Regulation of Dietary Protein Solubility Improves Ruminal Nitrogen Metabolism In Vitro: Role of Bacteria–Protozoa Interactions
Abstract
1. Introduction
2. Materials and Method
2.1. Ethical Statement
2.2. Substrate Preparation and Determination of Protein Fractions
2.3. Animal Management and Experimental Design
2.4. Rumen Fluid Inoculation and In Vitro Fermentation
2.5. Sample Collection and Analysis
2.6. DNA Extraction, Library Construction, 16S/18S rRNA Sequencing and Data Processing
2.7. Statistical Analysis
3. Results
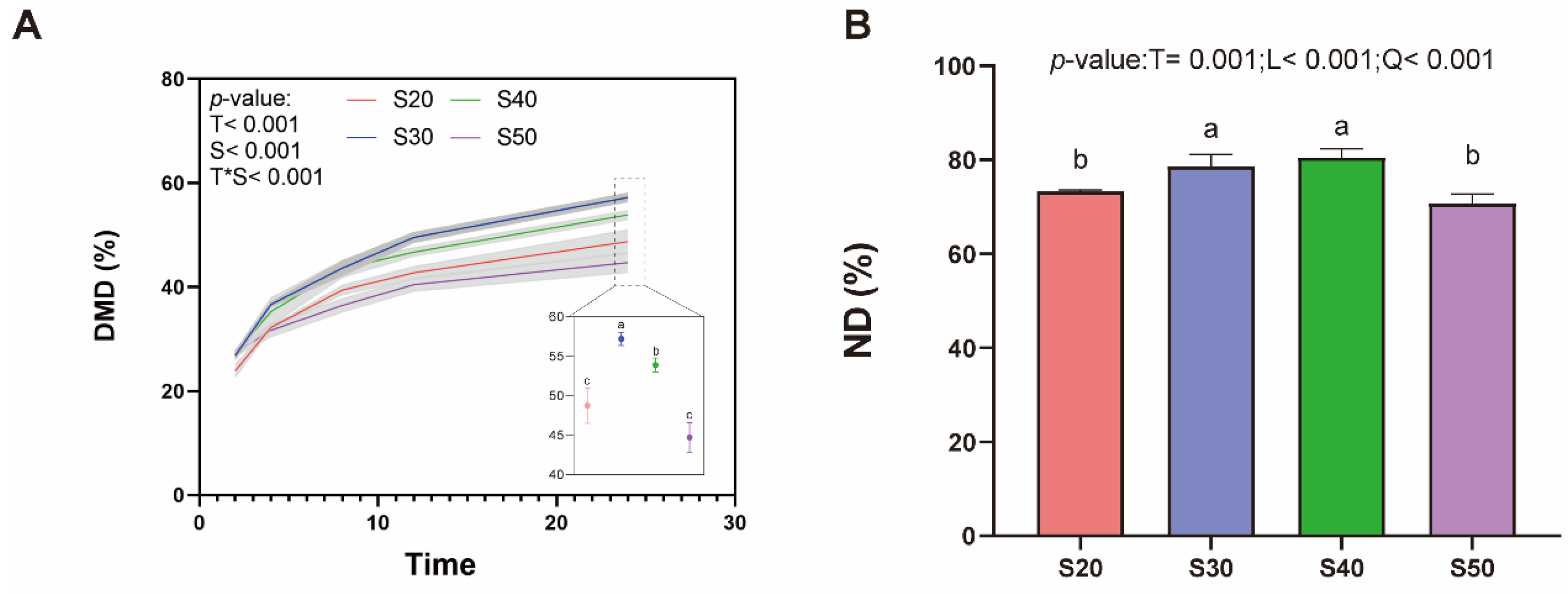
3.1. In Vitro Rumen Fermentation and Digestibility
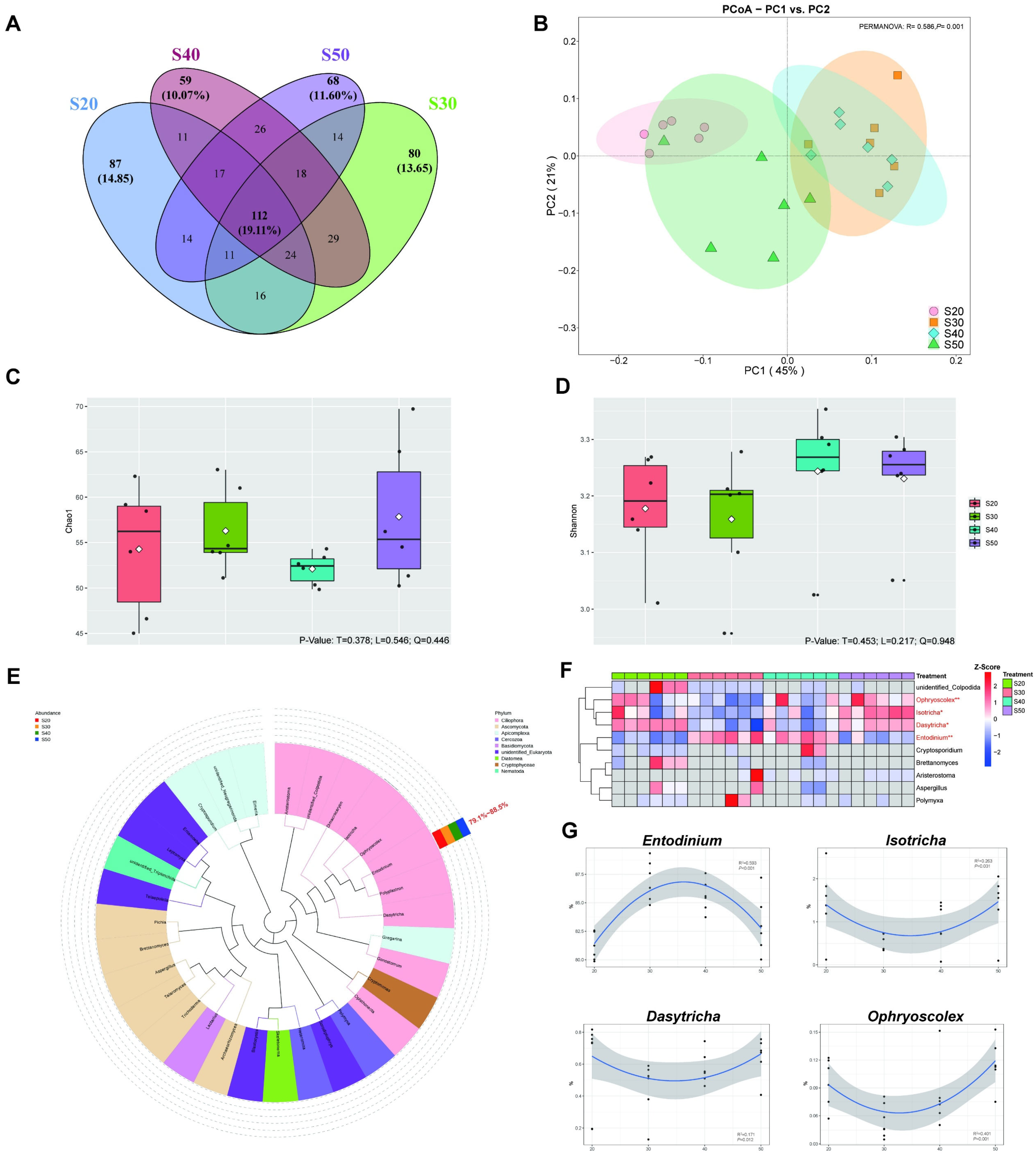
3.2. Bacterial Diversity and Taxonomic Differences In Vitro
3.3. Predicted Ruminal Microbial Functions via PICRUSt2
3.4. Protozoal Diversity and Taxonomic Differences In Vitro
3.5. The Interaction of Rumen Bacteria, Protozoa and Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.J.; Aveyard, P.; Garnett, T.; Hall, J.W.; Key, T.J.; Lorimer, J.; Pierrehumbert, R.T.; Scarborough, P.; Springmann, M.; Jebb, S.A. Meat consumption, health, and the environment. Science 2018, 361, eaam5324. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Clark, M. Global diets link environmental sustainability and human health. Nature 2014, 515, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Ge, Y.; Chang, J. Global Strategies to Minimize Environmental Impacts of Ruminant Production. Annu. Rev. Anim. Biosci. 2022, 10, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Ripple, W.; Smith, P.; Haberl, H.; Montzka, S.; McAlpine, C.; Boucher, D. COMMENTARY: Ruminants, climate change and climate policy. Nat. Clim. Change 2013, 4, 2–5. [Google Scholar] [CrossRef]
