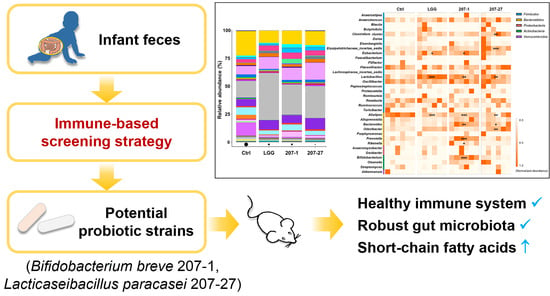
Potential Health-Promoting Effects of Two Candidate Probiotics Isolated from Infant Feces Using an Immune-Based Screening Strategy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Preparation of Bacterial Cultures and Suspensions
2.3. Isolation and Treatment of Primary Murine Splenic Cells
2.4. Animal Experimentation
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. RNA Isolation and RT-qPCR
2.7. Western Blotting
2.8. Cecal SCFA Detection
2.9. Fecal DNA Extraction and 16S rRNA Sequencing
2.10. Bioinformatic Analysis
2.11. Statistical Analyses
3. Results
3.1. Immunomodulatory Effects of the Tested Strains on Primary Murine Splenic Cells
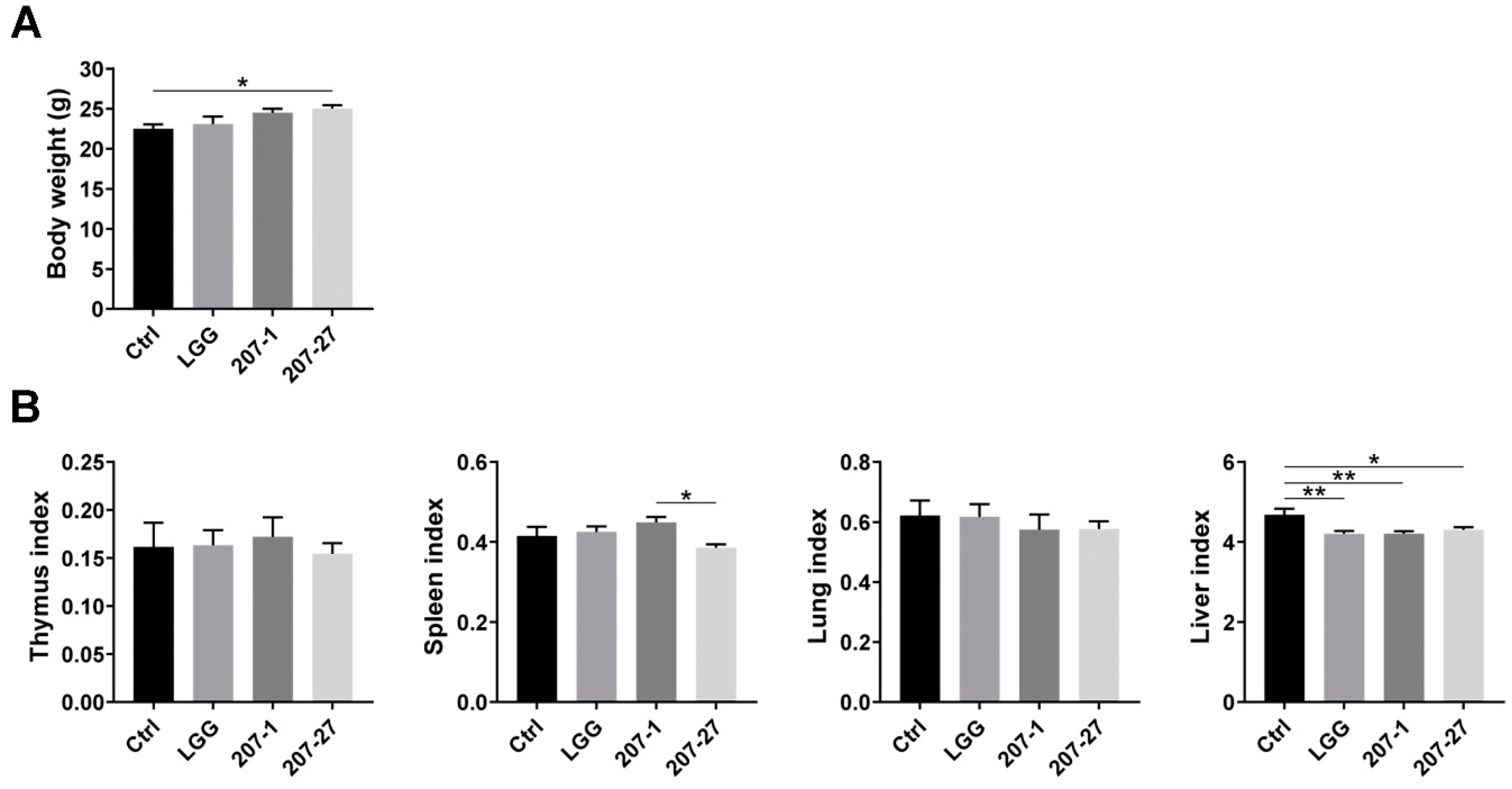
3.2. Bodyweight and Organ Indices of Mice
3.3. Splenic and Serum Cytokine Levels in Mice
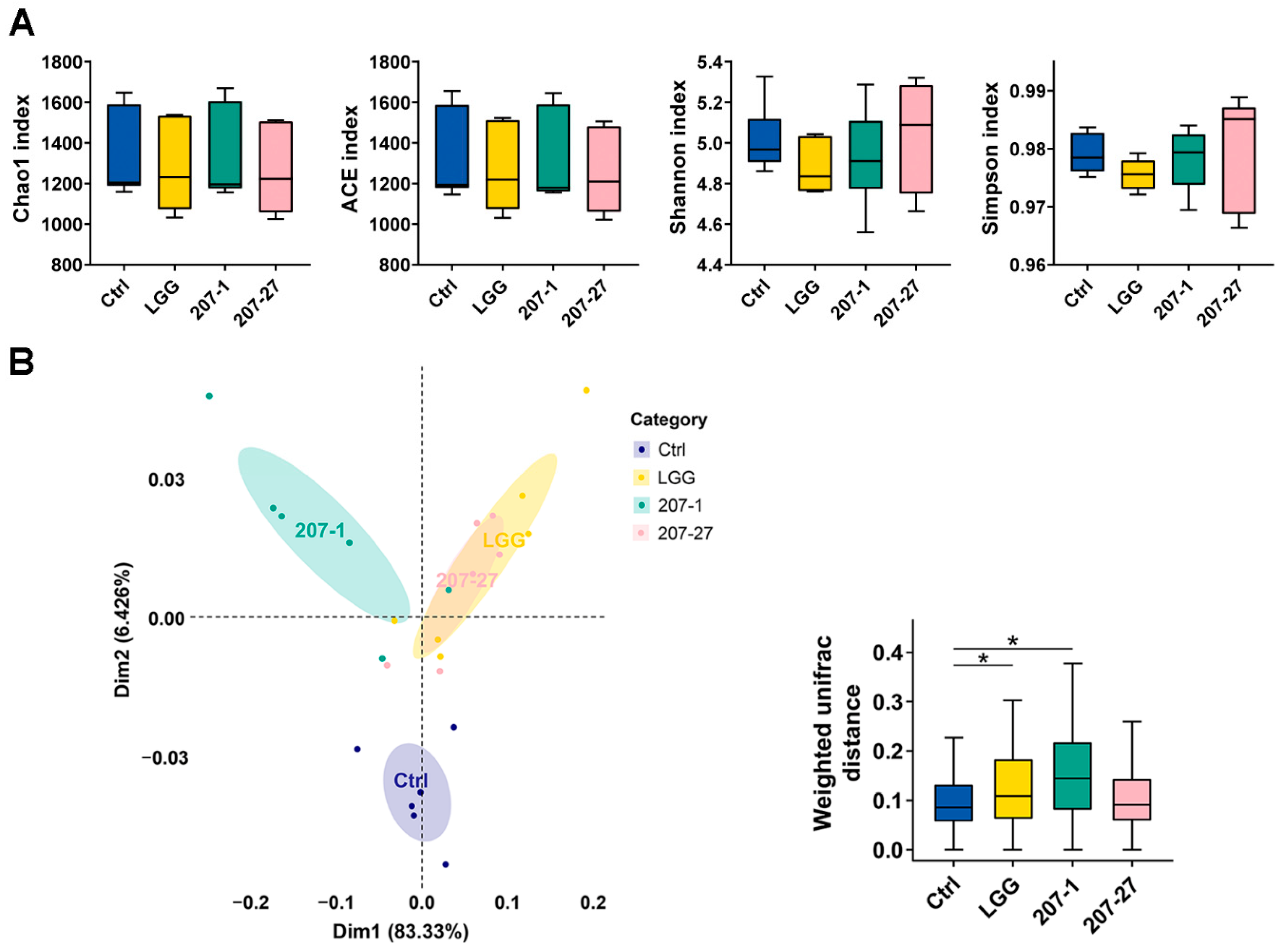
3.4. Changes in Fecal Microbial Diversity
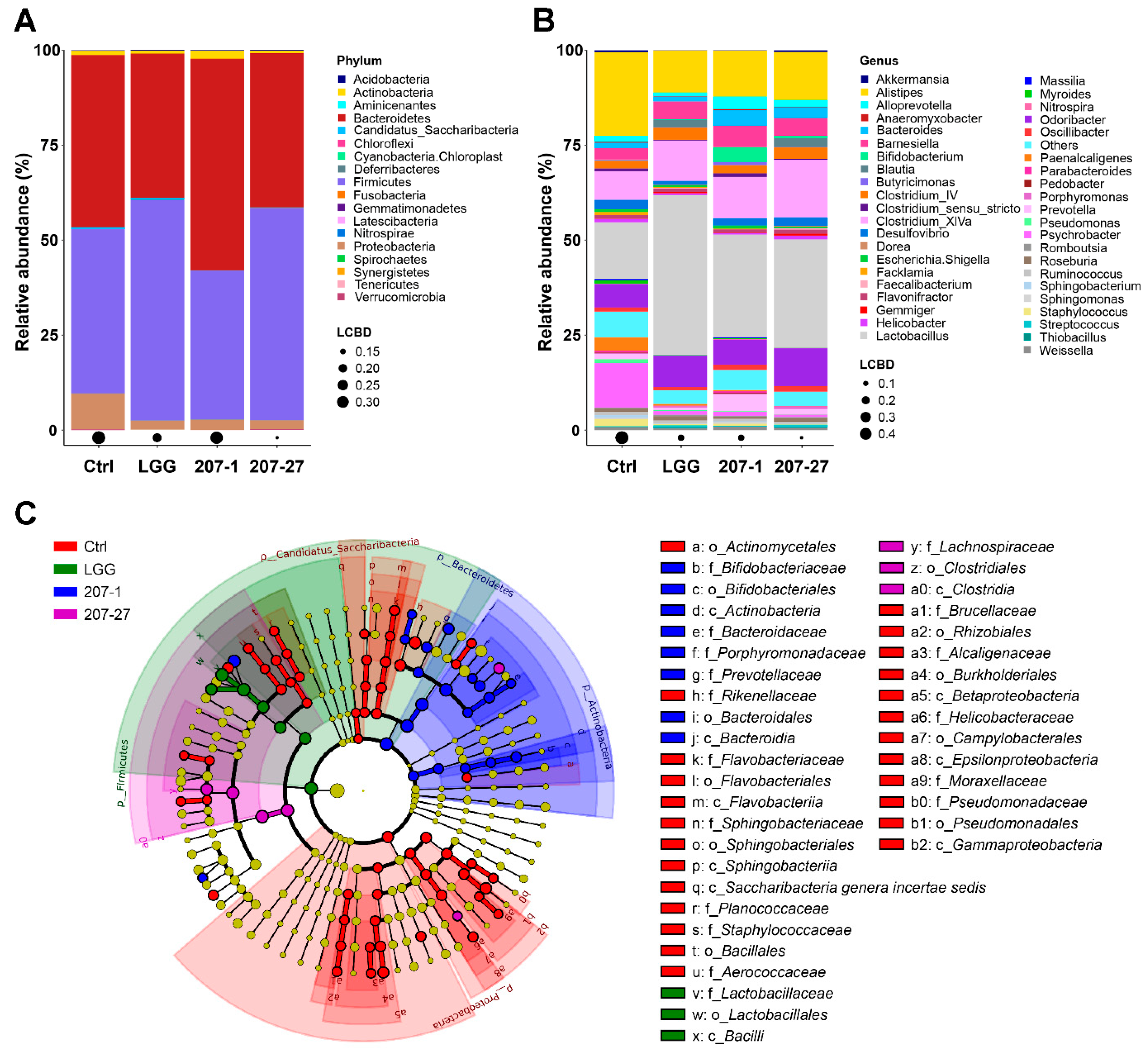
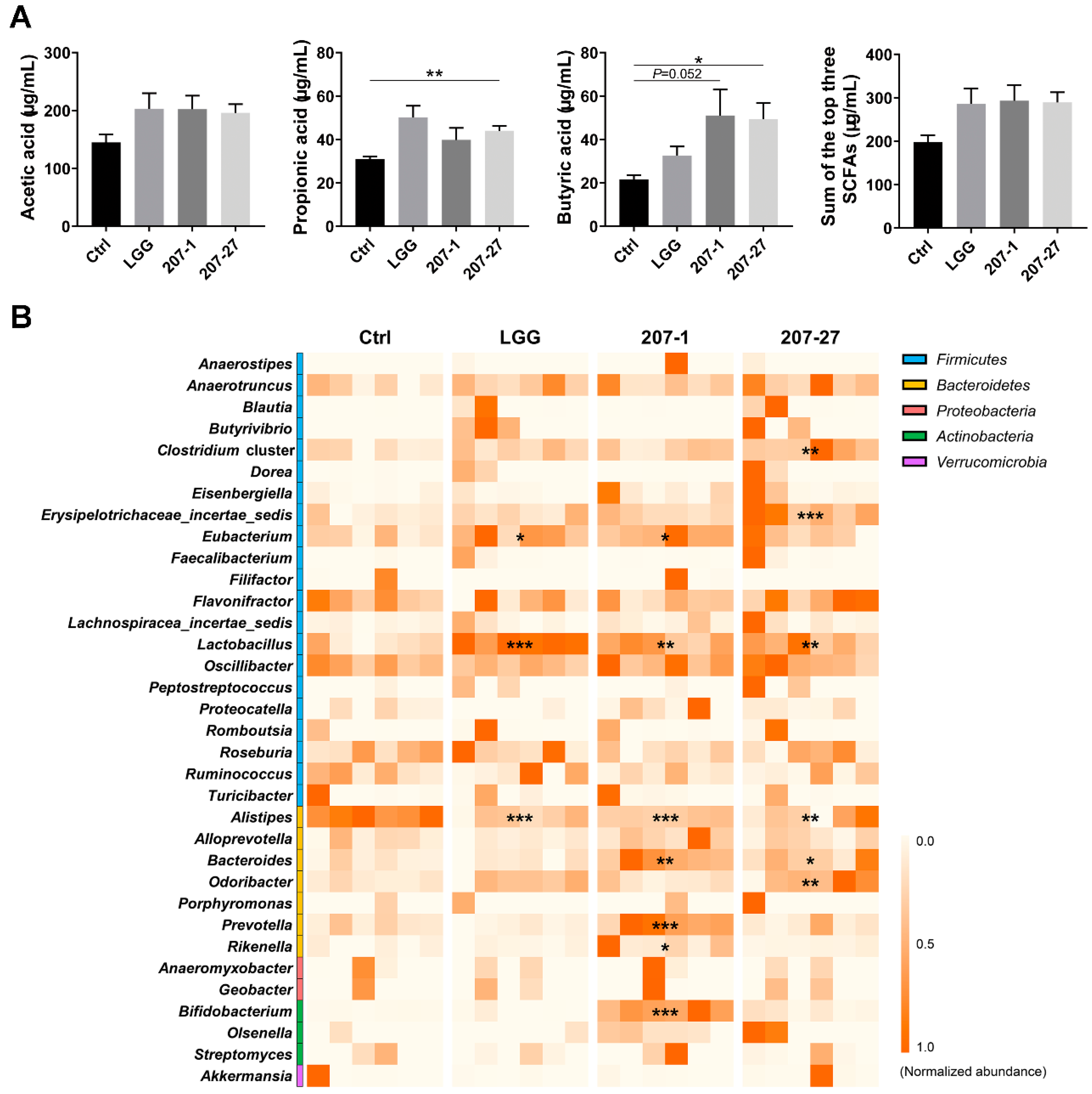
3.5. Changes in Fecal Microbial Composition
3.6. SCFA Levels Were Increased by Altered Gut Microbiota
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.T.; Chan, G.C.; Li, J.C. Systemic effects of gut microbiota and its relationship with disease and modulation. BMC Immunol. 2015, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Shokryazdan, P.; Faseleh Jahromi, M.; Liang, J.B.; Ho, Y.W. Probiotics: From Isolation to Application. J. Am. Coll. Nutr. 2017, 36, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations; World Health Organization. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2002; pp. 1–11. [Google Scholar]
