Anticancer Effects of Thymoquinone through the Antioxidant Activity, Upregulation of Nrf2, and Downregulation of PD-L1 in Triple-Negative Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
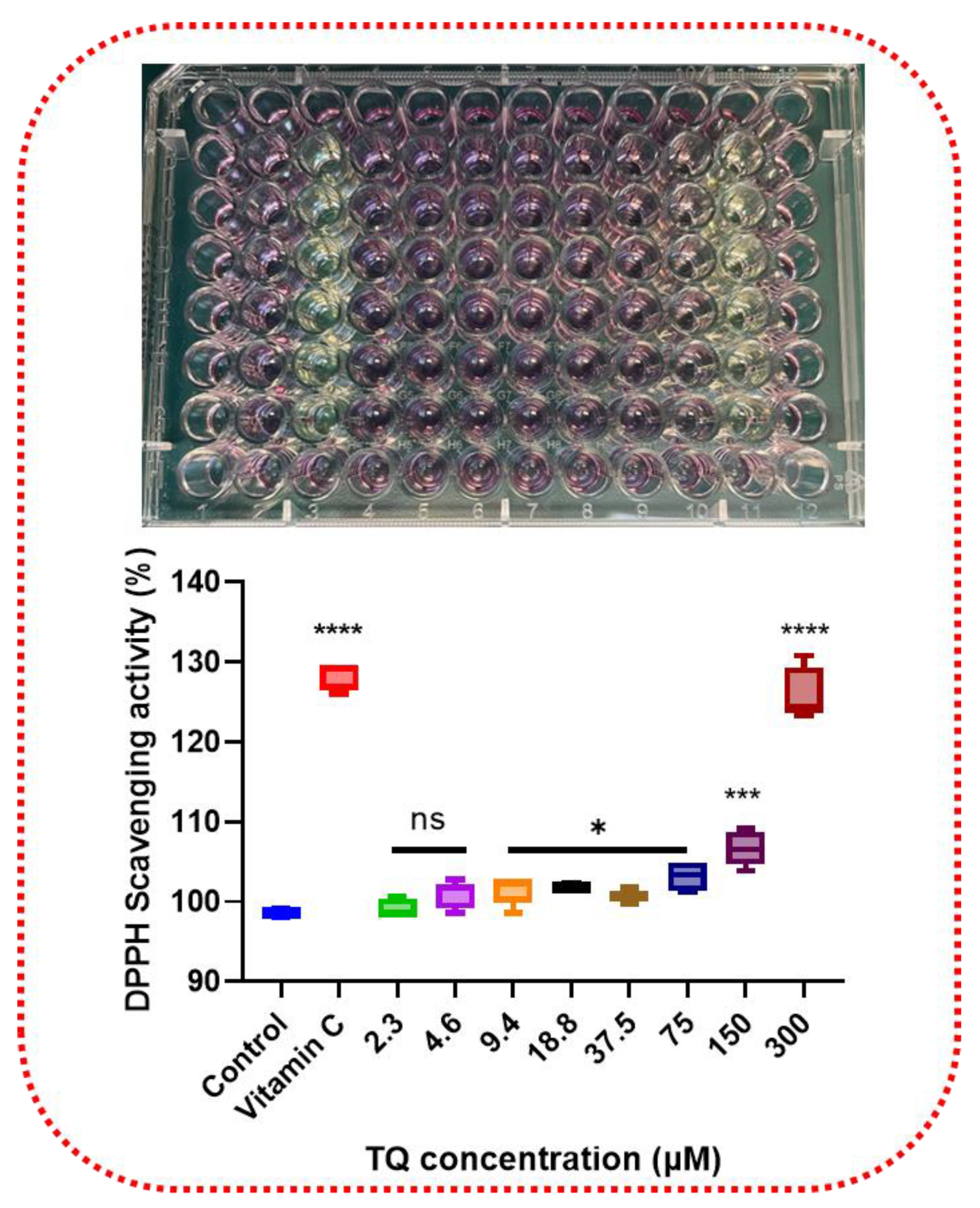
2.3. Assay for Scavenging DPPH Radicals
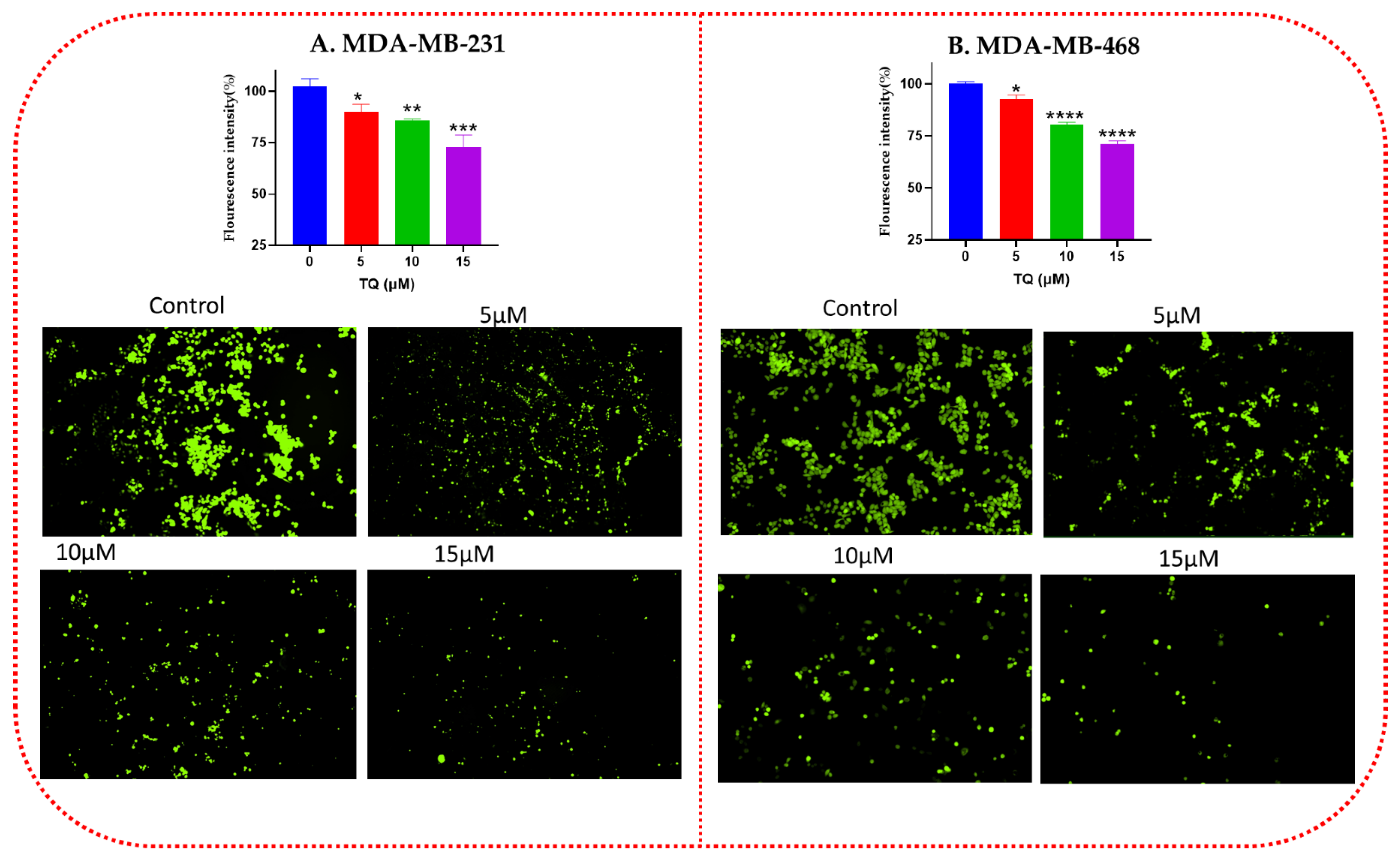
2.4. Detection of Intracellular Reactive Oxygen Species Level (ROS)
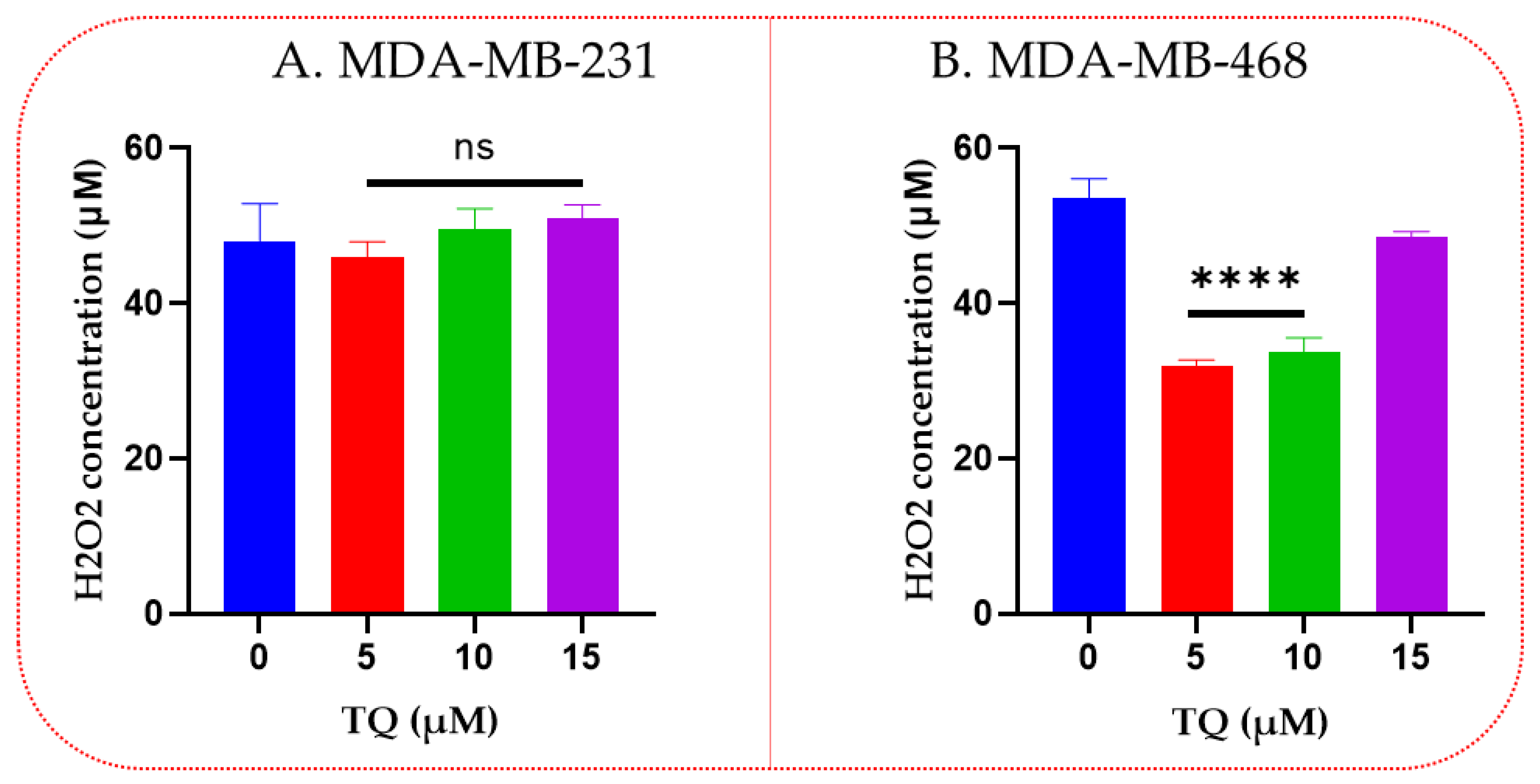
2.5. Hydrogen Peroxide Cell-Based Assay
2.6. Catalase Enzyme Activity Assay
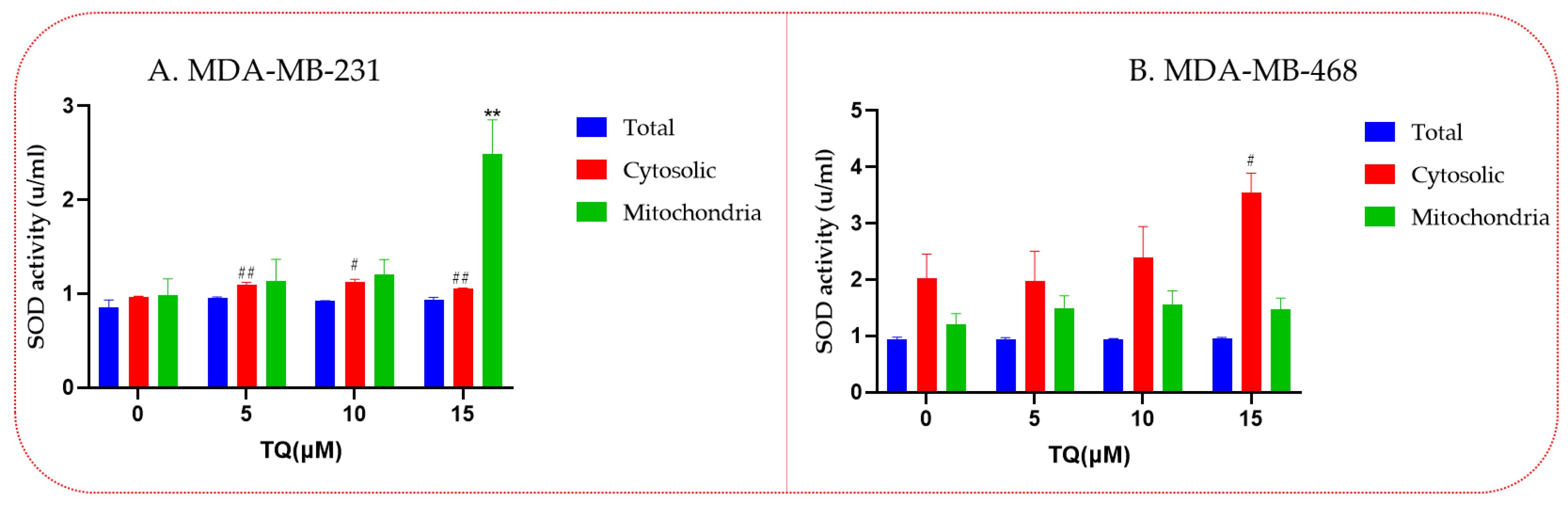
2.7. Superoxide Dismutase Enzyme Activity Assay
2.8. Glutathione Assay
2.9. Gene Expression Assay
2.9.1. RNA Extraction and cDNA Synthesis
2.9.2. Quantitative Real-Time PCR Arrays
2.9.3. Quantitative RT-PCR
2.10. Capillary Electrophoresis Western Analysis
2.11. Statistical Analysis
3. Results
3.1. Antioxidative Effect of Thymoquinone by Scavenging DPPH
3.2. Effect of TQ on the Intracellular ROS Level of TNBC Cells
3.3. Thymoquinone Decreases Hydrogen Peroxide Levels in TNBC
3.4. Thymoquinone Increased Catalase Enzyme Activities in TNBC Cells
3.5. Increased Superoxide Dismutase (SOD) Enzyme Activities in Thymoquinone-Treated TNBC Cells
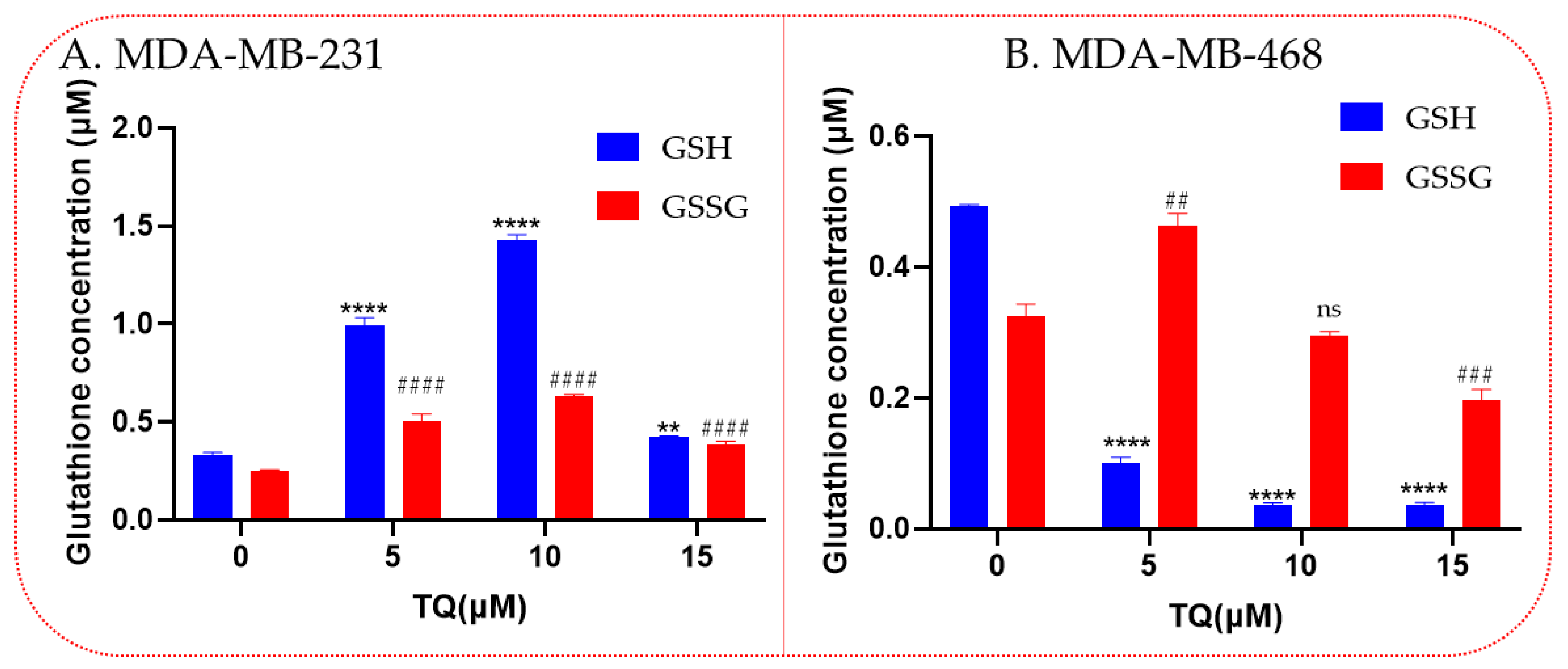
3.6. Thymoquinone Increases Levels of the Antioxidant Glutathione
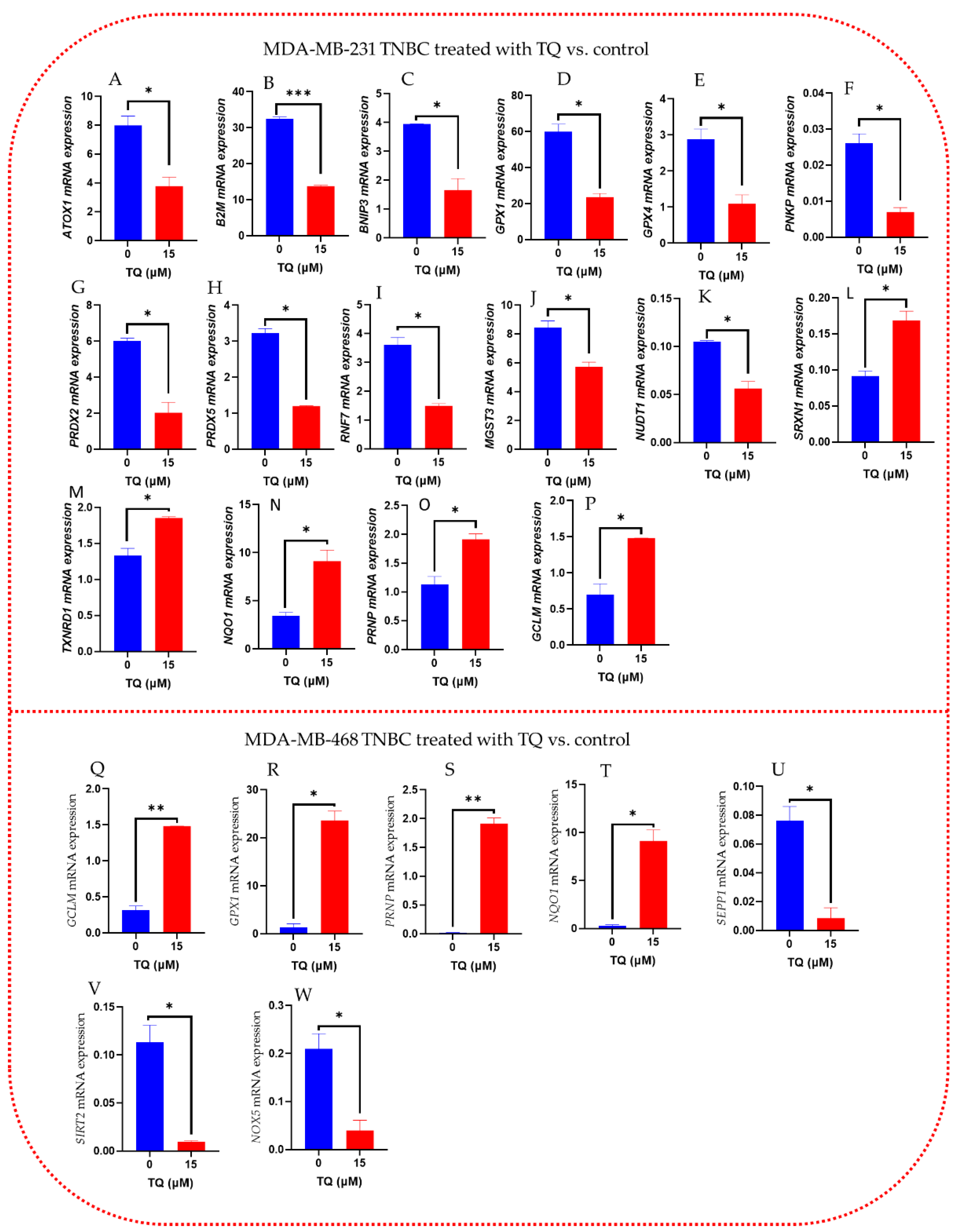
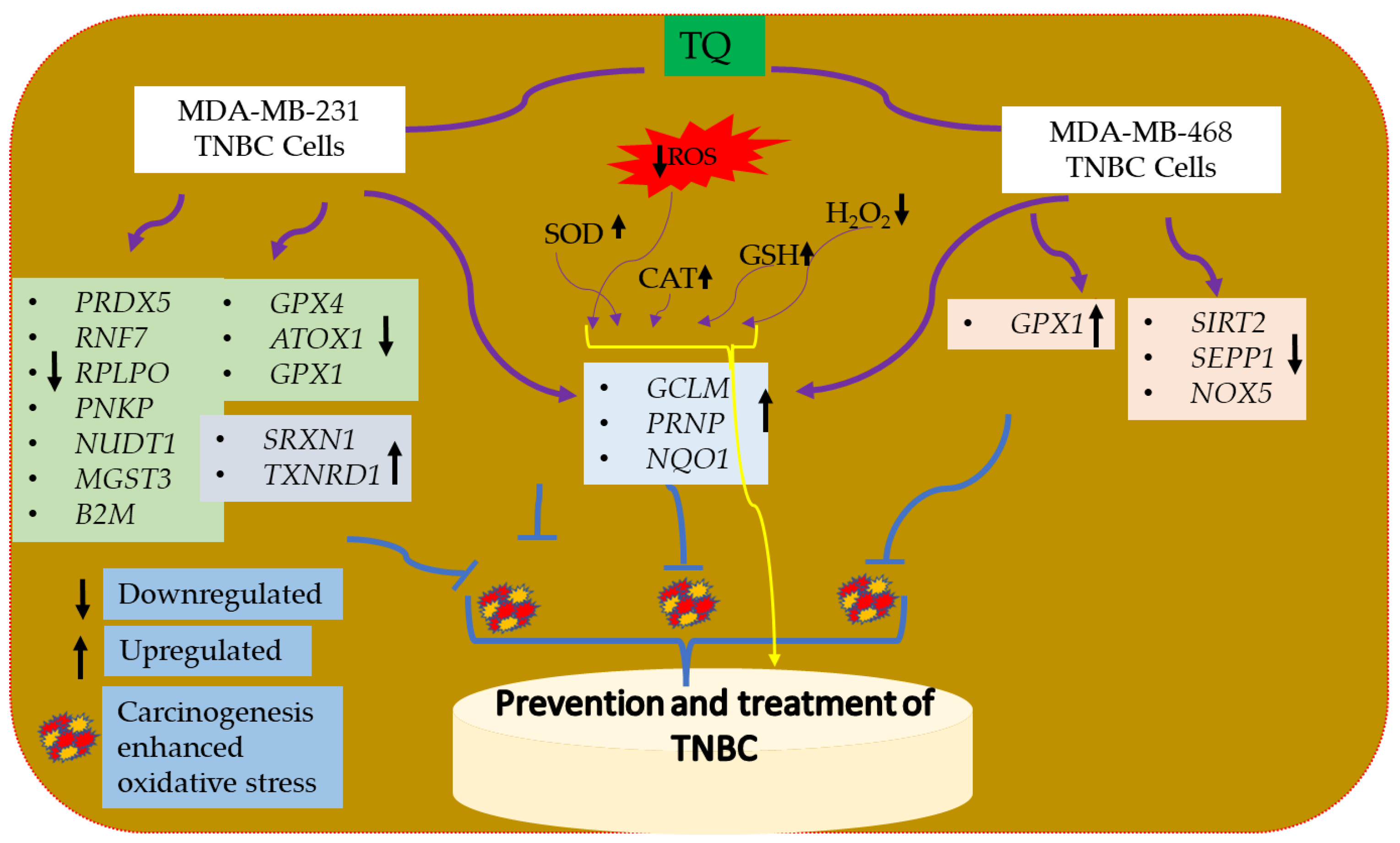
3.7. Oxidative Stress-Antioxidant Defense Gene Expression Alteration in TQ-Treated Triple-Negative Breast Cancer Cells
3.8. TQ Increases mRNA Expression of Nrf2 in TNBC Cells
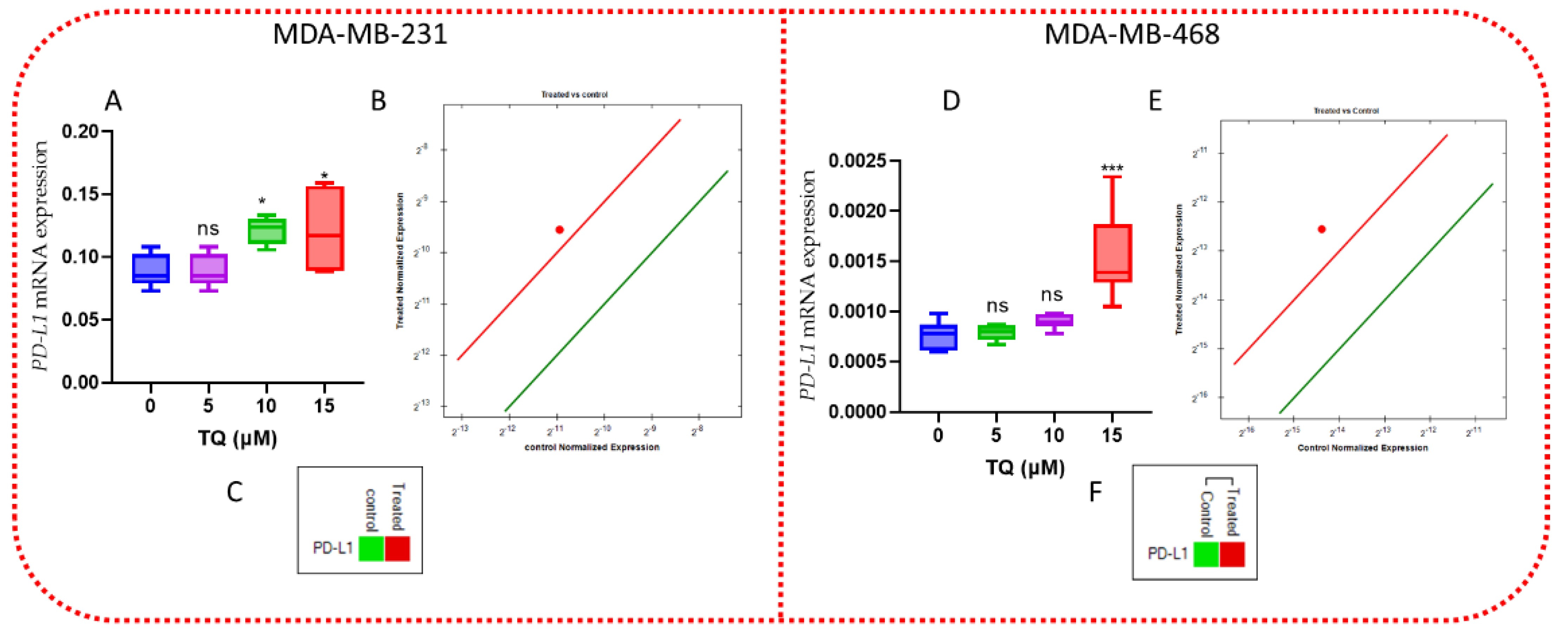
3.9. TQ Increases the Expression of PD-L1 mRNA
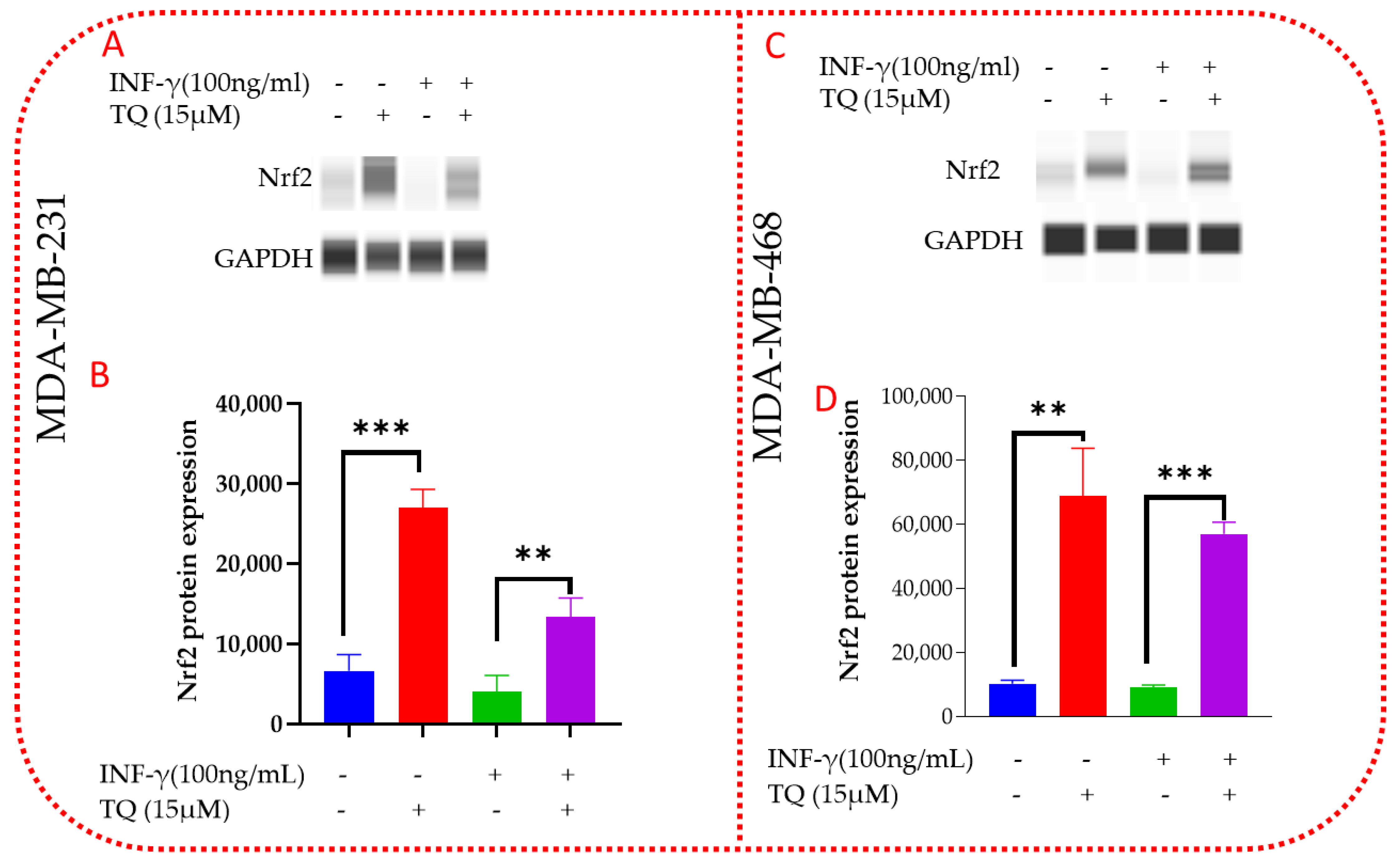
3.10. TQ Induces Nrf2 Protein Expression in MDA-MB-231 and MDA-MB-468 TNBC Cells
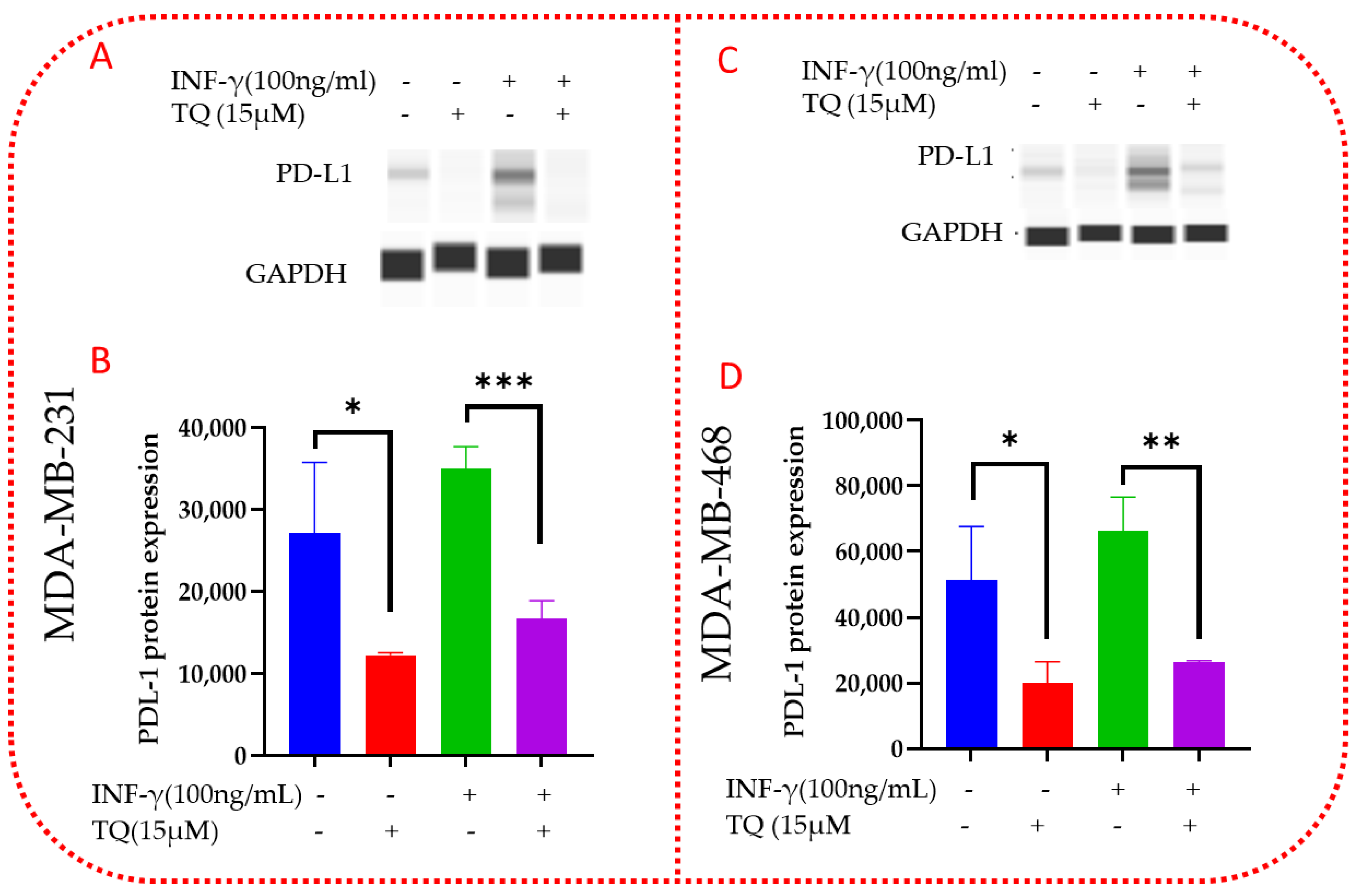
3.11. TQ Inhibits PD-L1 Protein Expression in MDA-MB-231 and MDA-MB-468 TNBC Cells
4. Discussion
5. Summary
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- NBCF (National Breast Cancer Foundation, Inc.). Breast Cancer Facts. 2022. Available online: https://www.nationalbreastcancer.org/breast-cancer-facts (accessed on 21 May 2022).
- Adinew, G.M.; Messeha, S.S.; Taka, E.; Badisa, R.B.; Antonie, L.M.; Soliman, K.F.A. Thymoquinone Alterations of the Apoptotic Gene Expressions and Cell Cycle Arrest in Genetically Distinct Triple-Negative Breast Cancer Cells. Nutrients 2022, 14, 2120. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Hossain, F.; Danos, D.; Lassak, A.; Scribner, R.; Miele, L. Racial Disparities in Triple Negative Breast Cancer: A Review of the Role of Biologic and Non-biologic Factors. Front. Public Health 2020, 8, 576964. [Google Scholar] [CrossRef] [PubMed]
- Unal, T.D.; Hamurcu, Z.; Delibasi, N.; Cinar, V.; Guler, A.; Gokce, S.; Nurdinov, N.; Ozpolat, B. Thymoquinone Inhibits Proliferation and Migration of MDA-MB-231 Triple Negative Breast Cancer Cells by Suppressing Autophagy, Beclin-1 and LC3. Anticancer Agents Med. Chem. 2021, 21, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Adinew, G.M.; Messeha, S.; Taka, E.; Soliman, K.F.A. The Prognostic and Therapeutic Implications of the Chemoresistance Gene BIRC5 in Triple-Negative Breast Cancer. Cancers 2022, 14, 5180. [Google Scholar] [CrossRef] [PubMed]
