Prediction, Discovery, and Characterization of Plant- and Food-Derived Health-Beneficial Bioactive Peptides
Highlights
- Natural bioactives provide molecular answers to many questions about human health and planetary sustainability.
- Due to the current mainly serendipitous research and high-throughput screenings, the exploitation and leverage of these bioactives has so far been limited.
- AI and design-first thinking can significantly enhance the efficiency of discovery, validation, and translation of natural bioactives for human health and planetary sustainability.
- Sustainable generation and use of food and energy are two major challenges to be met for preserving a healthy human life on a healthy planet.
- The efficient and sustainable leveraging of nature's treasure of bioactive compounds can contribute to a better food and health care system.
Abstract
1. Natural Bioactives from Plants and Foods
- -
- Peptides can be regarded as the “vocabulary of nature”: living systems use peptides to communicate and to regulate and fine-tune their functions. Peptides have co-evolved with humans as modulators of physiology and therefore exert highly specific biological functions. The presence in natural (e.g., plant and food) sources and the biological function of peptides can be predicted in silico by blasting peptide sequences against the plant and food genomes and by the computational and human interpretation of metabolic and signalling pathways. Orally administered peptides often suffer from a short half-life across ingestion and digestion, as well as in the blood circulation. The challenges of using peptides as orally delivered bioactives lie in their stability, bioavailability, and bioefficacy, rather than in their safety. From a food perspective, peptides can be considered as nutrients, and they are the only nutrients that are directly encoded in the genomes of their sources [8]. Food peptides are components of long-term consumed food sources. Food protein hydrolysates can therefore be “generally recognized as safe” (GRAS) [8].
2. Artificial Intelligence in (Life) Science and Technology
3. Artificial Intelligence for Prediction and Discovery of Bioactive Peptides
3.1. Concept and Computation
3.2. Prediction of Peptide Properties
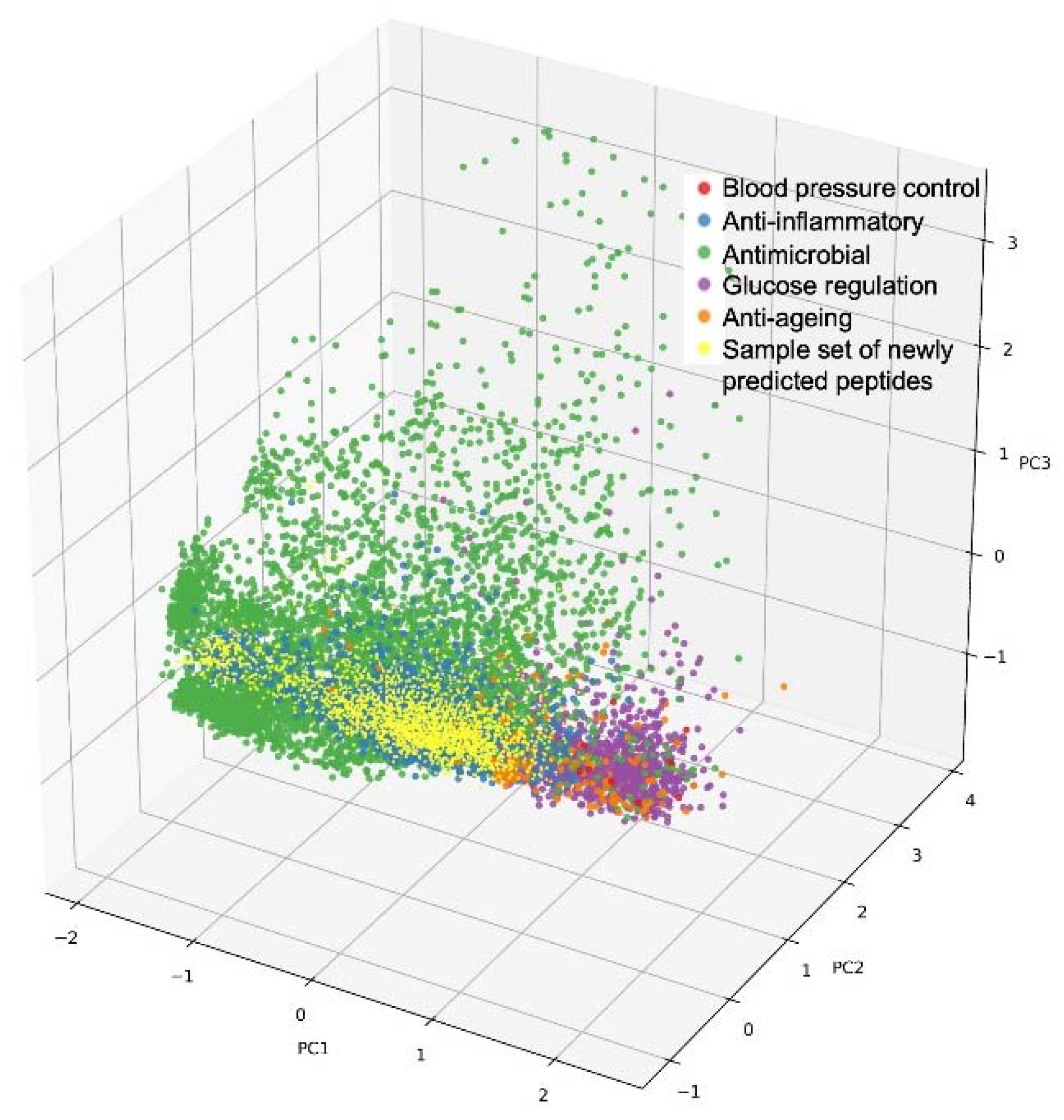
3.3. Natural Peptide Network (NPN) Design and Validation of Predicted Peptides and Designed Hydrolysates
3.4. Natural Peptide Network (NPN) Manufacturing and Analysis
4. Mass Spectrometric Characterization of Bioactive Peptides
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Monteiro, J.P.; Kussmann, M.; Kaput, J. The genomics of micronutrient requirements. Genes Nutr. 2015, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Howes, M.-J.R.; Simmonds, M.S.J. The role of phytochemicals as micronutrients in health and disease. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Reyes, P.K.; de Lara, J.C.-F.; Poquet, L.; Redeuil, K.; Kussmann, M.; Silva-Zolezzi, I.; Tejero, E.M. Circulating Structurally Related (-)-Epicatechin Metabolite Species and Levels after Sustained Intake of a Cocoa Powder High in Polyphenols Are Comparable to Those Achieved after a Single Dose. Nutrients 2021, 13, 3829. [Google Scholar] [CrossRef]
- Vyas, U.; Ranganathan, N. Probiotics, Prebiotics, and Synbiotics: Gut and Beyond. Gastroenterol. Res. Pract. 2012, 2012, 872716. [Google Scholar] [CrossRef] [PubMed]