- Kincaid, H.J.; Nagpal, R.; Yadav, H. Microbiome-immune-metabolic axis in the epidemic of childhood obesity: Evidence and opportunities. Obes. Rev. 2020, 21, e12963. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Microbes, Immunity, and Behavior: Psychoneuroimmunology Meets the Microbiome. Neuropsychopharmacology 2017, 42, 178–192. [Google Scholar] [CrossRef]
- Hand, T.W.; Vujkovic-Cvijin, I.; Ridaura, V.K.; Belkaid, Y. Linking the Microbiota, Chronic Disease, and the Immune System. Trends Endocrinol. Metab. 2016, 27, 831–843. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Sarker, M.; Wan, D. Immunomodulatory Effects of Probiotics on Cytokine Profiles. BioMed Res. Int. 2018, 2018, 8063647. [Google Scholar] [CrossRef]
- Li, S.; Wan, Q.; Cheng, R.; Ge, L.; Wu, Z.; Li, M.; Zhang, X.; Xiao, J.; Huang, Y.; He, F. Intestine lactobacilli of 1–4 months infant and characterize their biochemical properties in Chengdu City in 2015–2016. J. Hyg. Res. 2018, 47, 776–781. [Google Scholar]
- Liang, H.; Wan, Q.; Luo, Z.; Shen, X.; Zhang, X.; He, F. Composition and immunomodulatory effects of intestinal bifidobacteria in infants from Chengdu. Acta Nutr. Sin. 2019, 41, 568–575. [Google Scholar]
