High Dietary Folic Acid Intake Is Associated with Genomic Instability in Peripheral Lymphocytes of Healthy Adults
Abstract
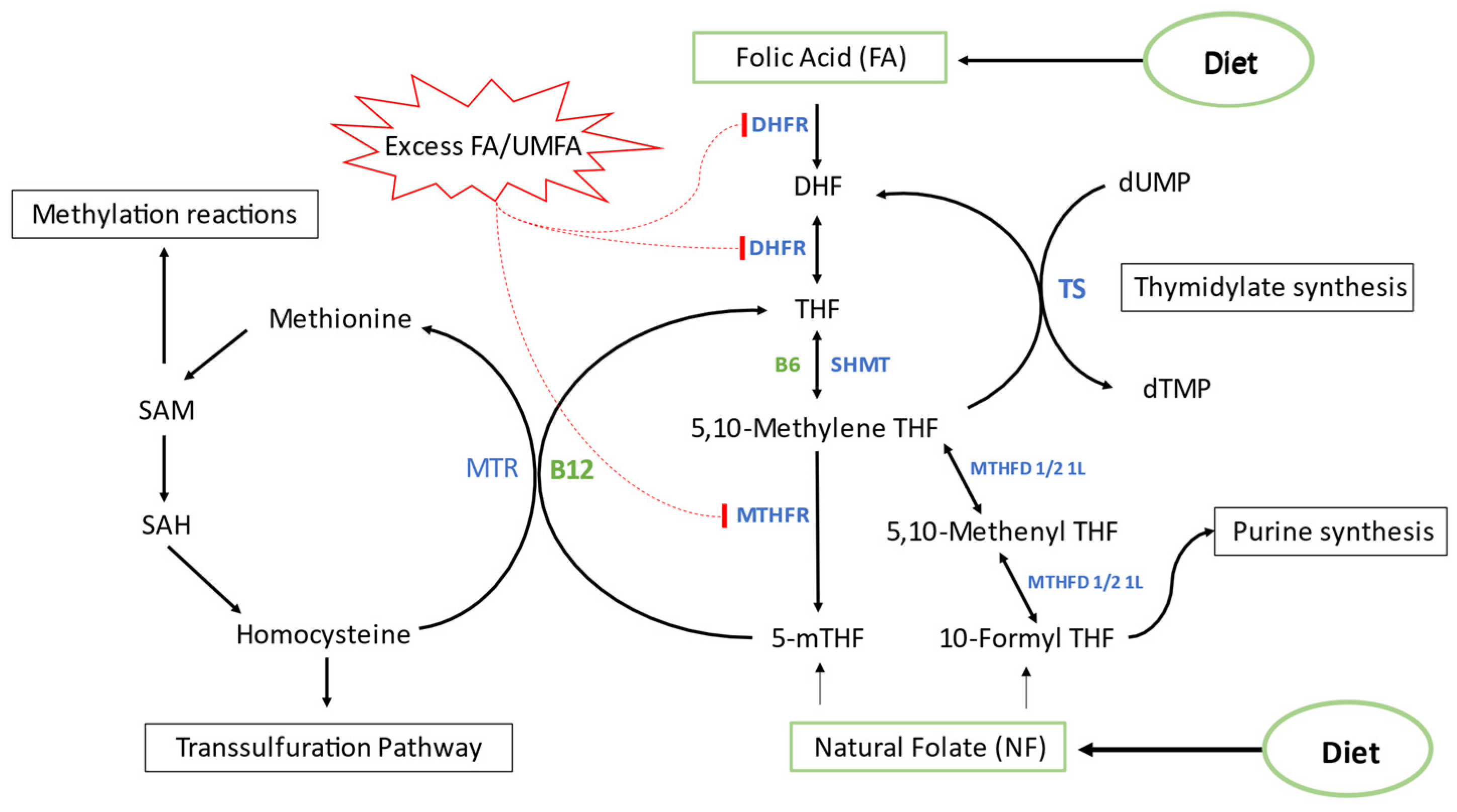
1. Introduction
2. Materials and Methods
2.1. Recruitment and Data Collection
2.2. Dietary Intake Assessment and Folic Acid Intake Analysis
2.3. Blood Samples Collection and Analysis
2.4. Cytokinesis-Block Micronucleus (CBMN) Assay
2.5. LINE-1 Methylation Assay
2.6. Gene Expression Profiling
2.7. Statistical Analysis
3. Results
3.1. General Participants Characteristics
3.2. Systemic Markers
3.3. Nutrient Intake
3.4. Comparative Data Analysis
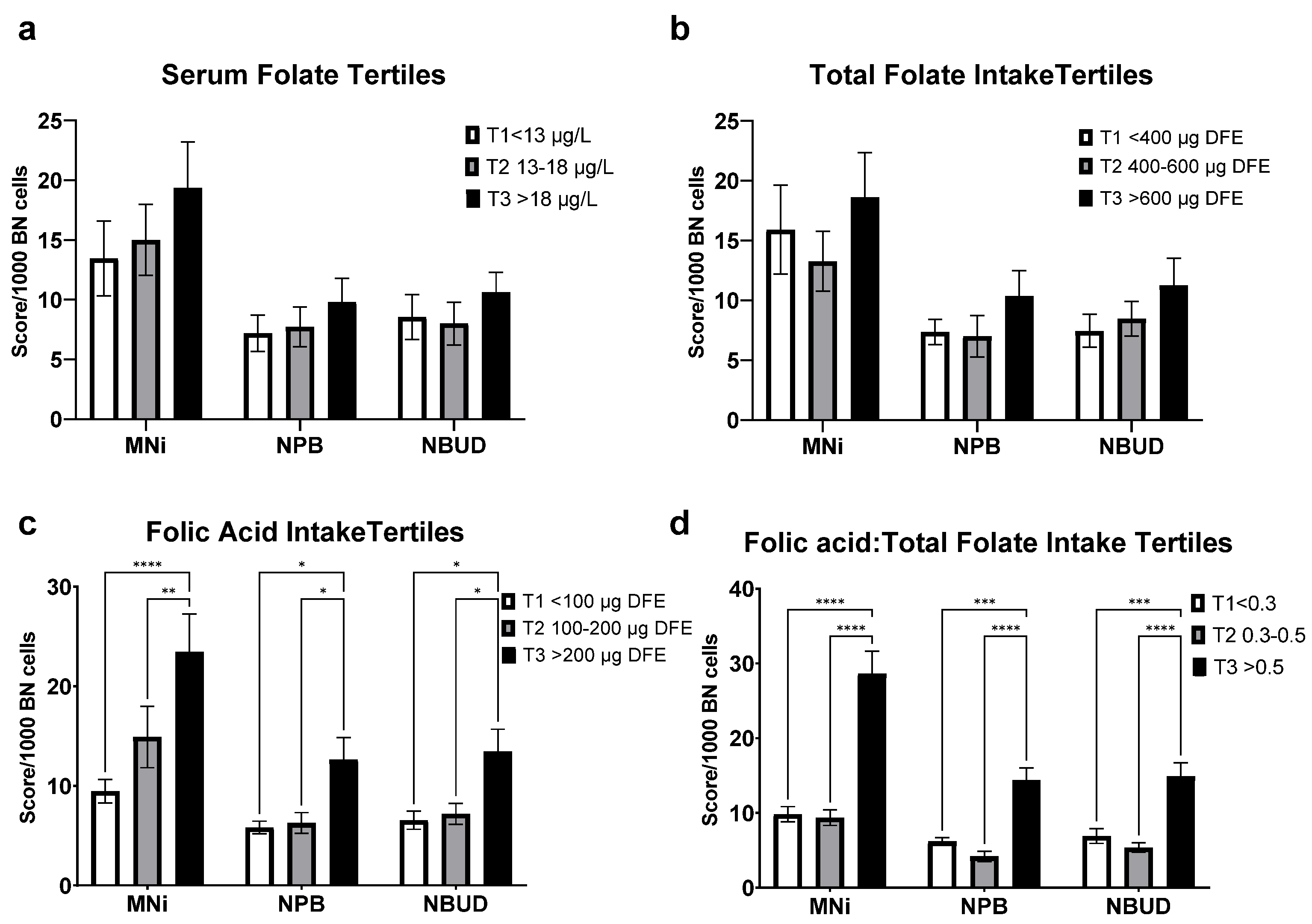
3.5. Cytome Biomarkers Analyses
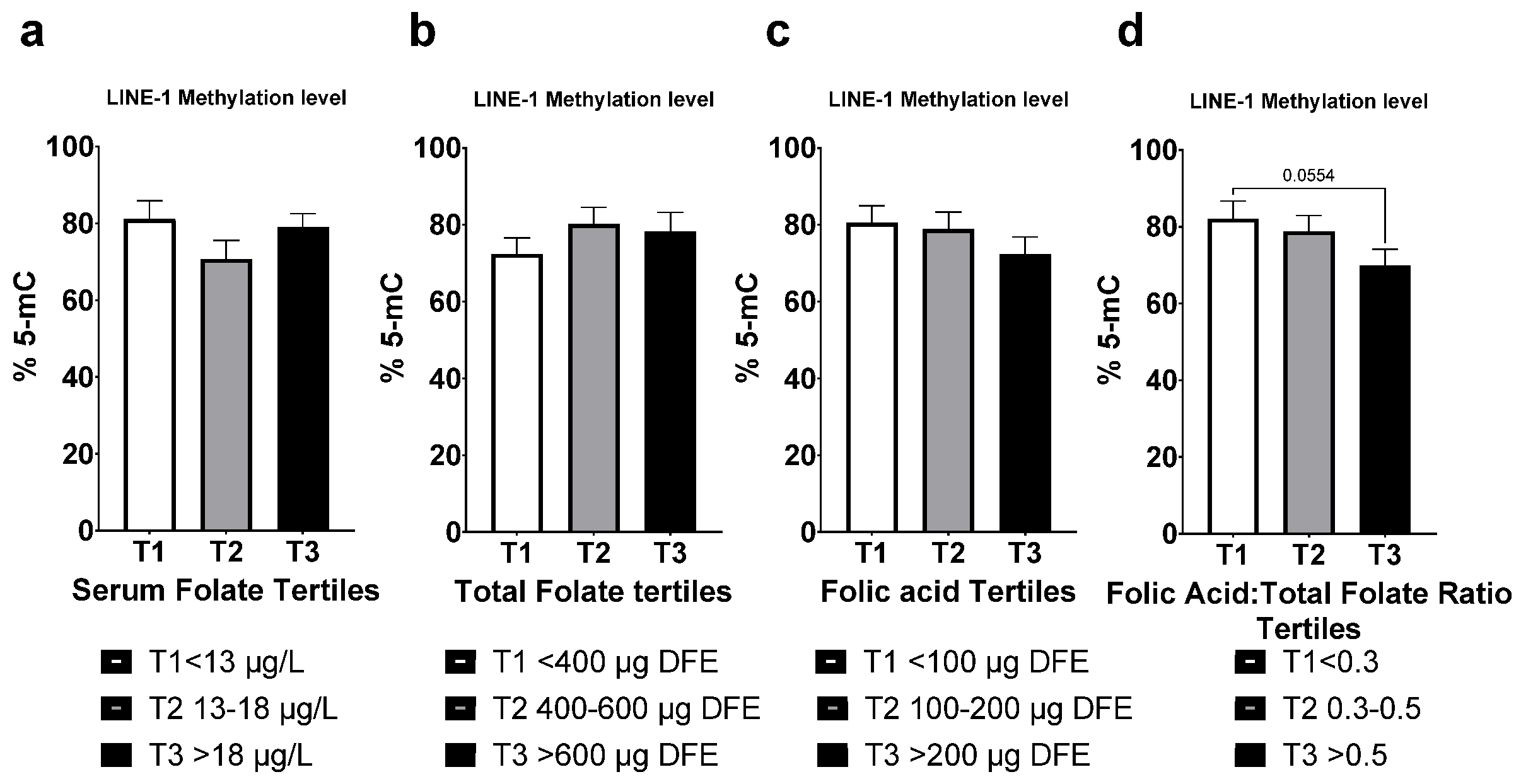
3.6. LINE-1 Methylation
3.7. Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-fTHF | 5-formyl-tetrahydrofolate |
| 5-mTHF | 5-methyl-tetrahydrofolate |
| CBMN | Cytokinesis block micronuclei assay |
| DHF | Dihydrofolate |
| DHFR | Dihydrofolate reductase |
| FA | Folic acid |
| FAR | Folic acid to total folate intake ratio |
| Hcy | Homocysteine |
| LCL | Lymphoblastoid cell line |
| LINE-1 | Long interspersed nuclear elements |
| MGMT | O-6-methylguanine-DNA methyltransferase |
| MLH1 | mutL homolog 1 |
| MNi | Micronuclei |
| MMA | Methylmalonic acid |
| MTHFR | Methylene tetrahydrofolate reductase |
| MTR | 5-methyltetrahydrofolate-homocysteine methyltransferase |
| NBUD | Nuclear buds |
| NPB | Nucleoplasmic bridges |
| NTD | Neural tube defects |
| RBC | Red blood cells |
| RDA | Recommended daily allowance |
| TS | Thymidylate synthase |
| UL | Upper limit |
| UMFA | Unmetabolized folic acid |
| UNG | Uracil DNA glycosylase |
References
- Beaudin, A.E.; Stover, P.J. Insights into metabolic mechanisms underlying folate-responsive neural tube defects: A minireview. Birth Defects Res. A Clin. Mol. Teratol. 2009, 85, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y. Folate and carcinogenesis: Evidence, mechanisms, and implications. J. Nutr. Biochem. 1999, 10, 66–88. [Google Scholar] [CrossRef]
- Wien, T.N.; Pike, E.; Wisloff, T.; Staff, A.; Smeland, S.; Klemp, M. Cancer risk with folic acid supplements: A systematic review and meta-analysis. BMJ Open 2012, 2, e000653. [Google Scholar] [CrossRef] [PubMed]
- Duthie, S.J. Folic acid deficiency and cancer: Mechanisms of DNA instability. Br. Med. Bull. 1999, 55, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Yates, Z.; Veysey, M.; Heo, Y.R.; Lucock, M. Contemporary issues surrounding folic Acid fortification initiatives. Prev. Nutr. Food Sci. 2014, 19, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Bailey, L.B.; Berry, R.J. Folic acid food fortification-its history, effect, concerns, and future directions. Nutrients 2011, 3, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Boulet, S.L.; Yang, Q.; Mai, C.; Kirby, R.S.; Collins, J.S.; Robbins, J.M.; Meyer, R.; Canfield, M.A.; Mulinare, J.; National Birth Defects Prevention, N. Trends in the postfortification prevalence of spina bifida and anencephaly in the United States. Birth Defects Res. A Clin. Mol. Teratol. 2008, 82, 527–532. [Google Scholar] [CrossRef]