- Adinew, G.M.; Taka, E.; Mochona, B.; Badisa, R.B.; Mazzio, E.A.; Elhag, R.; Soliman, K.F.A. Therapeutic Potential of Thymoquinone in Triple-Negative Breast Cancer Prevention and Progression through the Modulation of the Tumor Microenvironment. Nutrients 2021, 14, 79. [Google Scholar] [CrossRef] [PubMed]
- Qing, X.; Shi, D.; Lv, X.; Wang, B.; Chen, S.; Shao, Z. Prognostic significance of 8-hydroxy-2’-deoxyguanosine in solid tumors: A meta-analysis. BMC Cancer 2019, 19, 997. [Google Scholar] [CrossRef] [PubMed]
- Labani-Motlagh, A.; Ashja-Mahdavi, M.; Loskog, A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front. Immunol. 2020, 11, 940. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, G.; Yin, J.; Li, L.; Tan, Y.; Wei, H.; Liu, B.; Deng, L.; Tang, J.; Chen, Y.; et al. GSTP1 and cancer: Expression, methylation, polymorphisms and signaling (Review). Int. J. Oncol. 2020, 56, 867–878. [Google Scholar] [CrossRef]
- Azmanova, M.; Pitto-Barry, A. Oxidative Stress in Cancer Therapy: Friend or Enemy? Chembiochem 2022, 23, e202100641. [Google Scholar] [CrossRef]
- Al Haq, A.T.; Tseng, H.Y.; Chen, L.M.; Wang, C.C.; Hsu, H.L. Targeting prooxidant MnSOD effect inhibits triple-negative breast cancer (TNBC) progression and M2 macrophage functions under the oncogenic stress. Cell Death Dis. 2022, 13, 49. [Google Scholar] [CrossRef] [PubMed]
- Danesh, H.; Ziamajidi, N.; Mesbah-Namin, S.A.; Nafisi, N.; Abbasalipourkabir, R. Association between Oxidative Stress Parameters and Hematological Indices in Breast Cancer Patients. Int. J. Breast Cancer 2022, 2022, 1459410. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, C. Nrf2-mediated anti-inflammatory polarization of macrophages as therapeutic targets for osteoarthritis. Front. Immunol. 2022, 13, 967193. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Angarita, F.A.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. High Expression of NRF2 Is Associated with Increased Tumor-Infiltrating Lymphocytes and Cancer Immunity in ER-Positive/HER2-Negative Breast Cancer. Cancers 2020, 12, 3856. [Google Scholar] [CrossRef]
- Tascioglu Aliyev, A.; Panieri, E.; Stepanic, V.; Gurer-Orhan, H.; Saso, L. Involvement of NRF2 in Breast Cancer and Possible Therapeutical Role of Polyphenols and Melatonin. Molecules 2021, 26, 1853. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Payandeh, Z.; Pirpour Tazehkand, A.; Mansoori, B.; Khaze, V.; Asadi, M.; Baradaran, B.; Samadi, N. The Impact of Nrf2 Silencing on Nrf2-PD-L1 Axis to Overcome Oxaliplatin Resistance and Migration in Colon Cancer Cells. Avicenna J. Med. Biotechnol. 2021, 13, 116–122. [Google Scholar] [CrossRef]
- Li, F.; Ren, Y.; Wang, Z. Programmed death 1 Ligand 1 expression in breast cancer and its association with patients’ clinical parameters. J. Cancer Res. Ther. 2018, 14, 150–154. [Google Scholar] [CrossRef]
- Muenst, S.; Schaerli, A.R.; Gao, F.; Däster, S.; Trella, E.; Droeser, R.A.; Muraro, M.G.; Zajac, P.; Zanetti, R.; Gillanders, W.E.; et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2014, 146, 15–24. [Google Scholar] [CrossRef]