- Tan, P.; Liu, H.; Zhao, J.; Gu, X.; Wei, X.; Zhang, X.; Ma, N.; Johnston, L.J.; Bai, Y.; Zhang, W.; et al. Amino acids metabolism by rumen microorganisms: Nutrition and ecology strategies to reduce nitrogen emissions from the inside to the outside. Sci. Total Environ. 2021, 800, 149596. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Ge, Y.; Ren, Y.; Fan, X.; Pan, K.; Lin, L.; Wu, X.; Min, Y.; Meyerson, L.A.; Heino, M.; et al. A global strategy to mitigate the environmental impact of China’s ruminant consumption boom. Nat. Commun. 2018, 9, 4133. [Google Scholar] [CrossRef]
- Xian, C.; Zhang, X.; Zhang, J.; Fan, Y.; Zheng, H.; Salzman, J.; Ouyang, Z. Recent patterns of anthropogenic reactive nitrogen emissions with urbanization in China: Dynamics, major problems, and potential solutions. Sci. Total Environ. 2019, 656, 1071–1081. [Google Scholar] [CrossRef]
- Zhu, W.; Xu, W.; Wei, C.; Zhang, Z.; Jiang, C.; Chen, X. Effects of Decreasing Dietary Crude Protein Level on Growth Performance, Nutrient Digestion, Serum Metabolites, and Nitrogen Utilization in Growing Goat Kids (Capra hircus). Animals 2020, 10, 151. [Google Scholar] [CrossRef]
- López-Soto, M.A.; Rivera-Méndez, C.R.; Aguilar-Hernández, J.A.; Barreras, A.; Calderón-Cortés, J.F.; Plascencia, A.; Dávila-Ramos, H.; Estrada-Angulo, A.; Valdes-García, Y.S. Effects of Combining Feed Grade Urea and a Slow-release Urea Product on Characteristics of Digestion, Microbial Protein Synthesis and Digestible Energy in Steers Fed Diets with Different Starch:ADF Ratios. Asian-Australas. J. Anim. Sci. 2014, 27, 187–193. [Google Scholar] [CrossRef]
- Zhang, Z.; Shahzad, K.; Shen, S.; Dai, R.; Lu, Y.; Lu, Z.; Li, C.; Chen, Y.; Qi, R.; Gao, P.; et al. Altering Dietary Soluble Protein Levels with Decreasing Crude Protein May Be a Potential Strategy to Improve Nitrogen Efficiency in Hu Sheep Based on Rumen Microbiome and Metabolomics. Front. Nutr. 2021, 8, 815358. [Google Scholar] [CrossRef]
- Stefanski, T.; Ahvenjarvi, S.; Vanhatalo, A.; Huhtanen, P. Ruminal metabolism of ammonia N and rapeseed meal soluble N fraction. J. Dairy Sci. 2020, 103, 7081–7093. [Google Scholar] [CrossRef] [PubMed]