- Williams, L.J.; Mai, C.T.; Edmonds, L.D.; Shaw, G.M.; Kirby, R.S.; Hobbs, C.A.; Sever, L.E.; Miller, L.A.; Meaney, F.J.; Levitt, M. Prevalence of spina bifida and anencephaly during the transition to mandatory folic acid fortification in the United States. Teratology 2002, 66, 33–39. [Google Scholar] [CrossRef]
- Selhub, J.; Rosenberg, I.H. Excessive folic acid intake and relation to adverse health outcome. Biochimie 2016, 126, 71–78. [Google Scholar] [CrossRef]
- Choumenkovitch, S.F.; Selhub, J.; Wilson, P.W.; Rader, J.I.; Rosenberg, I.H.; Jacques, P.F. Folic acid intake from fortification in United States exceeds predictions. J. Nutr. 2002, 132, 2792–2798. [Google Scholar] [CrossRef]
- Kalmbach, R.D.; Choumenkovitch, S.F.; Troen, A.M.; D’Agostino, R.; Jacques, P.F.; Selhub, J. Circulating folic acid in plasma: Relation to folic acid fortification. Am. J. Clin. Nutr. 2008, 88, 763–768. [Google Scholar] [CrossRef]
- Shane, B. Folate fortification: Enough already? Am. J. Clin. Nutr. 2003, 77, 8–9. [Google Scholar] [CrossRef]
- Quinlivan, E.P.; Gregory, J.F., 3rd. Reassessing folic acid consumption patterns in the United States (1999 2004): Potential effect on neural tube defects and overexposure to folate. Am. J. Clin. Nutr. 2007, 86, 1773–1779. [Google Scholar] [CrossRef]
- Quinlivan, E.P.; Gregory, J.F. Effect of food fortification on folic acid intake in the United States. Am. J. Clin. Nutr. 2003, 77, 221–225. [Google Scholar] [CrossRef]
- Wright, A.J.A.; Dainty, J.R.; Finglas, P.M. Folic acid metabolism in human subjects revisited: Potential implications for proposed mandatory folic acid fortification in the UK. Br. J. Nutr. 2007, 98, 667–675. [Google Scholar] [CrossRef]
- Obeid, R.; Herrmann, W. The emerging role of unmetabolized folic acid in human diseases: Myth or reality? Curr. Drug Metab. 2012, 13, 1184–1195. [Google Scholar] [CrossRef]
- Kim, Y.I. Folic acid fortification and supplementation-Good for some but not so good for others. Nutr. Rev. 2007, 65, 504–511. [Google Scholar] [CrossRef]
- Kelly, P.; McPartlin, J.; Goggins, M.; Weir, D.G.; Scott, J.M. Unmetabolized folic acid in serum: Acute studies in subjects consuming fortified food and supplements. Am. J. Clin. Nutr. 1997, 65, 1790–1795. [Google Scholar] [CrossRef]
- Hirsch, S.; Sanchez, H.; Albala, C.; Maza, M.P.d.l.; Barrera, G.; Leiva, L.; Bunout, D. Colon cancer in Chile before and after the start of the flour fortification program with folic acid. Eur. J. Gastroenterol. Hepatol. 2009, 21, 436–439. [Google Scholar] [CrossRef]
