1067–1070. [Google Scholar] [CrossRef]
- Ning, M.; Jeong, H. High-Fat Diet Feeding Alters Expression of Hepatic Drug-Metabolizing Enzymes in Mice. Drug Metab. Dispos. 2017, 45, 707–711. [Google Scholar] [CrossRef]
- Volinsky, R.; Kinnunen, P.K.J. Oxidized phosphatidylcholines in membrane-level cellular signaling: From biophysics to physiology and molecular pathology. FEBS J. 2013, 280, 2806–2816. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Ahn, T.; Yun, C.H. Membrane properties induced by anionic phospholipids and phosphatidylethanolamine are critical for the membrane binding and catalytic activity of human cytochrome p450 3A4. Biochemistry 2003, 42, 15377–15387. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Marques, M.; Cunha, I.; Reis-Henriques, M.A.; Santos, M.M.; Castro, L.F.C. Diversity and history of the long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates. BMC Evol. Biol. 2013, 13, 271. [Google Scholar] [CrossRef]
- Tabernilla, A.; Rodrigues, B.D.S.; Pieters, A.; Caufriez, A.; Leroy, K.; Van Campenhout, R.; Cooreman, A.; Gomes, A.; Arnesdotter, E.; Gijbels, E.; et al. In Vitro Liver Toxicity Testing of Chemicals: A Pragmatic Approach. Int. J. Mol. Sci. 2021, 22, 5038. [Google Scholar] [CrossRef]
- Jha, P.; Claudel, T.; Baghdasaryan, A.; Mueller, M.; Halilbasic, E.; Das, S.K.; Lass, A.; Zimmermann, R.; Zechner, R.; Hoefler, G.; et al. Role of adipose triglyceride lipase (PNPLA2) in protection from hepatic inflammation in mouse models of steatohepatitis and endotoxemia. Hepatology 2014, 59, 858–869. [Google Scholar] [CrossRef]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef]
- Nshimiyimana, P.; Liu, L.; Du, G. Engineering of L-amino acid deaminases for the production of α-keto acids from L-amino acids. Bioengineered 2019, 10, 43–51. [Google Scholar] [CrossRef] [PubMed]
- El-Hattab, A.W. Serine biosynthesis and transport defects. Mol. Genet. Metab. 2016, 118, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, S.; Arsic, A.; Ristic-Medic, D.; Cvetkovic, Z.; Vucic, V. Lipid Peroxidation and Antioxidant Supplementation in Neurodegenerative Diseases: A Review of Human Studies. Antioxidants 2020, 9, 1128. [Google Scholar] [CrossRef]
- Adermark, L.; Gutierrez, S.; Lagström, O.; Hammarlund, M.; Licheri, V.; Johansson, M.E. Weight gain and neuroadaptations elicited by high fat diet depend on fatty acid composition. Psychoneuroendocrinology 2021, 126, 105143. [Google Scholar] [CrossRef]
- Basson, A.; Trotter, A.; Rodriguez-Palacios, A.; Cominelli, F. Mucosal Interactions between Genetics, Diet, and Microbiome in Inflammatory Bowel Disease. Front. Immunol. 2016, 7, 290. [Google Scholar] [CrossRef]
- Yuan, T.; Yang, T.; Chen, H.; Fu, D.; Hu, Y.; Wang, J.; Yuan, Q.; Yu, H.; Xu, W.; Xie, X. New insights into oxidative stress and inflammation during diabetes mellitus-accelerated atherosclerosis. Redox Biol. 2018, 20, 247–260. [Google Scholar] [CrossRef]
- Noh, H.; King, G. The role of protein kinase C activation in diabetic nephropathy. Kidney Int. 2007, 72, S49–S53. [Google Scholar] [CrossRef]
- Hennigs, J.K.; Lüneburg, N.; Stage, A.; Schmitz, M.; Körbelin, J.; Harbaum, L.; Matuszcak, C.; Mienert, J.; Bokemeyer, C.; Böger, R.H.; et al. The P2-receptor-mediated Ca2+ signalosome of the human pulmonary endothelium—Implications for pulmonary arterial hypertension. Purinergic Signal. 2019, 15, 299–311. [Google Scholar] [CrossRef]
- Sugden, P.H. An overview of endothelin signaling in the cardiac myocyte. J. Mol. Cell. Cardiol. 2003, 35, 871–886. [Google Scholar] [CrossRef]
- Bhat, N.; Narayanan, A.; Fathzadeh, M.; Kahn, M.; Zhang, D.; Goedeke, L.; Neogi, A.; Cardone, R.L.; Kibbey, R.G.; Fernandez-Hernando, C.; et al. Dyrk1b promotes hepatic lipogenesis by bypassing canonical insulin signaling and directly activating mTORC2 in mice. J. Clin. Investig. 2022, 132, e153724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Su, Q. PKC in developmental hypothyroid rat brain. Neurol. Sci. 2014, 35, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Picard, F.; Carter, S.; Caron, A.; Richard, D. Role of leptin resistance in the development of obesity in older patients. Clin. Interv. Aging 2013, 8, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Yamaguchi, T.; Shoji, K.; Hibi, M.; Sugita, T.; Takase, H. Effect of low concentration of diacylglycerol on mildly postprandial hypertriglyceridemia. Atherosclerosis 2010, 213, 539–544. [Google Scholar] [CrossRef]