- Liang, H.; Luo, Z.; Miao, Z.; Shen, X.; Li, M.; Zhang, X.; Chen, J.; Ze, X.; Chen, Q.; He, F. Lactobacilli and bifidobacteria derived from infant intestines may activate macrophages and lead to different IL-10 secretion. Biosci. Biotechnol. Biochem. 2020, 84, 2558–2568. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Jiang, F.; Cheng, R.; Luo, Y.; Wang, J.; Luo, Z.; Li, M.; Shen, X.; He, F. A high-fat diet and high-fat and high-cholesterol diet may affect glucose and lipid metabolism differentially through gut microbiota in mice. Exp. Anim. 2021, 70, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 2016. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Vital, M.; Karch, A.; Pieper, D.H. Colonic Butyrate-Producing Communities in Humans: An Overview Using Omics Data. mSystems 2017, 2, e00130-17. [Google Scholar] [CrossRef]
- Singhal, R.; Donde, H.; Ghare, S.; Stocke, K.; Zhang, J.; Vadhanam, M.; Reddy, S.; Gobejishvili, L.; Chilton, P.; Joshi-Barve, S.; et al. Decrease in acetyl-CoA pathway utilizing butyrate-producing bacteria is a key pathogenic feature of alcohol-induced functional gut microbial dysbiosis and development of liver disease in mice. Gut Microbes 2021, 13, 1946367. [Google Scholar] [CrossRef]
- Ratajczak, W.; Ryl, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczynska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef]
