Epidemiological Studies of Children’s Gut Microbiota: Validation of Sample Collection and Storage Methods and Microbiota Analysis of Toddlers’ Feces Collected from Diapers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
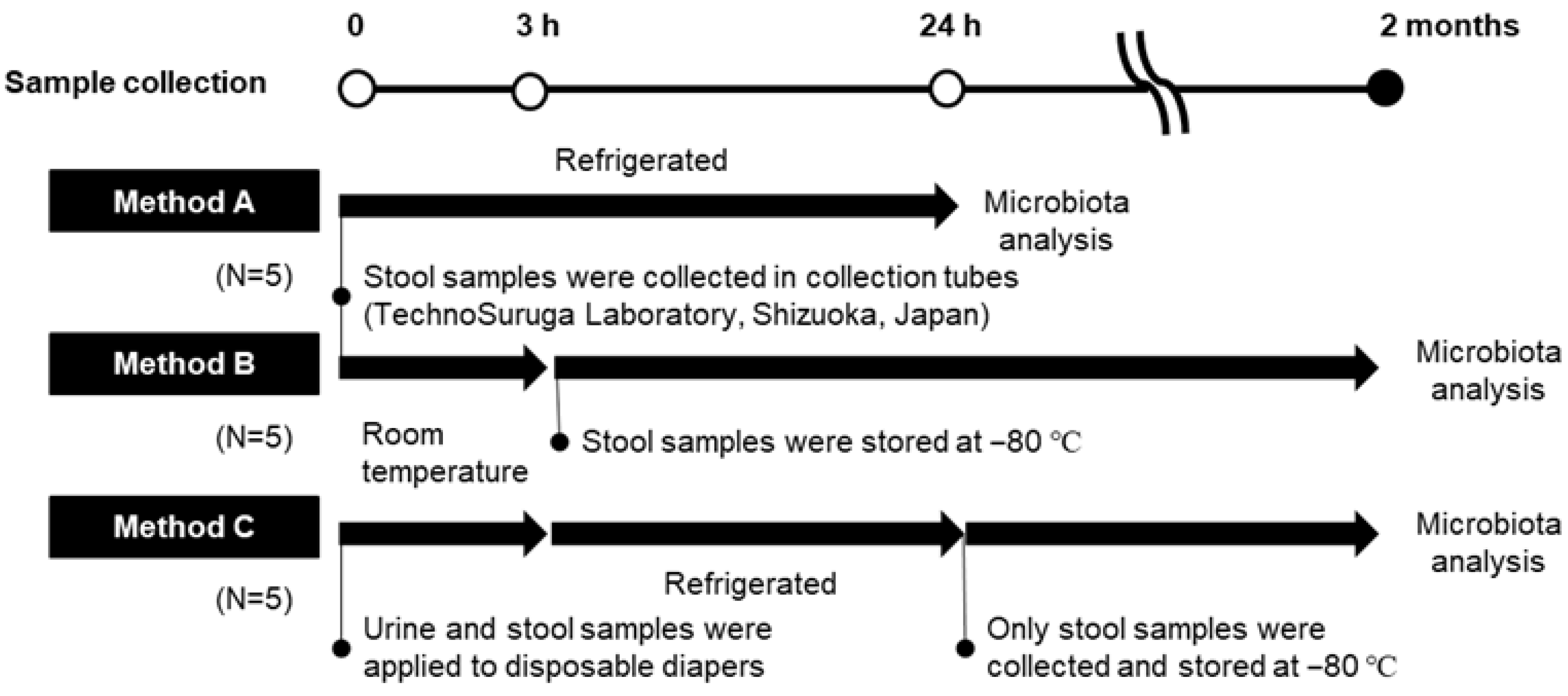
2.1.1. Research I: Gut Microbiota Analysis of Fecal Specimens Collected and Stored by Different Methods
2.1.2. Research II: Analysis of Toddlers’ Feces Excreted in Diapers
2.2. Gut Microbiota Analysis
2.3. Statistical Analysis
3. Results
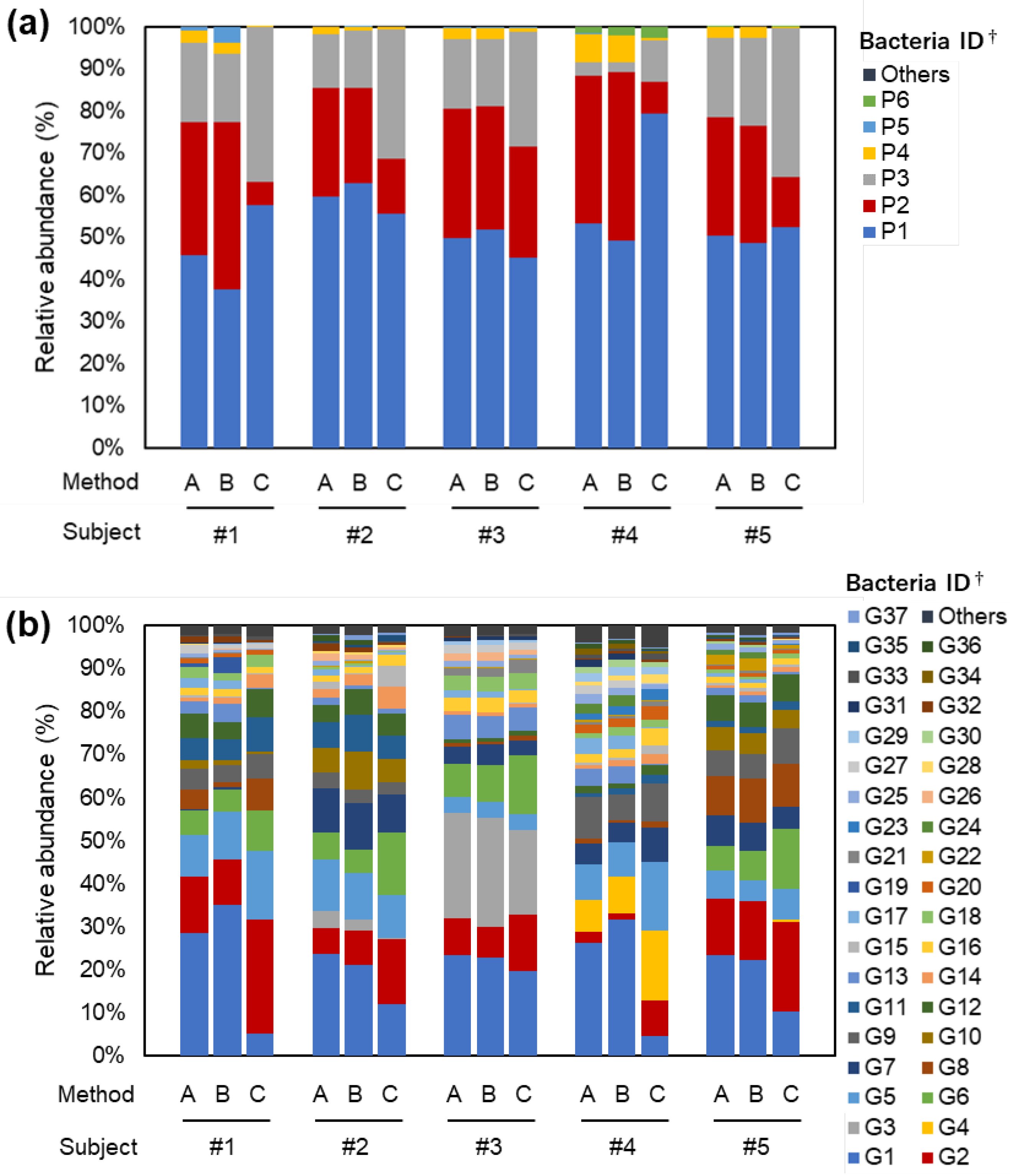
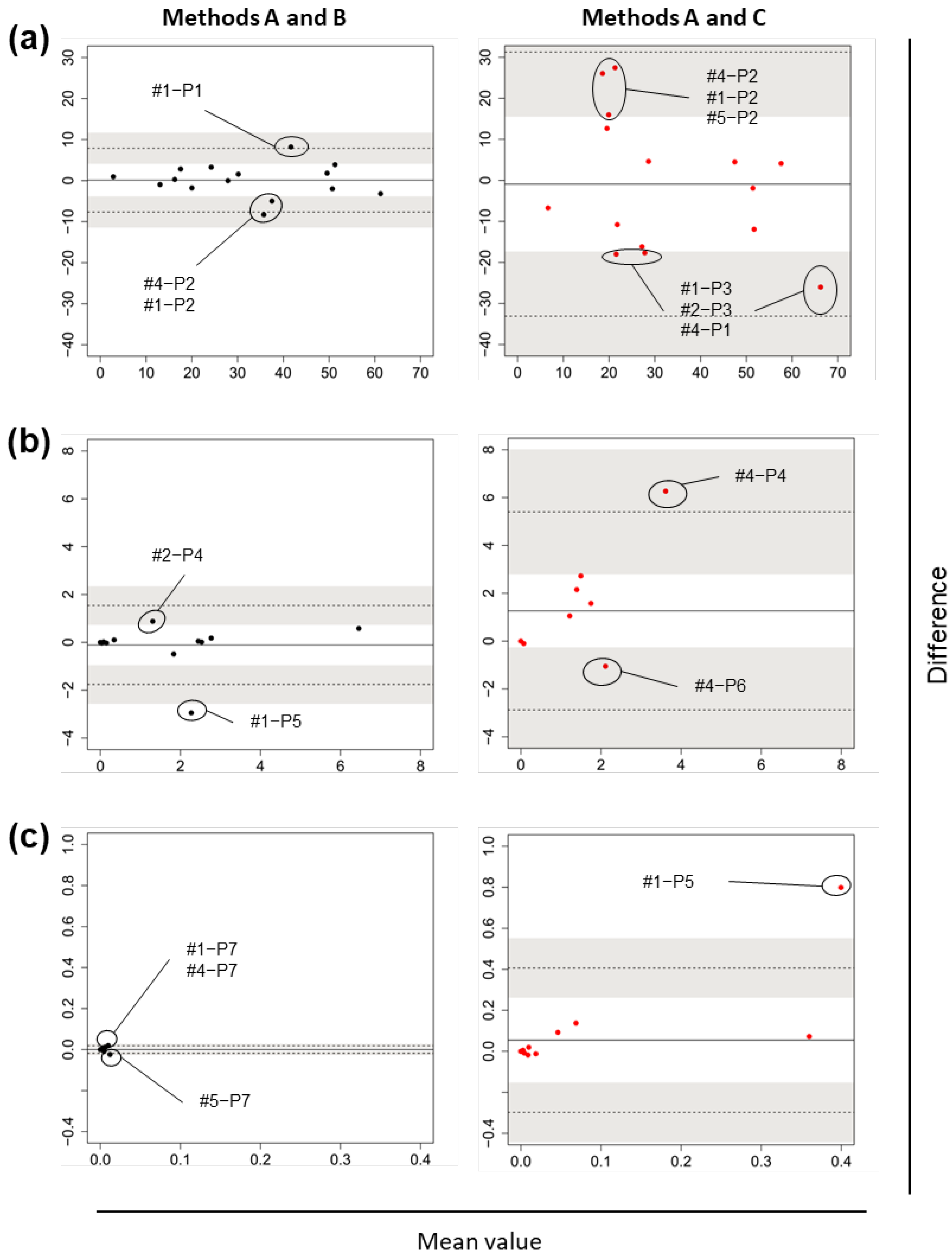
3.1. Research I: Gut Bacterial Compositions According to Three Different Methods
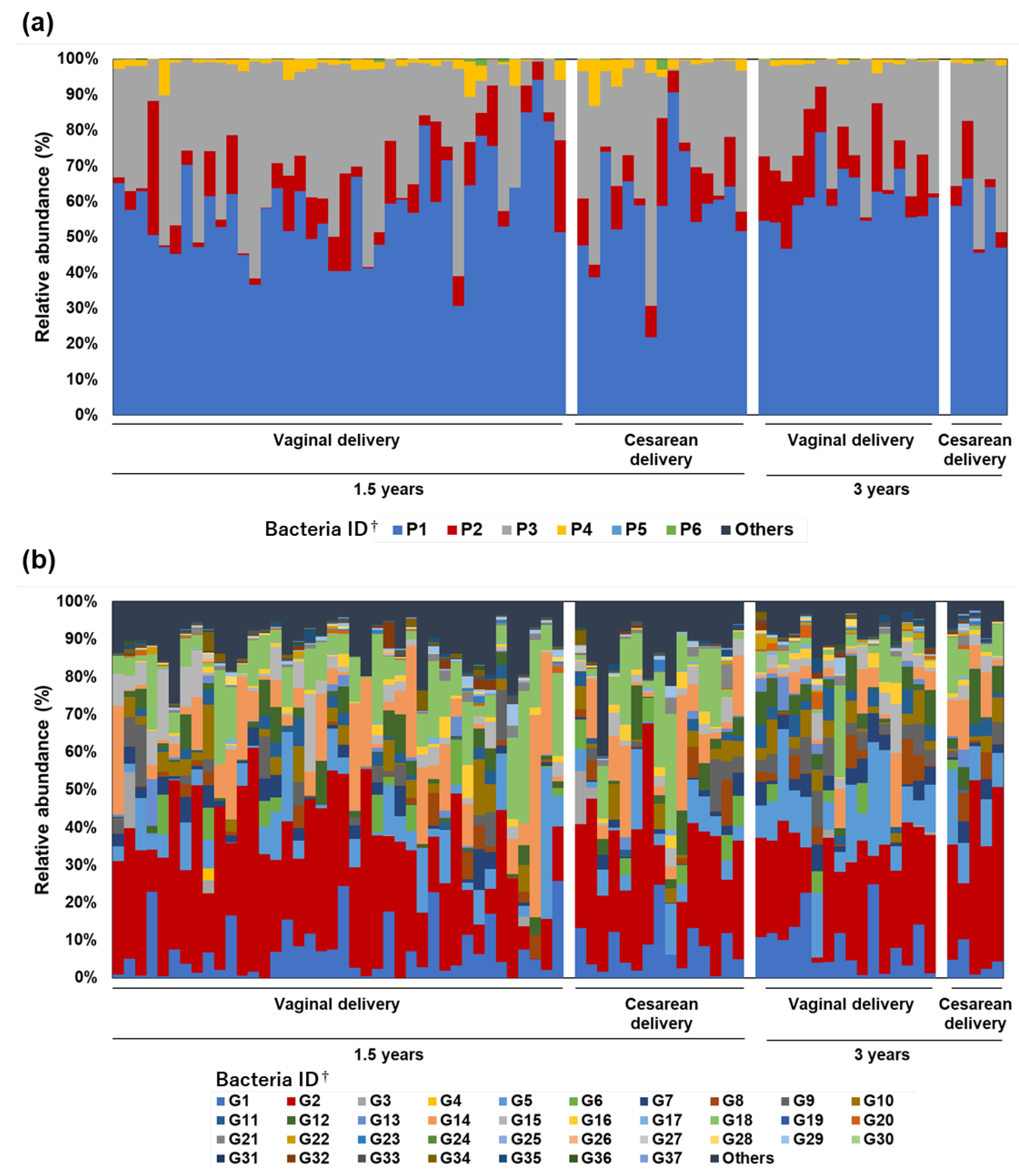
3.2. Research II: Bacterial Composition of Toddlers’ Feces Excreted in Diapers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Smidt, H.; de Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do? Front. Cell. Infect. Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.; Lee, S.M.; Shen, Y.; Khosravi, A.; Mazmanian, S.K. Host-bacterial symbiosis in health and disease. Adv. Immunol. 2010, 107, 243–274. [Google Scholar] [CrossRef]
- Biedermann, L.; Rogler, G. The intestinal microbiota: Its role in health and disease. Eur. J. Pediatrics 2015, 174, 151–167. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. MMBR 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. Off. J. Jpn. Soc. Allergol. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, Q.; Balakrishnan, B.; Belobrajdic, D.P.; Feng, Q.J.; Zhang, W. Role of dietary nutrients in the modulation of gut microbiota: A narrative review. Nutrients 2020, 12, 381. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- O’Callaghan, A.A.; Corr, S.C. Establishing Boundaries: The relationship that exists between intestinal epithelial cells and gut-dwelling bacteria. Microorganisms 2019, 7, 663. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Sinha, R.; Abu-Ali, G.; Vogtmann, E.; Fodor, A.A.; Ren, B.; Amir, A.; Schwager, E.; Crabtree, J.; Ma, S.; Abnet, C.C.; et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol. 2017, 35, 1077–1086. [Google Scholar] [CrossRef]