- Shahcheraghi, S.H.; Salemi, F.; Peirovi, N.; Ayatollahi, J.; Alam, W.; Khan, H.; Saso, L. Nrf2 Regulation by Curcumin: Molecular Aspects for Therapeutic Prospects. Molecules 2021, 27, 167. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, M.; Dong, H.; Miao, J.; Stagos, D.; Liu, M. Dietary prenylated flavonoid xanthohumol alleviates oxidative damage and accelerates diabetic wound healing via Nrf2 activation. Food Chem. Toxicol. 2022, 160, 112813. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Greven, J.; Qin, K.; Fragoulis, A.; Horst, K.; Blasius, F.; Wruck, C.; Pufe, T.; Kobbe, P.; Hildebrand, F.; et al. Sulforaphane Exerts Beneficial Immunomodulatory Effects on Liver Tissue via a Nrf2 Pathway-Related Mechanism in a Murine Model of Hemorrhagic Shock and Resuscitation. Front. Immunol. 2022, 13, 822895. [Google Scholar] [CrossRef] [PubMed]
- Kamisako, T.; Tanaka, Y. Oltipraz ameliorates the progression of steatohepatitis in Nrf2-null mice fed a high-fat diet. J. Clin. Biochem. Nutr. 2022, 70, 147–153. [Google Scholar] [CrossRef]
- Panda, H.; Wen, H.; Suzuki, M.; Yamamoto, M. Multifaceted Roles of the KEAP1-NRF2 System in Cancer and Inflammatory Disease Milieu. Antioxidants 2022, 11, 538. [Google Scholar] [CrossRef]
- Ryan, D.G.; Knatko, E.V.; Casey, A.M.; Hukelmann, J.L.; Dayalan Naidu, S.; Brenes, A.J.; Ekkunagul, T.; Baker, C.; Higgins, M.; Tronci, L.; et al. Nrf2 activation reprograms macrophage intermediary metabolism and suppresses the type I interferon response. iScience 2022, 25, 103827. [Google Scholar] [CrossRef]
- Holmstrom, K.M.; Kostov, R.V.; Dinkova-Kostova, A.T. The multifaceted role of Nrf2 in mitochondrial function. Curr. Opin. Toxicol. 2016, 1, 80–91. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Abramov, A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015, 88, 179–188. [Google Scholar] [CrossRef]
- Imran, M.; Rauf, A.; Khan, I.A.; Shahbaz, M.; Qaisrani, T.B.; Fatmawati, S.; Abu-Izneid, T.; Imran, A.; Rahman, K.U.; Gondal, T.A. Thymoquinone: A novel strategy to combat cancer: A review. Biomed. Pharmacother. 2018, 106, 390–402. [Google Scholar] [CrossRef]
- Banerjee, S.; Padhye, S.; Azmi, A.; Wang, Z.; Philip, P.A.; Kucuk, O.; Sarkar, F.H.; Mohammad, R.M. Review on the molecular and therapeutic potential of thymoquinone in cancer. Nutr. Cancer 2010, 62, 938–946. [Google Scholar] [CrossRef]
- Rajput, S.; Kumar, B.N.; Dey, K.K.; Pal, I.; Parekh, A.; Mandal, M. Molecular targeting of Akt by thymoquinone promotes G(1) arrest through translation inhibition of cyclin D1 and induces apoptosis in breast cancer cells. Life Sci. 2013, 93, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, Y.K.; Abdelrazek, H.M.A. Cancer: Thymoquinone antioxidant/prooxidant effect as a potential anticancer remedy. Biomed. Pharmacother. 2019, 115, 108783. [Google Scholar] [CrossRef] [PubMed]
- Harzallah, H.J.; Grayaa, R.; Kharoubi, W.; Maaloul, A.; Hammami, M.; Mahjoub, T. Thymoquinone, the Nigella sativa bioactive compound, prevents circulatory oxidative stress caused by 1,2-dimethylhydrazine in erythrocyte during colon postinitiation carcinogenesis. Oxid. Med. Cell. Longev. 2012, 2012, 854065. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tonarova, P.; Lochovska, K.; Pytlik, R.; Hubalek Kalbacova, M. The Impact of Various Culture Conditions on Human Mesenchymal Stromal Cells Metabolism. Stem. Cells Int. 2021, 2021, 6659244. [Google Scholar] [CrossRef]