- Mason, J.B.; Tang, S.Y. Folate status and colorectal cancer risk: A 2016 update. Mol. Aspects Med. 2017, 53, 73–79. [Google Scholar] [CrossRef]
- Yajnik, C.S.; Deshpande, S.S.; Jackson, A.A.; Refsum, H.; Rao, S.; Fisher, D.J.; Bhat, D.S.; Naik, S.S.; Coyaji, K.J.; Joglekar, C.V.; et al. Vitamin B12 and folate concentrations during pregnancy and insulin resistance in the offspring: The Pune Maternal Nutrition Study. Diabetologia 2008, 51, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Sawaengsri, H.; Wang, J.; Reginaldo, C.; Steluti, J.; Wu, D.; Meydani, S.N.; Selhub, J.; Paul, L. High folic acid intake reduces natural killer cell cytotoxicity in aged mice. J. Nutr. Biochem. 2016, 30, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Troen, A.M.; Mitchell, B.; Sorensen, B.; Wener, M.H.; Johnston, A.; Wood, B.; Selhub, J.; McTiernan, A.; Yasui, Y.; Oral, E.; et al. Unmetabolized folic acid in plasma is associated with reduced natural killer cell cytotoxicity among postmenopausal women. J. Nutr. 2006, 136, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Fardous, A.M.; Beydoun, S.; James, A.A.; Ma, H.; Cabelof, D.C.; Unnikrishnan, A.; Heydari, A.R. The Timing and Duration of Folate Restriction Differentially Impacts Colon Carcinogenesis. Nutrients 2021, 14, 16. [Google Scholar] [CrossRef]
- Kim, Y.I. Folate and colorectal cancer: An evidence-based critical review. Mol. Nutr. Food Res. 2007, 51, 267–292. [Google Scholar] [CrossRef]
- Wiens, D.; DeSoto, M.C. Is High Folic Acid Intake a Risk Factor for Autism?—A Review. Brain Sci. 2017, 7, 149. [Google Scholar] [CrossRef]
- Beard, C.M.; Panser, L.A.; Katusic, S.K. Is excess folic acid supplementation a risk factor for autism? Med. Hypotheses 2011, 77, 15–17. [Google Scholar] [CrossRef]
- Shane, B. Folate Chemistry and Metabolism*. Clin. Res. Regul. Aff. 2001, 18, 137–159. [Google Scholar] [CrossRef]
- Smith, A.D.; Kim, Y.I.; Refsum, H. Is folic acid good for everyone? Am. J. Clin. Nutr. 2008, 87, 517–533. [Google Scholar] [CrossRef]
- Bailey, L.B.; Stover, P.J.; McNulty, H.; Fenech, M.F.; Gregory, J.F., 3rd; Mills, J.L.; Pfeiffer, C.M.; Fazili, Z.; Zhang, M.; Ueland, P.M.; et al. Biomarkers of Nutrition for Development-Folate Review. J. Nutr. 2015, 145, 1636S–1680S. [Google Scholar] [CrossRef]
- Bailey, S.W.; Ayling, J.E. The extremely slow and variable activity of dihydrofolate reductase in human liver and its implications for high folic acid intake. Proc. Natl. Acad. Sci. USA 2009, 106, 15424–15429. [Google Scholar] [CrossRef]
- Patanwala, I.; King, M.J.; Barrett, D.A.; Rose, J.; Jackson, R.; Hudson, M.; Philo, M.; Dainty, J.R.; Wright, A.J.; Finglas, P.M.; et al. Folic acid handling by the human gut: Implications for food fortification and supplementation. Am. J. Clin. Nutr. 2014, 100, 593–599. [Google Scholar] [CrossRef]