- Tada, N.; Shoji, K.; Takeshita, M.; Watanabe, H.; Yoshida, H.; Hase, T.; Matsuo, N.; Tokimitsu, I. Effects of diacylglycerol ingestion on postprandial hyperlipidemia in diabetes. Clin. Chim. Acta 2005, 353, 87–94. [Google Scholar] [CrossRef]
- Ando, Y.; Saito, S.; Yamanaka, N.; Suzuki, C.; Ono, T.; Osaki, N.; Katsuragi, Y. Alpha Linolenic Acid-enriched Diacylglycerol Consumption Enhances Dietary Fat Oxidation in Healthy Subjects: A Randomized Double-blind Controlled Trial. J. Oleo Sci. 2017, 66, 181–185. [Google Scholar] [CrossRef]
- Hibi, M.; Takase, H.; Yasunaga, K.; Yamaguchi, T.; Harada, U.; Katsuragi, Y.; Tokimitsu, I. Fat utilization in healthy subjects consuming diacylglycerol oil diet: Dietary and whole body fat oxidation. Lipids 2008, 43, 517–524. [Google Scholar] [CrossRef]
- Devi, B.P.; Gangadhar, K.; Prasad, R.; Sugasini, D.; Rao, Y.P.C.; Lokesh, B. Nutritionally enriched 1,3-diacylglycerol-rich oil: Low calorie fat with hypolipidemic effects in rats. Food Chem. 2018, 248, 210–216. [Google Scholar] [CrossRef]
- Kim, H.; Choe, J.-H.; Choi, J.H.; Kim, H.J.; Park, S.H.; Lee, M.W.; Kim, W.; Go, G.-W. Medium-Chain Enriched Diacylglycerol (MCE-DAG) Oil Decreases Body Fat Mass in Mice by Increasing Lipolysis and Thermogenesis in Adipose Tissue. Lipids 2017, 52, 665–673. [Google Scholar] [CrossRef]



| Map_ID | Map Name | Gene_id | cpdName | Pathway Hierarchy |
|---|---|---|---|---|
| rno00591 | Linoleic acid metabolism * | Pla2g4c, Cyp1a2 | Linoleic acid, all cis-(6,9,12)-linolenic acid, PC(16:0/16:0) | Lipid metabolism |
| rno00564 | Glycerophospholipid metabolism | Pla2g4c | PC(16:0/16:0), Phosphorylcholine, | Lipid metabolism |
| rno00061 | Fatty acid biosynthesis | Acsbg1 | Oleic acid | Lipid metabolism |
| rno00592 | alpha-linolenic acid metabolism | Pla2g4c | PC(16:0/16:0) | Lipid metabolism |
| rno00071 | Fatty acid degradation | Acsbg1 | L-Palmitoylcarnitine | Lipid metabolism |
| rno00561 | Glycerolipid metabolism | Pnpla2 | Glyceric acid | Lipid metabolism |
| rno00590 | Arachidonic acid metabolism | Pla2g4c | PC(16:0/16:0) | Lipid metabolism |
| rno00260 | Glycine, serine and threonine metabolism * | Psat1, Cse, Phgdh | L-Threonine, Glyceric acid, L-Serine, Betaine | Amino acid metabolism |
| rno00270 | Cysteine and methionine metabolism | Psat1, Cse, Phgdh | L-Serine | Amino acid metabolism |
| rno00380 | Tryptophan metabolism | Cyp1a2, Acmsd | Formylanthranilic acid, Anthranilic acid (Vitamin L1) | Amino acid metabolism |
| rno00250 | Alanine, aspartate and glutamate metabolism | Asns | L-Glutamine | Amino acid metabolism |
| rno04724 | Glutamatergic synapse * | Pla2g4c, Gria4, Prkcg | L-Glutamine, 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04728 | Dopaminergic synapse | Gria4, Prkcg | L-Tyrosine, 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04723 | Retrograde endocannabinoid signaling | Gria4, Prkcg | PC(16:0/16:0), 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04730 | Long-term depression | Pla2g4c, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04726 | Serotonergic synapse | Pla2g4c, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04720 | Long-term potentiation | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04727 | GABAergic synapse | Prkcg | L-Glutamine | Nervous system |
| rno04725 | Cholinergic synapse | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Nervous system |
| rno04911 | Insulin secretion | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04912 | GnRH signaling pathway | Pla2g4c | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04925 | Aldosterone synthesis and secretion | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04921 | Oxytocin signaling pathway | Pla2g4c, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04918 | Thyroid hormone synthesis | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04919 | Thyroid hormone signaling pathway | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04920 | Adipocytokine signaling pathway | Acsbg1 | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04923 | Regulation of lipolysis in adipocytes | Pnpla2 | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04915 | Estrogen signaling pathway | Hsp90aa1 | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04916 | Melanogenesis | Prkcg | L-Tyrosine, 1-Stearoyl-2-arachidonoyl-sn-glycerol | Endocrine system |
| rno04650 | Natural killer cell mediated cytotoxicity | Gzmbl2, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Immune system |