- Darwish, A.G.; Samy, M.N.; Sugimoto, S.; Otsuka, H.; Abdel-Salam, H.; Matsunami, K. Effects of Hepatoprotective Compounds from the Leaves of Lumnitzera racemosa on Acetaminophen-Induced Liver Damage in Vitro. Chem. Pharm. Bull. (Tokyo) 2016, 64, 360–365. [Google Scholar] [CrossRef]
- Hozayen, W.G.; Mahmoud, A.M.; Desouky, E.M.; El-Nahass, E.S.; Soliman, H.A.; Farghali, A.A. Cardiac and pulmonary toxicity of mesoporous silica nanoparticles is associated with excessive ROS production and redox imbalance in Wistar rats. Biomed. Pharmacother. 2019, 109, 2527–2538. [Google Scholar] [CrossRef]
- Kurokawa, H.; Matsui, H. The Cytotoxicity of Doxorubicin Can Be Accelerated by a Combination of Hyperthermia and 5-Aminolevulinic Acid. Antioxidants 2021, 10, 1531. [Google Scholar] [CrossRef]
- Bai, J.; Lei, Y.; An, G.L.; He, L. Down-regulation of deacetylase HDAC6 inhibits the melanoma cell line A375.S2 growth through ROS-dependent mitochondrial pathway. PLoS ONE 2015, 10, e0121247. [Google Scholar] [CrossRef]
- Messeha, S.S.; Zarmouh, N.O.; Mendonca, P.; Alwagdani, H.; Cotton, C.; Soliman, K.F.A. Effects of gossypol on apoptosis-related gene expression in racially distinct triple-negative breast cancer cells. Oncol. Rep. 2019, 42, 467–478. [Google Scholar] [CrossRef]
- Sowrirajan, B.; Saito, Y.; Poudyal, D.; Chen, Q.; Sui, H.; DeRavin, S.S.; Imamichi, H.; Sato, T.; Kuhns, D.B.; Noguchi, N.; et al. Interleukin-27 Enhances the Potential of Reactive Oxygen Species Generation from Monocyte-derived Macrophages and Dendritic cells by Induction of p47(phox). Sci. Rep. 2017, 7, 43441. [Google Scholar] [CrossRef]
- Messeha, S.S.; Zarmouh, N.O.; Antonie, L.; Soliman, K.F.A. Sanguinarine Inhibition of TNF-alpha-Induced CCL2, IKBKE/NF-kappaB/ERK1/2 Signaling Pathway, and Cell Migration in Human Triple-Negative Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 8329. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.; Salamero-Boix, A.; Niesel, K.; Alekseeva, T.; Sevenich, L. Microenvironmental Regulation of Tumor Progression and Therapeutic Response in Brain Metastasis. Front. Immunol. 2019, 10, 1713. [Google Scholar] [CrossRef] [PubMed]
- Staurengo-Ferrari, L.; Badaro-Garcia, S.; Hohmann, M.S.N.; Manchope, M.F.; Zaninelli, T.H.; Casagrande, R.; Verri, W.A., Jr. Contribution of Nrf2 Modulation to the Mechanism of Action of Analgesic and Anti-inflammatory Drugs in Pre-clinical and Clinical Stages. Front. Pharmacol. 2018, 9, 1536. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. et Biophys. Acta (BBA)—Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Loubaki, L.; Rouabhia, M.; Zahrani, M.A.; Amri, A.A.; Semlali, A. Oxidative Stress and Autophagy Mediate Anti-Cancer Properties of Cannabis Derivatives in Human Oral Cancer Cells. Cancers 2022, 14, 4924. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Badana, A.K.; Malla, R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 1177271918755391. [Google Scholar] [CrossRef]