- Karami, Z.; Akbari-Adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Doherty, A.; Wall, A.; Khaldi, N.; Kussmann, M. Artificial Intelligence in Functional Food Ingredient Discovery and Characterisation: A Focus on Bioactive Plant and Food Peptides. Front. Genet. 2021, 12, 768979. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Lee, B.H.; Oh, D.H. Current trends and perspectives of bioactive peptides. Crit. Rev. Food Sci. Nutr. 2018, 58, 2273–2284. [Google Scholar] [CrossRef]
- Schaafsma, G. Safety of protein hydrolysates, fractions thereof and bioactive peptides in human nutrition. Eur. J. Clin. Nutr. 2009, 63, 1161–1168. [Google Scholar] [CrossRef]
- Hayes, M. Food Proteins and Bioactive Peptides: New and Novel Sources, Characterisation Strategies and Applications. Foods 2018, 7, 38. [Google Scholar] [CrossRef]
- Mohan, N.M.; Zorgani, A.; Earley, L.; Chauhan, S.; Trajkovic, S.; Savage, J.; Adelfio, A.; Khaldi, N.; Martins, M. Preservatives from food—For food: Pea protein hydrolysate as a novel bio-preservative against Escherichia coli O157:H7 on a lettuce leaf. Food Sci. Nutr. 2021, 9, 5946–5958. [Google Scholar] [CrossRef]
- Conway, A.R.A.; Kovacs, K. New and emerging models of human intelligence. Wiley Interdiscip. Rev. Cogn. Sci. 2015, 6, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Coen, E. The storytelling arms race: Origin of human intelligence and the scientific mind. Heredity 2019, 123, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Adotey, S.; Thomford, N.E.; Dzobo, W. Integrating Artificial and Human Intelligence: A Partnership for Responsible Innovation in Biomedical Engineering and Medicine. OMICS 2020, 24, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Abdelhalim, H.; Berber, A.; Lodi, M.; Jain, R.; Nair, A.; Pappu, A.; Patel, K.; Venkat, V.; Venkatesan, C.; Wable, R.; et al. Artificial Intelligence, Healthcare, Clinical Genomics, and Pharmacogenomics Approaches in Precision Medicine. Front. Genet. 2022, 13, 929736. [Google Scholar] [CrossRef]
- Limketkai, B.N.; Mauldin, K.; Manitius, N.; Jalilian, L.; Salonen, B.R. The Age of Artificial Intelligence: Use of Digital Technology in Clinical Nutrition. Curr. Surg. Rep. 2021, 9, 20. [Google Scholar] [CrossRef]
- Walter, W.; Haferlach, C.; Nadarajah, N.; Schmidts, I.; Kühn, C.; Kern, W.; Haferlach, T. How artificial intelligence might disrupt diagnostics in hematology in the near future. Oncogene 2021, 40, 4271–4280. [Google Scholar] [CrossRef]
- Harfouche, A.L.J.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Mugnozza, G.S.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.B.; Altman, A. Accelerating Climate Resilient Plant Breeding by Applying Next-Generation Artificial Intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef]
- Zhao, S.; Xie, C. Spatial Difference of China’s Regional Logistics Development and Construction of Information Network Platform Based on Artificial Intelligence Technology Under the Background of New Economy. Front. Psychol. 2022, 13, 871538. [Google Scholar] [CrossRef]
- Sarker, S.; Jamal, L.; Ahmed, S.F.; Irtisam, N. Robotics and artificial intelligence in healthcare during COVID-19 pandemic: A systematic review. Robot. Auton. Syst. 2021, 146, 103902. [Google Scholar] [CrossRef]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A., Jr.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2019, 19, 353–364. [Google Scholar] [CrossRef]