- Van Amburgh, M.E.; Collao-Saenz, E.A.; Higgs, R.J.; Ross, D.A.; Recktenwald, E.B.; Raffrenato, E.; Chase, L.E.; Overton, T.R.; Mills, J.K.; Foskolos, A. The Cornell Net Carbohydrate and Protein System: Updates to the model and evaluation of version 6.5. J. Dairy Sci. 2015, 98, 6361–6380. [Google Scholar] [CrossRef] [PubMed]
- Hartinger, T.; Gresner, N.; Südekum, K.-H. Does intra-ruminal nitrogen recycling waste valuable resources? A review of major players and their manipulation. J. Anim. Sci. Biotechnol. 2018, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, G.; Shitut, S.; Preussger, D.; Yousif, G.; Waschina, S.; Kost, C. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 2018, 35, 455–488. [Google Scholar] [CrossRef]
- Gast, R.; Sanders, R.; Caron, D. Ecological strategies of protists and their symbiotic relationships with prokaryotic microbes. Trends Microbiol. 2009, 17, 563–569. [Google Scholar] [CrossRef]
- Ushida, K.; Jouany, J.P. Effect of protozoa on rumen protein degradation in sheep. Reprod. Nutr. Dev. 1985, 25, 1075–1081. [Google Scholar] [CrossRef]
- Solomon, R.; Wein, T.; Levy, B.; Eshed, S.; Dror, R.; Reiss, V.; Zehavi, T.; Furman, O.; Mizrahi, I.; Jami, E. Protozoa populations are ecosystem engineers that shape prokaryotic community structure and function of the rumen microbial ecosystem. ISME J. 2022, 16, 1187–1197. [Google Scholar] [CrossRef]
- Licitra, G.; Hernandez, T.M.; Van Soest, P.J. Standardization of procedures for nitrogen fractionation of ruminant feeds. Anim. Feed Sci. Technol. 1996, 57, 347–358. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, S.; Wang, M.; Shahzad, K.; Zhang, X.; Qi, R.; Shi, L. Effects of Urtica cannabina to Leymus chinensis Ratios on Ruminal Microorganisms and Fiber Degradation In Vitro. Animals 2020, 10, 335. [Google Scholar] [CrossRef]