| rno04666 | Fc gamma R-mediated phagocytosis | Pla2g4c, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Immune system |
| rno04611 | Platelet activation | Pla2g4c, P2rx1 | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Immune system |
| rno04664 | Fc epsilon RI signaling pathway | Pla2g4c | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Immune system |
| rno04062 | Chemokine signaling pathway | Ccl17 | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Immune system |
| rno04014 | Ras signaling pathway | Pla2g4c, Fgf21, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04010 | MAPK signaling pathway | Pla2g4c, Fgf21, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04071 | Sphingolipid signaling pathway | Kng1, Prkcg | L-Serine, 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04066 | HIF-1 signaling pathway | LOC100911372, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04370 | VEGF signaling pathway | Pla2g4c, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04015 | Rap1 signaling pathway | Fgf21, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04020 | Calcium signaling pathway | P2rx1, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04012 | ErbB signaling pathway | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04072 | Phospholipase D signaling pathway | Pla2g4c | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04024 | cAMP signaling pathway | Gria4 | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Signal transduction |
| rno04970 | Salivary secretion | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Digestive system |
| rno04971 | Gastric acid secretion | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Digestive system |
| rno04972 | Pancreatic secretion | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Digestive system |
| rno05200 | Pathways in cancer | Kng1, Zbtb16, Hsp90aa1, Fgf21, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Cancers: Overview |
| rno05231 | Choline metabolism in cancer | Pla2g4c, Prkcg | PC(16:0/16:0), 1-Stearoyl-2-arachidonoyl-sn-glycerol, Phosphorylcholine | Cancers: Overview |
| rno05214 | Glioma | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Cancers: Specific types |
| rno05223 | Non-small cell lung cancer | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Cancers: Specific types |
| rno05143 | African trypanosomiasis | Kng1, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Infectious diseases: Parasitic |
| rno05146 | Amoebiasis | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Infectious diseases: Parasitic |
| rno04270 | Vascular smooth muscle contraction | Pla2g4c, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Circulatory system |
| rno04540 | Gap junction | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Cellular community |
| rno04750 | Inflammatory mediator regulation of TRP channels | Pla2g4c, Kng1, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Sensory system |
| rno04713 | Circadian entrainment | Gria4, Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Environmental adaptation |
| rno04961 | Endocrine and other factor-regulated calcium reabsorption | Prkcg | 1-Stearoyl-2-arachidonoyl-sn-glycerol | Excretory system |
| rno05031 | Amphetamine addiction | Gria4, Prkcg | L-Tyrosine | Substance dependence |
| rno00920 | Sulfur metabolism | LOC103689947, Impad1 | L-Serine | Energy metabolism |
| rno00910 | Nitrogen metabolism | Car6 | L-Glutamine | Energy metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, H.; Zong, L.; Sang, J.; Wa, Y.; Chen, D.; Huang, Y.; Chen, X.; Gu, R. Effect of Lactobacillus rhamnosus hsryfm 1301 Fermented Milk on Lipid Metabolism Disorders in High-Fat-Diet Rats. Nutrients 2022, 14, 4850. https://doi.org/10.3390/nu14224850
Qu H, Zong L, Sang J, Wa Y, Chen D, Huang Y, Chen X, Gu R. Effect of Lactobacillus rhamnosus hsryfm 1301 Fermented Milk on Lipid Metabolism Disorders in High-Fat-Diet Rats. Nutrients. 2022; 14(22):4850. https://doi.org/10.3390/nu14224850
Chicago/Turabian StyleQu, Hengxian, Lina Zong, Jian Sang, Yunchao Wa, Dawei Chen, Yujun Huang, Xia Chen, and Ruixia Gu. 2022. "Effect of Lactobacillus rhamnosus hsryfm 1301 Fermented Milk on Lipid Metabolism Disorders in High-Fat-Diet Rats" Nutrients 14, no. 22: 4850. https://doi.org/10.3390/nu14224850
APA StyleQu, H., Zong, L., Sang, J., Wa, Y., Chen, D., Huang, Y., Chen, X., & Gu, R. (2022). Effect of Lactobacillus rhamnosus hsryfm 1301 Fermented Milk on Lipid Metabolism Disorders in High-Fat-Diet Rats. Nutrients, 14(22), 4850. https://doi.org/10.3390/nu14224850