- Nam, D.; Chapiro, J.; Paradis, V.; Seraphin, T.P.; Kather, J.N. Artificial intelligence in liver diseases: Improving diagnostics, prognostics and response prediction. JHEP Rep. 2022, 4, 100443. [Google Scholar] [CrossRef] [PubMed]
- Luccioni, A.; Schmidt, V.; Vardanyan, V.; Bengio, Y.; Rhyne, T.-M. Using Artificial Intelligence to Visualize the Impacts of Climate Change. IEEE Comput. Graph. Appl. 2021, 41, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Doherty, A.; Wall, A.; Khaldi, N. Using Artificial Intelligence to Reduce Global Healthcare Costs through Discovery and Development of Nutritional Interventions. Int. J. Nurs. Didact. 2020, 10, 1–5. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, Z.; Zhang, M.; Mais, D.E.; Wang, M.-W. High Throughput Screening for Bioactive Components from Traditional Chinese Medicine. Comb. Chem. High Throughput Screen. 2010, 13, 837–848. [Google Scholar] [CrossRef]
- Kussmann, M.; van Bladeren, P. The Extended Nutrigenomics—Understanding the Interplay between the Genomes of Food, Gut Microbes, and Human Host. Front. Genet. 2011, 2, 21. [Google Scholar] [CrossRef]
- Beutler, J.A. Natural Products as a Foundation for Drug Discovery. Curr. Protoc. Pharmacol. 2019, 86, e67. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, P.; Ma, Y.; Wang, J.; Chen, Y. Artificial intelligence accelerates the mining of bioactive small molecules from human microbiome. Clin. Transl. Med. 2022, 12, e1011. [Google Scholar] [CrossRef]
- Casey, R.; Adelfio, A.; Connolly, M.; Wall, A.; Holyer, I.; Khaldi, N. Discovery through Machine Learning and Preclinical Validation of Novel Anti-Diabetic Peptides. Biomedicines 2021, 9, 276. [Google Scholar] [CrossRef]
- Corrochano, A.R.; Cal, R.; Kennedy, K.; Wall, A.; Murphy, N.; Trajkovic, S.; O’Callaghan, S.; Adelfio, A.; Khaldi, N. Characterising the efficacy and bioavailability of bioactive peptides identified for attenuating muscle atrophy within a Vicia faba-derived functional ingredient. Curr. Res. Food Sci. 2021, 4, 224–232. [Google Scholar] [CrossRef]
- Kennedy, K.; Keogh, B.; Lopez, C.; Adelfio, A.; Molloy, B.; Kerr, A.; Wall, A.M.; Jalowicki, G.; Holton, T.A.; Khaldi, N. An Artificial Intelligence Characterised Functional Ingredient, Derived from Rice, Inhibits TNF-α and Significantly Improves Physical Strength in an Inflammaging Population. Foods 2020, 9, 1147. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.; Kerr, A.; Keogh, B.; Nolan, S.; Casey, R.; Adelfio, A.; Murphy, N.; Doherty, A.; Davis, H.; Wall, A.; et al. An Artificial-Intelligence-Discovered Functional Ingredient, NRT_N0G5IJ, Derived from Pisum sativum, Decreases HbA1c in a Prediabetic Population. Nutrients 2021, 13, 1635. [Google Scholar] [CrossRef]
- van Erp, M.; Reynolds, C.; Maynard, D.; Starke, A.; Martín, R.I.; Andres, F.; Leite, M.C.A.; de Toledo, D.A.; Rivera, X.S.; Trattner, C.; et al. Using Natural Language Processing and Artificial Intelligence to Explore the Nutrition and Sustainability of Recipes and Food. Front. Artif. Intell. 2020, 3, 621577. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ruiz, A.; Colmenarejo, G. Systematic Analysis and Prediction of the Target Space of Bioactive Food Compounds: Filling the Chemobiological Gaps. J. Chem. Inf. Model. 2022, 62, 3734–3751. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Shuli, Z.; Linlin, L.; Yinghu, Z.; Nan, S.; Haibin, W.; Hongyu, X. Bioinformatics and Computer Simulation approaches to the discovery and analysis of Bioactive Peptides. Curr. Pharm. Biotechnol. 2022, 23, 1541–1555. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Sokołowska, J.; Starowicz, P.; Bucholska, J.; Hrynkiewicz, M. Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science. Int. J. Mol. Sci. 2015, 16, 20748–20773. [Google Scholar] [CrossRef]