- Gorzelak, M.A.; Gill, S.K.; Tasnim, N.; Ahmadi-Vand, Z.; Jay, M.; Gibson, D.L. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS ONE 2015, 10, e0134802. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Bik, E.M.; Costello, E.K.; Dethlefsen, L.; Haque, R.; Relman, D.A.; Singh, U. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS ONE 2013, 8, e53838. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Nakayama, J.; Yamamoto, A.; Palermo-Conde, L.A.; Higashi, K.; Sonomoto, K.; Tan, J.; Lee, Y.K. Impact of westernized diet on gut microbiota in children on Leyte Island. Front. Microbiol. 2017, 8, 197. [Google Scholar] [CrossRef]
- Nakayama, J.; Watanabe, K.; Jiang, J.; Matsuda, K.; Chao, S.H.; Haryono, P.; La-Ongkham, O.; Sarwoko, M.A.; Sujaya, I.N.; Zhao, L.; et al. Diversity in gut bacterial community of school-age children in Asia. Sci. Rep. 2015, 5, 8397. [Google Scholar] [CrossRef]
- Wong, W.S.W.; Clemency, N.; Klein, E.; Provenzano, M.; Iyer, R.; Niederhuber, J.E.; Hourigan, S.K. Collection of non-meconium stool on fecal occult blood cards is an effective method for fecal microbiota studies in infants. Microbiome 2017, 5, 114. [Google Scholar] [CrossRef]
- Oya, N.; Ito, Y.; Hioki, K.; Asai, Y.; Aoi, A.; Sugiura, Y.; Ueyama, J.; Oguri, T.; Kato, S.; Ebara, T.; et al. Quantitative analysis of organophosphate insecticide metabolites in urine extracted from disposable diapers of toddlers in Japan. Int. J. Hyg. Environ. Health 2017, 220, 209–216. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Mizutani, S.; Nakajima, T.; Hosoda, F.; Watanabe, H.; Saito, Y.; Shibata, T.; Yachida, S.; Yamada, T. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut 2016, 65, 1574–1575. [Google Scholar] [CrossRef]
- Ebara, T.; Yamada, Y.; Shoji, N.; Ito, Y.; Nakagawa, A.; Miyachi, T.; Ozaki, Y.; Omori, T.; Suzuki, S.; Kojima, M.; et al. Cohort profile: Aichi regional sub-cohort of the Japan Environment and Children’s Study (JECS-A). BMJ Open 2019, 9, e028105. [Google Scholar] [CrossRef]
- Kawamoto, T.; Nitta, H.; Murata, K.; Toda, E.; Tsukamoto, N.; Hasegawa, M.; Yamagata, Z.; Kayama, F.; Kishi, R.; Ohya, Y.; et al. Rationale and study design of the Japan environment and children’s study (JECS). BMC Public Health 2014, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef]