- Sarmiento-Salinas, F.L.; Perez-Gonzalez, A.; Acosta-Casique, A.; Ix-Ballote, A.; Diaz, A.; Trevino, S.; Rosas-Murrieta, N.H.; Millan-Perez-Pena, L.; Maycotte, P. Reactive oxygen species: Role in carcinogenesis, cancer cell signaling, and tumor progression. Life Sci. 2021, 284, 119942. [Google Scholar] [CrossRef]
- Arfin, S.; Jha, N.K.; Jha, S.K.; Kesari, K.K.; Ruokolainen, J.; Roychoudhury, S.; Rathi, B.; Kumar, D. Oxidative Stress in Cancer Cell Metabolism. Antioxidants 2021, 10, 642. [Google Scholar] [CrossRef]
- Wu, M.; Ma, L.; Xue, L.; Ye, W.; Lu, Z.; Li, X.; Jin, Y.; Qin, X.; Chen, D.; Tang, W.; et al. Resveratrol alleviates chemotherapy-induced oogonial stem cell apoptosis and ovarian aging in mice. Aging (Albany NY) 2019, 11, 1030–1044. [Google Scholar] [CrossRef]
- Sharifi-Rad, M.; Anil Kumar, N.V.; Zucca, P.; Varoni, E.M.; Dini, L.; Panzarini, E.; Rajkovic, J.; Tsouh Fokou, P.V.; Azzini, E.; Peluso, I.; et al. Lifestyle, Oxidative Stress, and Antioxidants: Back and Forth in the Pathophysiology of Chronic Diseases. Front. Physiol. 2020, 11, 694. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Divya, M.; Vaseeharan, B.; Chen, J.; Biruntha, M.; Silva, L.P.; Durán-Lara, E.F.; Shreema, K.; Ranjan, S.; Dasgupta, N. Biological Compound Capping of Silver Nanoparticle with the Seed Extracts of Blackcumin (Nigella sativa): A Potential Antibacterial, Antidiabetic, Anti-inflammatory, and Antioxidant. J. Inorg. Organomet. Polym. Mater. 2021, 31, 624–635. [Google Scholar] [CrossRef]
- Kassab, R.B.; El-Hennamy, R.E. The role of thymoquinone as a potent antioxidant in ameliorating the neurotoxic effect of sodium arsenate in female rat. Egypt. J. Basic Appl. Sci. 2017, 4, 160–167. [Google Scholar] [CrossRef]
- Alexander, H.R.; Syed Alwi, S.S.; Yazan, L.S.; Zakarial Ansar, F.H.; Ong, Y.S. Migration and Proliferation Effects of Thymoquinone-Loaded Nanostructured Lipid Carrier (TQ-NLC) and Thymoquinone (TQ) on In Vitro Wound Healing Models. Evid.-Based Complement. Altern. Med. 2019, 2019, 9725738. [Google Scholar] [CrossRef] [PubMed]
- Alrafiah, A. Thymoquinone Protects Neurons in the Cerebellum of Rats through Mitigating Oxidative Stress and Inflammation Following High-Fat Diet Supplementation. Biomolecules 2021, 11, 165. [Google Scholar] [CrossRef]
- Ashley, R.E.; Osheroff, N. Natural Products as Topoisomerase II Poisons: Effects of Thymoquinone on DNA Cleavage Mediated by Human Topoisomerase IIα. Chem. Res. Toxicol. 2014, 27, 787–793. [Google Scholar] [CrossRef]
- Kazutaka, H. Biomolecules Oxidation by Hydrogen Peroxide and Singlet Oxygen. In Reactive Oxygen Species (ROS) in Living Cells; Cristiana, F., Elena, A., Eds.; IntechOpen: Rijeka, Croatia, 2017; Chapter 9. [Google Scholar] [CrossRef]
- Di Marzo, N.; Chisci, E.; Giovannoni, R. The Role of Hydrogen Peroxide in Redox-Dependent Signaling: Homeostatic and Pathological Responses in Mammalian Cells. Cells 2018, 7, 156. [Google Scholar] [CrossRef]
- Doskey, C.M.; Buranasudja, V.; Wagner, B.A.; Wilkes, J.G.; Du, J.; Cullen, J.J.; Buettner, G.R. Tumor cells have decreased ability to metabolize H(2)O(2): Implications for pharmacological ascorbate in cancer therapy. Redox Biol. 2016, 10, 274–284. [Google Scholar] [CrossRef]