- Pfeiffer, C.M.; Sternberg, M.R.; Fazili, Z.; Yetley, E.A.; Lacher, D.A.; Bailey, R.L.; Johnson, C.L. Unmetabolized Folic Acid Is Detected in Nearly All Serum Samples from US Children, Adolescents, and Adults. J. Nutr. 2015, 145, 520–531. [Google Scholar] [CrossRef]
- Obeid, R.; Kasoha, M.; Kirsch, S.H.; Munz, W.; Herrmann, W. Concentrations of unmetabolized folic acid and primary folate forms in pregnant women at delivery and in umbilical cord blood. Am. J. Clin. Nutr. 2010, 92, 1416–1422. [Google Scholar] [CrossRef]
- Williams, B.A.; Mayer, C.; McCartney, H.; Devlin, A.M.; Lamers, Y.; Vercauteren, S.M.; Wu, J.K.; Karakochuk, C.D. Detectable Unmetabolized Folic Acid and Elevated Folate Concentrations in Folic Acid-Supplemented Canadian Children With Sickle Cell Disease. Front. Nutr. 2021, 8, 642306. [Google Scholar] [CrossRef]
- Sowers, R.; Toguchida, J.; Qin, J.; Meyers, P.A.; Healey, J.H.; Huvos, A.; Banerjee, D.; Bertino, J.R.; Gorlick, R. mRNA expression levels of E2F transcription factors correlate with dihydrofolate reductase, reduced folate carrier, and thymidylate synthase mRNA expression in osteosarcoma. Mol. Cancer 2003, 2, 535–541. [Google Scholar]
- Hjortoe, G.M.; Weilguny, D.; Willumsen, B.M. Elk3 from hamster—A ternary complex factor with strong transcriptional repressor activity. DNA Cell Biol. 2005, 24, 35–42. [Google Scholar] [CrossRef]
- Matthews, R.G.; Baugh, C.M. Interactions of pig liver methylenetetrahydrofolate reductase with methylenetetrahydropteroylpolyglutamate substrates and with dihydropteroylpolyglutamate inhibitors. Biochemistry 1980, 19, 2040–2045. [Google Scholar] [CrossRef]
- Tam, C.; O’Connor, D.; Koren, G. Circulating unmetabolized folic Acid: Relationship to folate status and effect of supplementation. Obs. Gynecol. Int. 2012, 2012, 485179. [Google Scholar] [CrossRef]
- Kao, T.T.; Wang, K.C.; Chang, W.N.; Lin, C.Y.; Chen, B.H.; Wu, H.L.; Shi, G.Y.; Tsai, J.N.; Fu, T.F. Characterization and comparative studies of zebrafish and human recombinant dihydrofolate reductases--inhibition by folic acid and polyphenols. Drug Metab. Dispos. 2008, 36, 508–516. [Google Scholar] [CrossRef]
- Alnabbat, K.I.; Fardous, A.M.; Cabelof, D.C.; Heydari, A.R. Excessive Folic Acid Mimics Folate Deficiency in Human Lymphocytes. Curr. Issues Mol. Biol. 2022, 44, 1452–1462. [Google Scholar] [CrossRef]
- Koseki, K.; Maekawa, Y.; Bito, T.; Yabuta, Y.; Watanabe, F. High-dose folic acid supplementation results in significant accumulation of unmetabolized homocysteine, leading to severe oxidative stress in Caenorhabditis elegans. Redox Biol. 2020, 37, 101724. [Google Scholar] [CrossRef]
- Ortbauer, M.; Ripper, D.; Fuhrmann, T.; Lassi, M.; Auernigg-Haselmaier, S.; Stiegler, C.; Konig, J. Folate deficiency and over-supplementation causes impaired folate metabolism: Regulation and adaptation mechanisms in Caenorhabditis elegans. Mol. Nutr. Food Res. 2016, 60, 949–956. [Google Scholar] [CrossRef]