- Hisada, T.; Endoh, K.; Kuriki, K. Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch. Microbiol. 2015, 197, 919–934. [Google Scholar] [CrossRef]
- Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Cancello, R.; Turroni, S.; Rampelli, S.; Cattaldo, S.; Candela, M.; Cattani, L.; Mai, S.; Vietti, R.; Scacchi, M.; Brigidi, P.; et al. Effect of short-term dietary intervention and probiotic mix supplementation on the gut microbiota of elderly obese women. Nutrients 2019, 11, 3011. [Google Scholar] [CrossRef]
- Kowalska-Duplaga, K.; Gosiewski, T.; Kapusta, P.; Sroka-Oleksiak, A.; Wędrychowicz, A.; Pieczarkowski, S.; Ludwig-Słomczyńska, A.H.; Wołkow, P.P.; Fyderek, K. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci. Rep. 2019, 9, 18880. [Google Scholar] [CrossRef]
- Alhinai, E.A.; Walton, G.E.; Commane, D.M. The role of the gut microbiota in colorectal cancer causation. Int. J. Mol. Sci. 2019, 20, 5295. [Google Scholar] [CrossRef]
- Indiani, C.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.F.C.; Darrieux, M.; Parisotto, T.M. Childhood obesity and firmicutes/bacteroidetes ratio in the gut microbiota: A systematic review. Child. Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Laursen, M.F. Gut microbiota development: Influence of diet from infancy to toddlerhood. Ann. Nutr. Metab. 2021, 77, 21–34. [Google Scholar] [CrossRef]
- Albenberg, L.G.; Wu, G.D. Diet and the intestinal microbiome: Associations, functions, and implications for health and disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Sugino, K.Y.; Ma, T.; Paneth, N.; Comstock, S.S. Effect of environmental exposures on the gut microbiota from early infancy to two years of age. Microorganisms 2021, 9, 2140. [Google Scholar] [CrossRef]
- Christian, L.M.; Galley, J.D.; Hade, E.M.; Schoppe-Sullivan, S.; Kamp Dush, C.; Bailey, M.T. Gut microbiome composition is associated with temperament during early childhood. Brain Behav. Immun. 2015, 45, 118–127. [Google Scholar] [CrossRef]