- Farsiabi, R.; Khodadadi, I.; Karimi, J.; Shafiee, G. Evaluation of the Effects of Thymoquinone on Oxidative Stress in A549 Lung Cancer Cell Line. Middle East J. Cancer 2021. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell. Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell. Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Cobourne-Duval, M.K.; Taka, E.; Mendonca, P.; Bauer, D.; Soliman, K.F. The Antioxidant Effects of Thymoquinone in Activated BV-2 Murine Microglial Cells. Neurochem. Res. 2016, 41, 3227–3238. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.T.; Park, H.J. Implications of NQO1 in cancer therapy. BMB Rep. 2015, 48, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Bendavit, G.; Aboulkassim, T.; Hilmi, K.; Shah, S.; Batist, G. Nrf2 Transcription Factor Can Directly Regulate mTOR: Linking cytoprotective gene expression to a major metabolic regulator that generates redox activity*. J. Biol. Chem. 2016, 291, 25476–25488. [Google Scholar] [CrossRef]
- Di Francesco, A.; Choi, Y.; Bernier, M.; Zhang, Y.; Diaz-Ruiz, A.; Aon, M.A.; Kalafut, K.; Ehrlich, M.R.; Murt, K.; Ali, A.; et al. NQO1 protects obese mice through improvements in glucose and lipid metabolism. npj Aging Mech. Dis. 2020, 6, 13. [Google Scholar] [CrossRef]
- Pereyra, C.E.; Dantas, R.F.; Ferreira, S.B.; Gomes, L.P.; Silva, F.P., Jr. The diverse mechanisms and anticancer potential of naphthoquinones. Cancer Cell Int. 2019, 19, 207. [Google Scholar] [CrossRef]
- Castle, A.R.; Gill, A.C. Physiological Functions of the Cellular Prion Protein. Front. Mol. Biosci. 2017, 4, 19. [Google Scholar] [CrossRef]
- Cichon, A.-C.; Brown, D.R. Nrf-2 regulation of prion protein expression is independent of oxidative stress. Mol. Cell. Neurosci. 2014, 63, 31–37. [Google Scholar] [CrossRef]
- Gall Troselj, K.; Tomljanovic, M.; Jaganjac, M.; Matijevic Glavan, T.; Cipak Gasparovic, A.; Milkovic, L.; Borovic Sunjic, S.; Buttari, B.; Profumo, E.; Saha, S.; et al. Oxidative Stress and Cancer Heterogeneity Orchestrate NRF2 Roles Relevant for Therapy Response. Molecules 2022, 27, 1468. [Google Scholar] [CrossRef]
- Yun, J.W.; Zhao, Z.; Yan, X.; Vatamaniuk, M.Z.; Lei, X.G. Glutathione peroxidase-1 inhibits transcription of regenerating islet-derived protein-2 in pancreatic islets. Free Radic. Biol. Med. 2019, 134, 385–393. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohe, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Gharib, A.F.; Alaa Eldeen, M.; Khalifa, A.S.; Elsawy, W.H.; Eed, E.M.; Askary, A.E.; Eid, R.A.; Soltan, M.A.; Raafat, N. Assessment of Glutathione Peroxidase-1 (GPX1) Gene Expression as a Specific Diagnostic and Prognostic Biomarker in Malignant Pleural Mesothelioma. Diagnostics 2021, 11, 2285. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, E.; Gromadzinska, J.; Peplonska, B.; Fendler, W.; Reszka, E.; Krol, M.B.; Wieczorek, E.; Bukowska, A.; Gresner, P.; Galicki, M.; et al. Lipid peroxidation and glutathione peroxidase activity relationship in breast cancer depends on functional polymorphism of GPX1. BMC Cancer 2015, 15, 657. [Google Scholar] [CrossRef]
- Liu, Q.; Ma, J.Y.; Wu, G. Identification and validation of a ferroptosis-related gene signature predictive of prognosis in breast cancer. Aging (Albany NY) 2021, 13, 21385–21399. [Google Scholar] [CrossRef]