- Henry, C.J.; Nemkov, T.; Casas-Selves, M.; Bilousova, G.; Zaberezhnyy, V.; Higa, K.C.; Serkova, N.J.; Hansen, K.C.; D’Alessandro, A.; DeGregori, J. Folate dietary insufficiency and folic acid supplementation similarly impair metabolism and compromise hematopoiesis. Haematologica 2017, 102, 1985–1994. [Google Scholar] [CrossRef]
- Basten, G.P.; Hill, M.H.; Duthie, S.J.; Powers, H.J. Effect of folic Acid supplementation on the folate status of buccal mucosa and lymphocytes. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1244–1249. [Google Scholar] [CrossRef]
- Fenech, M.F.; Dreosti, I.E.; Rinaldi, J.R. Folate, vitamin B12, homocysteine status and chromosome damage rate in lymphocytes of older men. Carcinogenesis 1997, 18, 1329–1336. [Google Scholar] [CrossRef]
- Fenech, M.; Aitken, C.; Rinaldi, J. Folate, vitamin B12, homocysteine status and DNA damage in young Australian adults. Carcinogenesis 1998, 19, 1163–1171. [Google Scholar] [CrossRef]
- Thomas, P.; Fenech, M. Cytokinesis-block micronucleus cytome assay in lymphocytes. Methods Mol. Biol. 2011, 682, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Esha Research. Available online: https://esha.com (accessed on 1 October 2020).
- FoodDate Central. Available online: https://fdc.nal.usda.gov (accessed on 1 October 2020).
- Great Lakes Medical Laboratory, Inc. Available online: http://glmlinc.com (accessed on 1 October 2020).
- Fenech, M.; Rinaldi, J. The relationship between micronuclei in human lymphocytes and plasma levels of vitamin C, vitamin E, vitamin B12 and folic acid. Carcinogenesis 1994, 15, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Baghurst, P.; Luderer, W.; Turner, J.; Record, S.; Ceppi, M.; Bonassi, S. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability—Results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis 2005, 26, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007, 2, 1084–1104. [Google Scholar] [CrossRef]
- Fenech, M.; Chang, W.P.; Kirsch-Volders, M.; Holland, N.; Bonassi, S.; Zeiger, E. HUMN project: Detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2003, 534, 65–75. [Google Scholar] [CrossRef]
- Crott, J.W.; Mashiyama, S.T.; Ames, B.N.; Fenech, M.F. Methylenetetrahydrofolate reductase C677T polymorphism does not alter folic acid deficiency-induced uracil incorporation into primary human lymphocyte DNA in vitro. Carcinogenesis 2001, 22, 1019–1025. [Google Scholar] [CrossRef]
- Wang, X.; Fenech, M. A comparison of folic acid and 5-methyltetrahydrofolate for prevention of DNA damage and cell death in human lymphocytes in vitro. Mutagenesis 2003, 18, 81–86. [Google Scholar] [CrossRef]