- Galley, J.D.; Bailey, M.; Kamp Dush, C.; Schoppe-Sullivan, S.; Christian, L.M. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS ONE 2014, 9, e113026. [Google Scholar] [CrossRef]
- Ayeni, K.I.; Berry, D.; Wisgrill, L.; Warth, B.; Ezekiel, C.N. Early-life chemical exposome and gut microbiome development: African research perspectives within a global environmental health context. Trends Microbiol. 2022. [Google Scholar] [CrossRef]
- Lim, E.S.; Wang, D.; Holtz, L.R. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 2016, 24, 801–810. [Google Scholar] [CrossRef]



| Total | Age during Stool Collection | |||||
|---|---|---|---|---|---|---|
| 1.5 Years | 3 Years | |||||
| n | (%) | n | (%) | |||
| Sex | ||||||
| Male | 41 | 26 | (47) | 15 | (71) | |
| Female | 35 | 29 | (53) | 6 | (29) | |
| Mode of delivery | ||||||
| Spontaneous delivery | 33 | 23 | (42) | 10 | (48) | |
| Induced delivery | 18 | 13 | (24) | 5 | (24) | |
| Vacuum extraction | 5 | 4 | (7) | 1 | (5) | |
| Planned Cesarean delivery | 19 | 14 | (25) | 5 | (24) | |
| /Emergent Cesarean delivery | ||||||
| Missing | 1 | 1 | (2) | 0 | (0) | |
| Feeding method during the first month after birth | ||||||
| Breastfeeding only | 36 | 27 | (49) | 9 | (43) | |
| Mixed feeding | 37 | 25 | (45) | 12 | (57) | |
| Infant formula only | 2 | 2 | (4) | 0 | (0) | |
| Missing | 1 | 1 | (2) | 0 | (0) | |
| Starting date of feeding solid foods | ||||||
| 4 months old | 1 | 1 | (2) | 0 | (0) | |
| 5 months old | 38 | 26 | (47) | 12 | (57) | |
| 6 months old | 29 | 22 | (40) | 7 | (33) | |
| 7 months old | 3 | 2 | (4) | 1 | (5) | |
| 8 months old | 1 | 1 | (2) | 0 | (0) | |
| Missing | 4 | 3 | (5) | 1 | (5) | |
| (a) | |||||
|---|---|---|---|---|---|
| Maximum Relative Abundance | Bacteria ID † | Number of Specimens with the Bacteria Detection (/5) | Number of Specimens with the Bacteria Abundance Out of the LOA | ||
| Method A | Method B | SD € | CI ∫ | ||
| ≥10% | P1 | 5 | 5 | 1 | 1 |
| P2 | 5 | 5 | 1 | 2 | |
| P3 | 5 | 5 | 0 | 0 | |
| ≥1%, <10% | P4 | 5 | 5 | 0 | 1 |
| P5 | 4 | 4 | 1 | 1 | |
| P6 | 2 | 2 | 0 | 0 | |
| <1% | P7 | 2 | 1 | 2 | 3 |
| P9 | 0 | 1 | 0 | 0 | |
| P10 | 1 | 0 | 0 | 0 | |
| (b) | |||||