- Duffuler, P.; Bhullar, K.S.; de Campos Zani, S.C.; Wu, J. Bioactive Peptides: From Basic Research to Clinical Trials and Commercialization. J. Agric. Food Chem. 2022, 70, 3585–3595. [Google Scholar] [CrossRef]
- Fan, K.-T.; Hsu, C.-W.; Chen, Y.-R. Mass spectrometry in the discovery of peptides involved in intercellular communication: From targeted to untargeted peptidomics approaches. Mass Spectrom. Rev. 2022, 41, e21789. [Google Scholar] [CrossRef]
- Meissner, F.; Geddes-McAlister, J.; Mann, M.; Bantscheff, M. The emerging role of mass spectrometry-based proteomics in drug discovery. Nat. Rev. Drug Discov. 2022, 21, 637–654. [Google Scholar] [CrossRef]
- Kussmann, M.; Affolter, M.; Nagy, K.; Holst, B.; Fay, L.B. Mass spectrometry in nutrition: Understanding dietary health effects at the molecular level. Mass Spectrom. Rev. 2007, 26, 727–750. [Google Scholar] [CrossRef]
- Collins, S.L.; Koo, I.; Peters, J.M.; Smith, P.B.; Patterson, A.D. Current Challenges and Recent Developments in Mass Spectrometry–Based Metabolomics. Annu. Rev. Anal. Chem. 2021, 14, 467–487. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, X.; Cong, P.; Xu, J.; Xue, C. Mass spectrometry-based lipidomics in food science and nutritional health: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2530–2558. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.R.; Ruparel, H.; Ju, J. Mass-spectrometry DNA sequencing. Mutat. Res. Mol. Mech. Mutagen. 2005, 573, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Rozanova, S.; Barkovits, K.; Nikolov, M.; Schmidt, C.; Urlaub, H.; Marcus, K. Quantitative Mass Spectrometry-Based Proteomics: An Overview. Methods Mol. Biol. 2021, 2228, 85–116. [Google Scholar] [CrossRef]
- Omenn, G.S.; Lane, L.; Overall, C.M.; Corrales, F.J.; Schwenk, J.M.; Paik, Y.-K.; Van Eyk, J.E.; Liu, S.; Snyder, M.; Baker, M.S.; et al. Progress on Identifying and Characterizing the Human Proteome: 2018 Metrics from the HUPO Human Proteome Project. J. Proteome Res. 2018, 17, 4031–4041. [Google Scholar] [CrossRef]
- de Graaf, S.C.; Hoek, M.; Tamara, S.; Heck, A.J.R. A perspective toward mass spectrometry-based de novo sequencing of endogenous antibodies. MAbs 2022, 14, 2079449. [Google Scholar] [CrossRef]
- McCool, E.N.; Lubeckyj, R.A.; Chen, D.; Sun, L. Top-Down Proteomics by Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Large-Scale Characterization of Proteoforms in Complex Samples. Methods Mol. Biol. 2022, 2531, 107–124. [Google Scholar] [CrossRef]
- Manes, N.P.; Nita-Lazar, A. Application of targeted mass spectrometry in bottom-up proteomics for systems biology research. J. Proteom. 2018, 189, 75–90. [Google Scholar] [CrossRef]
- Rotello, R.J.; Veenstra, T.D. Mass Spectrometry Techniques: Principles and Practices for Quantitative Proteomics. Curr. Protein Pept. Sci. 2021, 22, 121–133. [Google Scholar] [CrossRef]