- Wang, X.; Wu, X.; Liang, Z.; Huang, Y.; Fenech, M.; Xue, J. A comparison of folic acid deficiency-induced genomic instability in lymphocytes of breast cancer patients and normal non-cancer controls from a Chinese population in Yunnan. Mutagenesis 2006, 21, 41–47. [Google Scholar] [CrossRef]
- Fenech, M. Folate (vitamin B9) and vitamin B12 and their function in the maintenance of nuclear and mitochondrial genome integrity. Mutat. Res. 2012, 733, 21–33. [Google Scholar] [CrossRef]
- Stopper, H.; Treutlein, A.T.; Bahner, U.; Schupp, N.; Schmid, U.; Brink, A.; Perna, A.; Heidland, A. Reduction of the genomic damage level in haemodialysis patients by folic acid and vitamin B12 supplementation. Nephrol. Dial. Transpl. 2008, 23, 3272–3279. [Google Scholar] [CrossRef]
- Ohrvik, V.E.; Witthoft, C.M. Human folate bioavailability. Nutrients 2011, 3, 475–490. [Google Scholar] [CrossRef]
- Pfeiffer, C.M.; Caudill, S.P.; Gunter, E.W.; Osterloh, J.; Sampson, E.J. Biochemical indicators of B vitamin status in the US population after folic acid fortification: Results from the National Health and Nutrition Examination Survey 1999–2000. Am. J. Clin. Nutr. 2005, 82, 442–450. [Google Scholar] [CrossRef]
- Maruvada, P.; Stover, P.J.; Mason, J.B.; Bailey, R.L.; Davis, C.D.; Field, M.S.; Finnell, R.H.; Garza, C.; Green, R.; Gueant, J.L.; et al. Knowledge gaps in understanding the metabolic and clinical effects of excess folates/folic acid: A summary, and perspectives, from an NIH workshop. Am. J. Clin. Nutr. 2020, 112, 1390–1403. [Google Scholar] [CrossRef]
- Obeid, R.; Kirsch, S.H.; Kasoha, M.; Eckert, R.; Herrmann, W. Concentrations of unmetabolized folic acid and primary folate forms in plasma after folic acid treatment in older adults. Metabolism 2011, 60, 673–680. [Google Scholar] [CrossRef]
- Berti, C.; Fekete, K.; Dullemeijer, C.; Trovato, M.; Souverein, O.W.; Cavelaars, A.; Dhonukshe-Rutten, R.; Massari, M.; Decsi, T.; Van’t Veer, P.; et al. Folate intake and markers of folate status in women of reproductive age, pregnant and lactating women: A meta-analysis. J. Nutr. Metab. 2012, 2012, 470656. [Google Scholar] [CrossRef]
- Ladeira, C.; Carolino, E.; Gomes, M.C.; Brito, M. Role of Macronutrients and Micronutrients in DNA Damage: Results From a Food Frequency Questionnaire. Nutr. Metab. Insights 2017, 10. [Google Scholar] [CrossRef]
- Caudill, M.A. Folate bioavailability: Implications for establishing dietary recommendations and optimizing status. Am. J. Clin. Nutr. 2010, 91, 1455S–1460S. [Google Scholar] [CrossRef]
- Madhyastha, S.; Prabhu, L.V.; Saralaya, V.; Rai, R. A comparison of vitamin A and leucovorin for the prevention of methotrexate-induced micronuclei production in rat bone marrow. Clinics 2008, 63, 821–826. [Google Scholar] [CrossRef]
- Cho, Y.H.; Woo, H.D.; Jang, Y.; Porter, V.; Christensen, S.; Hamilton, R.F., Jr.; Chung, H.W. The Association of LINE-1 Hypomethylation with Age and Centromere Positive Micronuclei in Human Lymphocytes. PLoS ONE 2015, 10, e0133909. [Google Scholar] [CrossRef]