| Maximum Relative Abundance | Bacteria ID † | Number of Specimens with the Bacteria Detection (/5) | Number of Specimens with the Bacteria Abundance Out of the LOA | ||
| Method A | Method C | SD € | CI ∫ | ||
| ≥10% | P1 | 5 | 5 | 0 | 1 |
| P2 | 5 | 5 | 0 | 3 | |
| P3 | 5 | 5 | 0 | 2 | |
| ≥1%, <10% | P4 | 5 | 5 | 1 | 1 |
| P6 | 2 | 2 | 0 | 1 | |
| <1% | P5 | 4 | 1 | 1 | 1 |
| P7 | 2 | 2 | 0 | 0 | |
| P8 | 0 | 1 | 0 | 0 | |
| P10 | 1 | 0 | 0 | 0 | |
| (a) | |||||
|---|---|---|---|---|---|
| Maximum Relative Abundance | Bacteria ID † | Number of Specimens with the Bacteria Detection (/5) | Number of Specimens with the Bacteria Abundance Out of the LOA | ||
| Method A | Method B | SD € | CI ∫ | ||
| ≥10% | G1 | 5 | 5 | 2 | 2 |
| G2 | 5 | 5 | 0 | 0 | |
| G3 | 2 | 2 | 0 | 0 | |
| G5 | 5 | 5 | 0 | 0 | |
| G7 | 5 | 5 | 0 | 0 | |
| G8 | 5 | 5 | 0 | 1 | |
| ≥1%, <10% | G4 | 1 | 1 | 0 | 1 |
| G6 | 4 | 4 | 0 | 1 | |
| G9 | 4 | 4 | 1 | 1 | |
| G10 | 3 | 4 | 1 | 1 | |
| G11 | 4 | 4 | 1 | 1 | |
| G12 | 5 | 5 | 2 | 2 | |
| G13 | 5 | 5 | 1 | 1 | |
| G14 | 5 | 5 | 0 | 0 | |
| G15 | 5 | 5 | 0 | 1 | |
| G16 | 5 | 5 | 0 | 0 | |
| G17 | 5 | 5 | 0 | 0 | |
| G18 | 5 | 5 | 0 | 0 | |
| G19 | 2 | 2 | 1 | 1 | |
| G20 | 3 | 4 | 0 | 0 | |
| G21 | 5 | 5 | 0 | 0 | |
| G22 | 4 | 5 | 0 | 0 | |
| G23 | 2 | 2 | 0 | 0 | |
| G24 | 2 | 2 | 0 | 0 | |
| G25 | 5 | 5 | 0 | 0 | |
| G26 | 2 | 3 | 0 | 0 | |
| G27 | 4 | 4 | 0 | 0 | |
| G28 | 5 | 4 | 0 | 0 | |
| G29 | 4 | 4 | 0 | 0 | |
| G30 | 4 | 3 | 0 | 0 | |
| G31 | 4 | 4 | 0 | 0 | |
| G32 | 5 | 5 | 0 | 0 | |
| G34 | 1 | 2 | 0 | 0 | |
| G36 | 3 | 4 | 0 | 0 | |
| G37 | 3 | 4 | 0 | 0 | |
| ≥0.1%, <1% | G33 | 5 | 5 | 0 | 1 |
| G35 | 3 | 3 | 0 | 1 | |
| G38 | 4 | 4 | 2 | 2 | |
| G39 | 2 | 3 | 1 | 1 | |
| G40 | 5 | 5 | 1 | 2 | |
| G41 | 0 | 1 | 1 | 1 | |
| G42 | 5 | 5 | 0 | 0 | |
| G43 | 3 | 2 | 1 | 1 | |
| G44 | 5 | 5 | 1 | 1 | |
| G45 | 2 | 2 | 0 | 0 | |
| G46 | 1 | 1 | 1 | 1 | |
| G47 | 4 | 4 | 0 | 0 | |
| G48 | 1 | 1 | 0 | 0 | |
| G49 | 2 | 2 | 0 | 0 | |
| G50 | 4 | 4 | 0 | 0 | |
| G51 | 4 | 4 | 0 | 0 | |
| G52 | 1 | 2 | 0 | 0 | |
| G54 | 5 | 5 | 0 | 1 | |
| G55 | 5 | 3 | 0 | 0 | |
| G57 | 1 | 1 | 0 | 0 | |
| G58 | 4 | 4 | 0 | 0 | |
| G59 | 2 | 0 | 0 | 0 | |