- Morita, H.; He, F.; Fuse, T.; Ouwehand, A.C.; Hashimoto, H.; Hosoda, M.; Mizumachi, K.; Kurisaki, J.-I. Cytokine production by the murine macrophage cell line J774.1 after exposure to lactobacilli. Biosci. Biotechnol. Biochem. 2002, 66, 1963–1966. [Google Scholar] [CrossRef]
- Miyazawa, K.; He, F.; Yoda, K.; Hiramatsu, M. Potent effects of, and mechanisms for, modification of crosstalk between macrophages and adipocytes by lactobacilli. Microbiol. Immunol. 2012, 56, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Li, M.; Miyazawa, K.; Li, Y.; Hiramatsu, M.; Xu, J.; Gong, C.; Jing, X.; He, F.; Huang, C. Effects of heat-inactivated Lactobacillus gasseri TMC0356 on metabolic characteristics and immunity of rats with the metabolic syndrome. Br. J. Nutr. 2013, 109, 263–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Fata, G.; Weber, P.; Mohajeri, M.H. Probiotics and the Gut Immune System: Indirect Regulation. Probiotics Antimicrob. Proteins 2018, 10, 11–21. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef] [PubMed]
- Mohr, A.E.; Basile, A.J.; Crawford, M.S.; Sweazea, K.L.; Carpenter, K.C. Probiotic Supplementation Has a Limited Effect on Circulating Immune and Inflammatory Markers in Healthy Adults: A Systematic Review of Randomized Controlled Trials. J. Acad. Nutr. Diet. 2020, 120, 548–564. [Google Scholar] [CrossRef] [PubMed]