- Li, Z.; Chen, L.; Chen, C.; Zhou, Y.; Hu, D.; Yang, J.; Chen, Y.; Zhuo, W.; Mao, M.; Zhang, X.; et al. Targeting ferroptosis in breast cancer. Biomark. Res. 2020, 8, 58. [Google Scholar] [CrossRef]
- Wang, N.; Gu, Y.; Li, L.; Chi, J.; Liu, X.; Xiong, Y.; Jiang, S.; Zhang, W.; Zhong, C. Identification of novel prognostic risk signature of breast cancer based on ferroptosis-related genes. Sci. Rep. 2022, 12, 13766. [Google Scholar] [CrossRef]
- Tonolo, F.; Folda, A.; Scalcon, V.; Marin, O.; Bindoli, A.; Rigobello, M.P. Nrf2-Activating Bioactive Peptides Exert Anti-Inflammatory Activity through Inhibition of the NF-kappaB Pathway. Int. J. Mol. Sci. 2022, 23, 4382. [Google Scholar] [CrossRef]
- Vomund, S.; Schafer, A.; Parnham, M.J.; Brune, B.; von Knethen, A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef]
- Ahuja, M.; Kaidery, N.A.; Dutta, D.; Attucks, O.C.; Kazakov, E.H.; Gazaryan, I.; Matsumoto, M.; Igarashi, K.; Sharma, S.M.; Thomas, B. Harnessing the Therapeutic Potential of the Nrf2/Bach1 Signaling Pathway in Parkinson’s Disease. Antioxidants 2022, 11, 1780. [Google Scholar] [CrossRef]
- Fock, E.M.; Parnova, R.G. Protective Effect of Mitochondria-Targeted Antioxidants against Inflammatory Response to Lipopolysaccharide Challenge: A Review. Pharmaceutics 2021, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Kahroba, H.; Ramezani, B.; Maadi, H.; Sadeghi, M.R.; Jaberie, H.; Ramezani, F. The role of Nrf2 in neural stem/progenitor cells: From maintaining stemness and self-renewal to promoting differentiation capability and facilitating therapeutic application in neurodegenerative disease. Ageing Res. Rev. 2021, 65, 101211. [Google Scholar] [CrossRef] [PubMed]
- Bryan, H.K.; Olayanju, A.; Goldring, C.E.; Park, B.K. The Nrf2 cell defense pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 2013, 85, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Gorrini, C.; Baniasadi, P.S.; Harris, I.S.; Silvester, J.; Inoue, S.; Snow, B.; Joshi, P.A.; Wakeham, A.; Molyneux, S.D.; Martin, B.; et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J. Exp. Med. 2013, 210, 1529–1544. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.; Li, W.; Li, H.; Bao, J.; Yang, C.; Wang, A.; Wei, J.; Chen, S.; Jin, H. Role of Nrf2 in the antioxidation and oxidative stress induced developmental toxicity of honokiol in zebrafish. Toxicol. Appl. Pharmacol. 2019, 373, 48–61. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Markman, J.L.; Shiao, S.L. Impact of the immune system and immunotherapy in colorectal cancer. J. Gastrointest. Oncol. 2015, 6, 208–223. [Google Scholar] [CrossRef]
- Inaguma, S.; Lasota, J.; Wang, Z.; Czapiewski, P.; Langfort, R.; Rys, J.; Szpor, J.; Waloszczyk, P.; Okon, K.; Biernat, W.; et al. Expression of ALCAM (CD166) and PD-L1 (CD274) independently predicts shorter survival in malignant pleural mesothelioma. Hum. Pathol. 2018, 71, 1–7. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, X.; Zhang, C.; Wang, Y.; Cheng, T.; Duan, L.; Tong, Z.; Tan, S.; Zhang, H.; Saw, P.E.; et al. Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc. Natl. Acad. Sci. USA 2020, 117, 6640–6650. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K.; Cross, N.; Jordan-Mahy, N.; Leyland, R. The Extrinsic and Intrinsic Roles of PD-L1 and Its Receptor PD-1: Implications for Immunotherapy Treatment. Front. Immunol. 2020, 11, 568931. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Regulation of PD-L1 expression on cancer cells with ROS-modulating drugs. Life Sci. 2020, 246, 117403. [Google Scholar] [CrossRef] [PubMed]