| <0.1% | G53 | 1 | 0 | 1 | 1 |
| G56 | 2 | 2 | 2 | 2 | |
| G60 | 4 | 4 | 0 | 1 | |
| G61 | 1 | 1 | 0 | 0 | |
| G62 | 0 | 1 | 0 | 0 | |
| G63 | 4 | 4 | 0 | 0 | |
| G64 | 1 | 1 | 0 | 0 | |
| G65 | 1 | 1 | 0 | 1 | |
| G66 | 3 | 1 | 0 | 0 | |
| G67 | 2 | 3 | 0 | 0 | |
| G68 | 2 | 1 | 1 | 1 | |
| G69 | 0 | 1 | 0 | 0 | |
| G70 | 2 | 2 | 1 | 1 | |
| G71 | 0 | 2 | 1 | 1 | |
| G72 | 1 | 1 | 1 | 1 | |
| G73 | 0 | 1 | 1 | 1 | |
| G74 | 1 | 2 | 0 | 0 | |
| G75 | 2 | 2 | 0 | 0 | |
| G76 | 3 | 3 | 0 | 1 | |
| G77 | 0 | 1 | 0 | 0 | |
| G78 | 1 | 0 | 0 | 0 | |
| G80 | 0 | 1 | 0 | 0 | |
| G81 | 2 | 0 | 0 | 0 | |
| G83 | 0 | 1 | 0 | 0 | |
| G84 | 1 | 1 | 0 | 0 | |
| G85 | 0 | 1 | 0 | 0 | |
| G86 | 2 | 0 | 0 | 0 | |
| G87 | 0 | 2 | 0 | 0 | |
| G88 | 0 | 1 | 0 | 0 | |
| G89 | 2 | 0 | 0 | 0 | |
| G91 | 1 | 0 | 0 | 0 | |
| G92 | 1 | 0 | 0 | 0 | |
| G93 | 0 | 1 | 0 | 0 | |
| (b) | |||||
| Maximum Relative Abundance | Bacteria ID † | Number of Specimens with the Bacteria Detection (/5) | Number of Specimens with the Bacteria Abundance Out of the LOA | ||
| Method A | Method C | SD € | CI ∫ | ||
| ≥10% | G1 | 5 | 5 | 2 | 4 |
| G2 | 5 | 5 | 0 | 1 | |
| G3 | 2 | 2 | 0 | 0 | |
| G4 | 1 | 2 | 0 | 0 | |
| G5 | 5 | 5 | 0 | 0 | |
| G6 | 4 | 4 | 0 | 0 | |
| G7 | 5 | 5 | 0 | 0 | |
| G8 | 5 | 5 | 0 | 0 | |
| ≥1%, <10% | G9 | 4 | 4 | 1 | 1 |
| G10 | 3 | 3 | 0 | 0 | |
| G11 | 4 | 4 | 1 | 1 | |
| G12 | 5 | 5 | 0 | 0 | |
| G13 | 5 | 5 | 2 | 2 | |
| G14 | 5 | 5 | 2 | 2 | |
| G15 | 5 | 5 | 1 | 1 | |
| G16 | 5 | 5 | 1 | 1 | |
| G17 | 5 | 5 | 2 | 2 | |
| G18 | 5 | 5 | 0 | 0 | |
| G20 | 3 | 3 | 0 | 0 | |
| G21 | 5 | 5 | 0 | 0 | |
| G22 | 4 | 5 | 0 | 0 | |
| G23 | 2 | 2 | 0 | 0 | |
| G24 | 2 | 2 | 1 | 1 | |
| G25 | 5 | 5 | 0 | 0 | |
| G26 | 2 | 2 | 0 | 0 | |
| G27 | 4 | 4 | 0 | 0 | |
| G28 | 5 | 4 | 0 | 0 | |
| G29 | 4 | 3 | 0 | 0 | |
| G30 | 4 | 2 | 0 | 0 | |
| G31 | 4 | 3 | 0 | 0 | |
| G32 | 5 | 5 | 0 | 0 | |
| G33 | 5 | 5 | 0 | 0 | |
| G34 | 1 | 1 | 0 | 0 | |
| G35 | 3 | 4 | 0 | 0 | |
| G36 | 3 | 4 | 0 | 0 | |
| ≥0.1%, <1% | G19 | 2 | 0 | 1 | 1 |
| G37 | 3 | 3 | 0 | 0 | |
| G38 | 4 | 4 | 2 | 2 | |
| G39 | 2 | 2 | 0 | 0 | |
| G40 | 5 | 5 | 0 | 1 | |
| G41 | 0 | 2 | 1 | 1 | |