- Meyer, J.G. Qualitative and Quantitative Shotgun Proteomics Data Analysis from Data-Dependent Acquisition Mass Spectrometry. Methods Mol. Biol. 2021, 2259, 297–308. [Google Scholar] [CrossRef]
- Zhang, F.; Ge, W.; Ruan, G.; Cai, X.; Guo, T. Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools: A Glimpse in 2020. Proteomics 2020, 20, e1900276. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.V.; Piersma, S.R.; Oudgenoeg, G.; Jimenez, C.R. Label-free mass spectrometry-based proteomics for biomarker discovery and validation. Expert Rev. Mol. Diagn. 2012, 12, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.A.; Aebersold, R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr. Opin. Biotechnol. 2003, 14, 110–118. [Google Scholar] [CrossRef]
- Itzhak, D.N.; Sacco, F.; Nagaraj, N.; Tyanova, S.; Mann, M.; Murgia, M. SILAC-based quantitative proteomics using mass spectrometry quantifies endoplasmic reticulum stress in whole HeLa cells. Dis. Model. Mech. 2019, 12, dmm040741. [Google Scholar] [CrossRef] [PubMed]
- Maes, E.; Oeyen, E.; Boonen, K.; Schildermans, K.; Mertens, I.; Pauwels, P.; Valkenborg, D.; Baggerman, G. The challenges of peptidomics in complementing proteomics in a clinical context. Mass Spectrom. Rev. 2019, 38, 253–264. [Google Scholar] [CrossRef]
- He, B.; Huang, Z.; Huang, C.; Nice, E.C. Clinical applications of plasma proteomics and peptidomics: Towards precision medicine. Proteom. Clin. Appl. 2022, 16, e2100097. [Google Scholar] [CrossRef]
- Fabre, B.; Combier, J.-P.; Plaza, S. Recent advances in mass spectrometry–based peptidomics workflows to identify short-open-reading-frame-encoded peptides and explore their functions. Curr. Opin. Chem. Biol. 2021, 60, 122–130. [Google Scholar] [CrossRef]
- Foreman, R.E.; George, A.L.; Reimann, F.; Gribble, F.M.; Kay, R.G. Peptidomics: A Review of Clinical Applications and Methodologies. J. Proteome Res. 2021, 20, 3782–3797. [Google Scholar] [CrossRef]
- Agyei, D.; Tsopmo, A.; Udenigwe, C.C. Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides. Anal. Bioanal. Chem. 2018, 410, 3463–3472. [Google Scholar] [CrossRef]
- Ibañez, C.; Simo, C.; Garcia-Cañas, V.; Cifuentes, A.; Castro-Puyana, M. Metabolomics, peptidomics and proteomics applications of capillary electrophoresis-mass spectrometry in Foodomics: A review. Anal. Chim. Acta 2013, 802, 1–13. [Google Scholar] [CrossRef]
- Magalhães, B.T.; Trindade, F.; Barros, A.; Klein, J.; Amado, F.; Ferreira, R.; Vitorino, R. Reviewing Mechanistic Peptidomics in Body Fluids Focusing on Proteases. Proteomics 2018, 18, e1800187. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kussmann, M. Prediction, Discovery, and Characterization of Plant- and Food-Derived Health-Beneficial Bioactive Peptides. Nutrients 2022, 14, 4810. https://doi.org/10.3390/nu14224810
Kussmann M. Prediction, Discovery, and Characterization of Plant- and Food-Derived Health-Beneficial Bioactive Peptides. Nutrients. 2022; 14(22):4810. https://doi.org/10.3390/nu14224810
Chicago/Turabian StyleKussmann, Martin. 2022. "Prediction, Discovery, and Characterization of Plant- and Food-Derived Health-Beneficial Bioactive Peptides" Nutrients 14, no. 22: 4810. https://doi.org/10.3390/nu14224810
APA StyleKussmann, M. (2022). Prediction, Discovery, and Characterization of Plant- and Food-Derived Health-Beneficial Bioactive Peptides. Nutrients, 14(22), 4810. https://doi.org/10.3390/nu14224810