- Foligne, B.; Nutten, S.; Grangette, C.; Dennin, V.; Goudercourt, D.; Poiret, S.; Dewulf, J.; Brassart, D.; Mercenier, A.; Pot, B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 2007, 13, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L. The gut microbiota and obesity: From correlation to causality. Nat. Rev. Microbiol. 2013, 11, 639–647. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Shao, L.; Ling, Z.; Chen, D.; Liu, Y.; Yang, F.; Li, L. Disorganized Gut Microbiome Contributed to Liver Cirrhosis Progression: A Meta-Omics-Based Study. Front. Microbiol. 2018, 9, 3166. [Google Scholar] [CrossRef]
- Sung, C.M.; Lin, Y.F.; Chen, K.F.; Ke, H.M.; Huang, H.Y.; Gong, Y.N.; Tsai, W.S.; You, J.F.; Lu, M.J.; Cheng, H.T.; et al. Predicting Clinical Outcomes of Cirrhosis Patients With Hepatic Encephalopathy From the Fecal Microbiome. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 301–318.e2. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.; Gui, S.; Zhou, C.; Chen, J.; Yang, C.; Hu, Z.; Wang, H.; Zhong, X.; Zeng, L.; et al. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. NeuroReport 2018, 29, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Y.; Zhang, X.; Yu, Z.H.; Zhang, Z.; Deng, M.; Zhao, J.H.; Ruan, B. Altered gut microbiota profile in patients with generalized anxiety disorder. J. Psychiatr. Res. 2018, 104, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Kang, Y.; Li, Y.; Du, Y.; Guo, L.; Chen, M.; Huang, X.; Yang, F.; Hong, J.; Kong, X. Konjaku flour reduces obesity in mice by modulating