| G42 | 5 | 5 | 1 | 1 | |
| G43 | 3 | 2 | 0 | 0 | |
| G44 | 5 | 4 | 2 | 2 | |
| G45 | 2 | 2 | 1 | 1 | |
| G46 | 1 | 2 | 1 | 1 | |
| G47 | 4 | 4 | 1 | 1 | |
| G48 | 1 | 1 | 0 | 0 | |
| G49 | 2 | 1 | 0 | 0 | |
| G50 | 4 | 3 | 0 | 0 | |
| G51 | 4 | 3 | 0 | 0 | |
| G52 | 1 | 2 | 0 | 0 | |
| G53 | 1 | 1 | 0 | 0 | |
| G54 | 5 | 5 | 0 | 0 | |
| G55 | 5 | 4 | 0 | 0 | |
| G56 | 2 | 3 | 0 | 0 | |
| G57 | 1 | 2 | 0 | 0 | |
| G58 | 4 | 3 | 0 | 0 | |
| G59 | 2 | 3 | 0 | 0 | |
| <0.1% | G60 | 4 | 4 | 1 | 1 |
| G61 | 1 | 2 | 1 | 1 | |
| G62 | 0 | 1 | 1 | 1 | |
| G63 | 4 | 4 | 3 | 3 | |
| G64 | 1 | 1 | 1 | 1 | |
| G65 | 1 | 0 | 1 | 1 | |
| G66 | 3 | 4 | 1 | 1 | |
| G67 | 2 | 3 | 0 | 1 | |
| G68 | 2 | 0 | 0 | 0 | |
| G69 | 0 | 1 | 1 | 1 | |
| G70 | 2 | 2 | 0 | 0 | |
| G71 | 0 | 1 | 1 | 1 | |
| G72 | 1 | 1 | 1 | 1 | |
| G74 | 1 | 3 | 0 | 0 | |
| G75 | 2 | 1 | 0 | 0 | |
| G76 | 3 | 3 | 0 | 0 | |
| G77 | 0 | 1 | 0 | 0 | |
| G78 | 1 | 2 | 0 | 0 | |
| G79 | 0 | 1 | 0 | 0 | |
| G80 | 0 | 1 | 0 | 0 | |
| G81 | 2 | 1 | 0 | 0 | |
| G82 | 0 | 1 | 0 | 0 | |
| G84 | 1 | 1 | 0 | 0 | |
| G85 | 0 | 1 | 0 | 0 | |
| G86 | 2 | 0 | 0 | 0 | |
| G89 | 2 | 0 | 0 | 0 | |
| G90 | 0 | 1 | 0 | 0 | |
| G91 | 1 | 0 | 0 | 0 | |
| G92 | 1 | 0 | 0 | 0 | |
| Adults (Research I, Method C) | 1.5 Years Group (Research II) | 3 Years Group (Research II) | |
|---|---|---|---|
| Chao1 | 131.2 ± 36.5 | 64.9 ± 12.8 ** | 83.9 ± 16.4 ** |
| Shannon | 5.0 ± 0.5 | 4.4 ± 0.5 * | 4.7 ± 0.5 |
| Simpson | 0.94 ± 0.02 | 0.91 ± 0.04 | 0.92 ± 0.03 |
| Observed operational taxonomic units | 129.4 ± 36.4 | 64.8 ± 12.8 ** | 83.8 ± 16.4 ** |
| Faith’s phylogenetic diversity | 10.1 ± 2.6 | 5.9 ± 1.0 ** | 7.0 ± 1.2 ** |
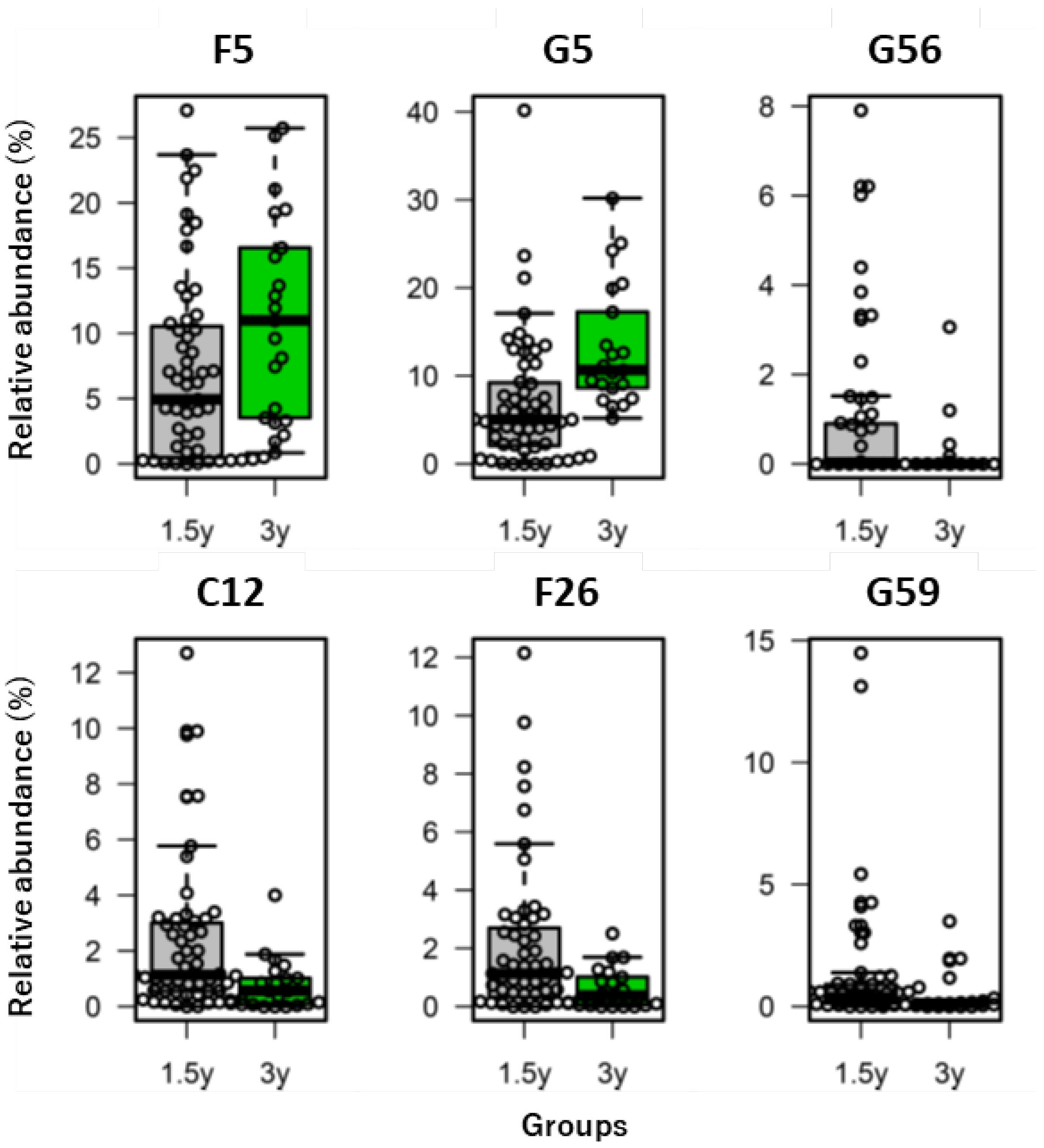
| Functionality | References | Category | ID † | p-Value ¶ | Coefficient of Variation § | |
|---|---|---|---|---|---|---|
| 1.5 Years Group | 3 Years Group | |||||
| Beneficial | [31] | Family | F5 | 0.018 | 1.011 | 0.697 |
| [7,31] | Genus | G5 | 0.000 | 1.051 | 0.529 | |
| [7,32] | G10 | 0.194 | 1.235 | 0.862 | ||
| [31,32] | G12 | 0.077 | 1.159 | 0.847 | ||
| [7] | G23 | 0.331 | 3.120 | 1.844 | ||
| [31] | G25 | 0.071 | 1.267 | 1.234 | ||