- Charles, M.A.; Johnson, I.T.; Belshaw, N.J. Supra-physiological folic acid concentrations induce aberrant DNA methylation in normal human cells in vitro. Epigenetics 2012, 7, 689–694. [Google Scholar] [CrossRef]
- Christensen, K.E.; Mikael, L.G.; Leung, K.Y.; Levesque, N.; Deng, L.; Wu, Q.; Malysheva, O.V.; Best, A.; Caudill, M.A.; Greene, N.D.; et al. High folic acid consumption leads to pseudo-MTHFR deficiency, altered lipid metabolism, and liver injury in mice. Am. J. Clin. Nutr. 2015, 101, 646–658. [Google Scholar] [CrossRef]
- Cosin-Tomas, M.; Luan, Y.; Leclerc, D.; Malysheva, O.V.; Lauzon, N.; Bahous, R.H.; Christensen, K.E.; Caudill, M.A.; Rozen, R. Moderate Folic Acid Supplementation in Pregnant Mice Results in Behavioral Alterations in Offspring with Sex-Specific Changes in Methyl Metabolism. Nutrients 2020, 12, 1716. [Google Scholar] [CrossRef]
- Steluti, J.; Palchetti, C.Z.; Miranda, A.M.; Fisberg, R.M.; Marchioni, D.M. DNA methylation and one-carbon metabolism related nutrients and polymorphisms: Analysis after mandatory flour fortification with folic acid. Br. J. Nutr. 2020, 123, 23–29. [Google Scholar] [CrossRef]
- Plumptre, L.; Tammen, S.A.; Sohn, K.J.; Masih, S.P.; Visentin, C.E.; Aufreiter, S.; Malysheva, O.; Schroder, T.H.; Ly, A.; Berger, H.; et al. Maternal and Cord Blood Folate Concentrations Are Inversely Associated with Fetal DNA Hydroxymethylation, but Not DNA Methylation, in a Cohort of Pregnant Canadian Women. J. Nutr. 2020, 150, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Yajnik, C.S.; Deshmukh, U.S. Maternal nutrition, intrauterine programming and consequential risks in the offspring. Rev. Endocr. Metab. Disord. 2008, 9, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Kintaka, Y.; Wada, N.; Shioda, S.; Nakamura, S.; Yamazaki, Y.; Mochizuki, K. Excessive folic acid supplementation in pregnant mice impairs insulin secretion and induces the expression of genes associated with fatty liver in their offspring. Heliyon 2020, 6, e03597. [Google Scholar] [CrossRef]
- Yadon, N.; Owen, A.; Cakora, P.; Bustamante, A.; Hall-South, A.; Smith, N.; Felder, M.R.; Vrana, P.B.; Shorter, K.R. A high methyl donor diet affects physiology and behavior in Peromyscus polionotus. Physiol. Behav. 2019, 209, 112615. [Google Scholar] [CrossRef]
- Tojal, A.; Neves, C.; Veiga, H.; Ferreira, S.; Rodrigues, I.; Martel, F.; Calhau, C.; Negrao, R.; Keating, E. Perigestational high folic acid: Impact on offspring’s peripheral metabolic response. Food Funct. 2019, 10, 7216–7226. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Li, L.; Jiang, Y.; Tan, J.; Ji, J.; Zhang, Y.; Jin, N.; Liu, F. Excess Folic Acid Supplementation Before and During Pregnancy and Lactation Activates Fos Gene Expression and Alters Behaviors in Male Mouse Offspring. Front. Neurosci. 2019, 13, 313. [Google Scholar] [CrossRef]