the composition of the gut microbiota. Int. J. Obes. 2019, 43, 1631–1643. [Google Scholar] [CrossRef]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; Qin, H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 2019, 60, 175–184. [Google Scholar] [CrossRef]
- Wan, Y.; Wang, F.; Yuan, J.; Li, J.; Jiang, D.; Zhang, J.; Li, H.; Wang, R.; Tang, J.; Huang, T.; et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: A 6-month randomised controlled-feeding trial. Gut 2019, 69, 1717. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Health-Promoting Effects. Microbiol. Spectr. 2017, 5, 73–98. [Google Scholar] [CrossRef]
- Heeney, D.D.; Gareau, M.G.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef]
- Ubeda, C.; Bucci, V.; Caballero, S.; Djukovic, A.; Toussaint, N.C.; Equinda, M.; Lipuma, L.; Ling, L.; Gobourne, A.; No, D.; et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 2013, 81, 965–973. [Google Scholar] [CrossRef]
- Anhê, F.F.; Nachbar, R.T.; Varin, T.V.; Vilela, V.; Dudonné, S.; Pilon, G.; Fournier, M.; Lecours, M.A.; Desjardins, Y.; Roy, D.; et al. A polyphenol-rich cranberry extract reverses insulin resistance and hepatic steatosis independently of body weight loss. Mol. Metab. 2017, 6, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Grigor’eva, I.N. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J. Pers. Med. 