| [7] | G56 | 0.019 | 1.980 | 3.000 | ||
| [32] | G57 | 0.551 | 3.839 | 4.583 | ||
| [7,8] | Species | S29 | 0.331 | 3.120 | 1.844 | |
| [32] | S40 | 0.079 | 2.266 | 1.384 | ||
| [7] | S43 | 0.917 | 0.774 | 0.701 | ||
| [7] | S127 | 0.286 | 5.204 | - | ||
| Both | [7,8,33] | Species | S73 | 0.896 | 1.993 | 2.452 |
| Detrimental | [8] | Class | C12 | 0.003 | 1.209 | 1.242 |
| [31] | Family | F7 | 0.515 | 1.352 | 1.333 | |
| [8] | F26 | 0.003 | 1.274 | 1.157 | ||
| [31] | Genus | G6 | 0.260 | 1.814 | 2.023 | |
| [7,32] | G59 | 0.015 | 2.178 | 1.869 | ||
| [7] | Species | S147 | 0.476 | 7.416 | 4.583 | |
| [7] | S167 | 0.139 | 7.416 | 4.583 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamada, H.; Ito, Y.; Ebara, T.; Kato, S.; Kaneko, K.; Matsuki, T.; Sugiura-Ogasawara, M.; Saitoh, S.; Kamijima, M. Epidemiological Studies of Children’s Gut Microbiota: Validation of Sample Collection and Storage Methods and Microbiota Analysis of Toddlers’ Feces Collected from Diapers. Nutrients 2022, 14, 3315. https://doi.org/10.3390/nu14163315
Tamada H, Ito Y, Ebara T, Kato S, Kaneko K, Matsuki T, Sugiura-Ogasawara M, Saitoh S, Kamijima M. Epidemiological Studies of Children’s Gut Microbiota: Validation of Sample Collection and Storage Methods and Microbiota Analysis of Toddlers’ Feces Collected from Diapers. Nutrients. 2022; 14(16):3315. https://doi.org/10.3390/nu14163315
Chicago/Turabian StyleTamada, Hazuki, Yuki Ito, Takeshi Ebara, Sayaka Kato, Kayo Kaneko, Taro Matsuki, Mayumi Sugiura-Ogasawara, Shinji Saitoh, and Michihiro Kamijima. 2022. "Epidemiological Studies of Children’s Gut Microbiota: Validation of Sample Collection and Storage Methods and Microbiota Analysis of Toddlers’ Feces Collected from Diapers" Nutrients 14, no. 16: 3315. https://doi.org/10.3390/nu14163315
APA StyleTamada, H., Ito, Y., Ebara, T., Kato, S., Kaneko, K., Matsuki, T., Sugiura-Ogasawara, M., Saitoh, S., & Kamijima, M. (2022). Epidemiological Studies of Children’s Gut Microbiota: Validation of Sample Collection and Storage Methods and Microbiota Analysis of Toddlers’ Feces Collected from Diapers. Nutrients, 14(16), 3315. https://doi.org/10.3390/nu14163315