- Faux, N.G.; Ellis, K.A.; Porter, L.; Fowler, C.J.; Laws, S.M.; Martins, R.N.; Pertile, K.K.; Rembach, A.; Rowe, C.C.; Rumble, R.L.; et al. Homocysteine, vitamin B12, and folic acid levels in Alzheimer’s disease, mild cognitive impairment, and healthy elderly: Baseline characteristics in subjects of the Australian Imaging Biomarker Lifestyle study. J. Alzheimers Dis. 2011, 27, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, H.; Hossain, M.B.; Lera, L.; Hirsch, S.; Albala, C.; Uauy, R.; Broberg, K.; Ronco, A.M. High levels of circulating folate concentrations are associated with DNA methylation of tumor suppressor and repair genes p16, MLH1, and MGMT in elderly Chileans. Clin. Epigenetics 2017, 9, 74. [Google Scholar] [CrossRef]
- Shan, M.; Yin, H.; Li, J.; Li, X.; Wang, D.; Su, Y.; Niu, M.; Zhong, Z.; Wang, J.; Zhang, X.; et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget 2016, 7, 18485–18494. [Google Scholar] [CrossRef]
- Blount, B.C.; Mack, M.M.; Wehr, C.M.; MacGregor, J.T.; Hiatt, R.A.; Wang, G.; Wickramasinghe, S.N.; Everson, R.B.; Ames, B.N. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. USA 1997, 94, 3290–3295. [Google Scholar] [CrossRef]
- Bahous, R.H.; Jadavji, N.M.; Deng, L.; Cosin-Tomas, M.; Lu, J.; Malysheva, O.; Leung, K.Y.; Ho, M.K.; Pallas, M.; Kaliman, P.; et al. High dietary folate in pregnant mice leads to pseudo-MTHFR deficiency and altered methyl metabolism, with embryonic growth delay and short-term memory impairment in offspring. Hum. Mol. Genet. 2017, 26, 888–900. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alnabbat, K.I.; Fardous, A.M.; Shahab, A.; James, A.A.; Bahry, M.R.; Heydari, A.R. High Dietary Folic Acid Intake Is Associated with Genomic Instability in Peripheral Lymphocytes of Healthy Adults. Nutrients 2022, 14, 3944. https://doi.org/10.3390/nu14193944
Alnabbat KI, Fardous AM, Shahab A, James AA, Bahry MR, Heydari AR. High Dietary Folic Acid Intake Is Associated with Genomic Instability in Peripheral Lymphocytes of Healthy Adults. Nutrients. 2022; 14(19):3944. https://doi.org/10.3390/nu14193944
Chicago/Turabian StyleAlnabbat, Khadijah I., Ali M. Fardous, Aiman Shahab, Andrew A. James, Manhel R. Bahry, and Ahmad R. Heydari. 2022. "High Dietary Folic Acid Intake Is Associated with Genomic Instability in Peripheral Lymphocytes of Healthy Adults" Nutrients 14, no. 19: 3944. https://doi.org/10.3390/nu14193944
APA StyleAlnabbat, K. I., Fardous, A. M., Shahab, A., James, A. A., Bahry, M. R., & Heydari, A. R. (2022). High Dietary Folic Acid Intake Is Associated with Genomic Instability in Peripheral Lymphocytes of Healthy Adults. Nutrients, 14(19), 3944. https://doi.org/10.3390/nu14193944