- Brion, C.; Lutz, S.M.; Albert, F.W. Simultaneous quantification of mRNA and protein in single cells reveals post-transcriptional effects of genetic variation. eLife 2020, 9, e60645. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Laham-Karam, N.; Pinto, G.P.; Poso, A.; Kokkonen, P. Transcription and Translation Inhibitors in Cancer Treatment. Front. Chem. 2020, 8, 276. [Google Scholar] [CrossRef]
- Crunkhorn, S. Deal watch: Abbott boosts investment in NRF2 activators for reducing oxidative stress. Nat. Rev. Drug Discov. 2012, 11, 96. [Google Scholar] [CrossRef]



| A. MDA-MB-231 TNBC Treated with TQ vs. Control | B. MDA-MB-468 TNBC Treated with TQ vs. Control | ||||
|---|---|---|---|---|---|
| GENE | Fold Change | p-Value | GENE | Fold Change | p-Value |
| B2M | −2.37 | 0.000986 | SIRT2 | −11.68 | 0.027194 |
| PRDX5 | −2.69 | 0.003623 | SEPP1 | −15.84 | 0.031466 |
| RNF7 | −2.42 | 0.014579 | NOX5 | −6.14 | 0.048628 |
| GPX1 | −2.54 | 0.015438 | PRNP | +157.65 | 0.002687 |
| RPLPO | −3.09 | 0.020751 | GCLM | +4.78 | 0.002548 |
| PNKP | −3.77 | 0.020877 | GPX1 | +20.87 | 0.008767 |
| NUDT1 | −1.88 | 0.022986 | NQO1 | +48.87 | 0.016668 |
| BNIP3 | −2.46 | 0.030008 | |||
| MGST3 | −1.48 | 0.039413 | |||
| GPX4 | −2.69 | 0.042205 | |||
| ATOX1 | −2.14 | 0.043778 | |||
| GCLM | +2.17 | 0.034524 | |||
| SRXN1 | +1.85 | 0.035065 | |||
| TXNRD1 | +1.40 | 0.036500 | |||
| NQO1 | +2.63 | 0.042095 | |||
| PRNP | +1.70 | 0.042419 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adinew, G.M.; Messeha, S.S.; Taka, E.; Badisa, R.B.; Soliman, K.F.A. Anticancer Effects of Thymoquinone through the Antioxidant Activity, Upregulation of Nrf2, and Downregulation of PD-L1 in Triple-Negative Breast Cancer Cells. Nutrients 2022, 14, 4787. https://doi.org/10.3390/nu14224787
Adinew GM, Messeha SS, Taka E, Badisa RB, Soliman KFA. Anticancer Effects of Thymoquinone through the Antioxidant Activity, Upregulation of Nrf2, and Downregulation of PD-L1 in Triple-Negative Breast Cancer Cells. Nutrients. 2022; 14(22):4787. https://doi.org/10.3390/nu14224787
Chicago/Turabian StyleAdinew, Getinet M., Samia S. Messeha, Equar Taka, Ramesh B. Badisa, and Karam F. A. Soliman. 2022. "Anticancer Effects of Thymoquinone through the Antioxidant Activity, Upregulation of Nrf2, and Downregulation of PD-L1 in Triple-Negative Breast Cancer Cells" Nutrients 14, no. 22: 4787. https://doi.org/10.3390/nu14224787
APA StyleAdinew, G. M., Messeha, S. S., Taka, E., Badisa, R. B., & Soliman, K. F. A. (2022). Anticancer Effects of Thymoquinone through the Antioxidant Activity, Upregulation of Nrf2, and Downregulation of PD-L1 in Triple-Negative Breast Cancer Cells. Nutrients, 14(22), 4787. https://doi.org/10.3390/nu14224787