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- May, K.S.; den Hartigh, L.J. Modulation of Adipocyte Metabolism by Microbial Short-Chain Fatty Acids. Nutrients 2021, 13, 3666. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, H.; Ze, X.; Wang, S.; Wang, Y.; Peng, C.; Cheng, R.; Jiang, F.; Wu, S.; He, R.; He, F.; et al. Potential Health-Promoting Effects of Two Candidate Probiotics Isolated from Infant Feces Using an Immune-Based Screening Strategy. Nutrients 2022, 14, 3651. https://doi.org/10.3390/nu14173651
Liang H, Ze X, Wang S, Wang Y, Peng C, Cheng R, Jiang F, Wu S, He R, He F, et al. Potential Health-Promoting Effects of Two Candidate Probiotics Isolated from Infant Feces Using an Immune-Based Screening Strategy. Nutrients. 2022; 14(17):3651. https://doi.org/10.3390/nu14173651
Chicago/Turabian StyleLiang, Huijing, Xiaolei Ze, Silu Wang, Yimei Wang, Chenrui Peng, Ruyue Cheng, Fengling Jiang, Simou Wu, Ruikun He, Fang He, and et al. 2022. "Potential Health-Promoting Effects of Two Candidate Probiotics Isolated from Infant Feces Using an Immune-Based Screening Strategy" Nutrients 14, no. 17: 3651. https://doi.org/10.3390/nu14173651
APA StyleLiang, H., Ze, X., Wang, S., Wang, Y., Peng, C., Cheng, R., Jiang, F., Wu, S., He, R., He, F., Zhang, X., & Shen, X. (2022). Potential Health-Promoting Effects of Two Candidate Probiotics Isolated from Infant Feces Using an Immune-Based Screening Strategy. Nutrients, 14(17), 3651. https://doi.org/10.3390/nu14173651