- Menke, K.H.; Steingass, H. Estimation of the energetic feed value obtained by chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Develop. 1988, 28, 7–55. [Google Scholar]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Firkins, J. Maximizing Microbial Protein Synthesis in the Rumen. J. Nutr. 1996, 126, 1347S–1354S. [Google Scholar] [CrossRef]
- Erwin, E.; Marco, G.; Emery, E. Volatile Fatty Acid Analyses of Blood and Rumen Fluid by Gas Chromatography. J. Dairy Sci. 1961, 44, 1768–1771. [Google Scholar] [CrossRef]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Caporaso, J.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platformsOpen. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, C.; Elmhadi, M.; Zhang, H.; Liu, F.; Gao, X.; Wang, H. Dietary supplementation of thiamine enhances colonic integrity and modulates mucosal inflammation injury in goats challenged by lipopolysaccharide and low pH. Br. J. Nutr. 2022, 1–11. [Google Scholar] [CrossRef]
- Tian, H.; Fotidis, I.A.; Kissas, K.; Angelidaki, I. Effect of different ammonia sources on aceticlastic and hydrogenotrophic methanogens. Bioresour. Technol. 2018, 250, 390–397. [Google Scholar] [CrossRef]
- Erdman, R.A.; Proctor, G.H.; Vandersall, J.H. Effect of rumen ammonia concentration on in situ rate and extent of digestion of feedstuffs. J. Dairy Sci. 1986, 69, 2312–2320. [Google Scholar] [CrossRef]
- Lobley, G.; Bremner, D.; Holtrop, G. Effects of diet quality on urea fates in sheep as assessed by refined, non-invasive [15N15N]urea kinetics. Br. J. Nutr. 2000, 84, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Overton, T.R.; Clark, J.H. Effects of Yucca shidigera extract and soluble protein on performance of cows and concentrations of urea nitrogen in plasma and milk. J. Dairy Sci. 1998, 81, 1022–1027. [Google Scholar] [CrossRef]
- Stern, M.D.; Hoover, W.H.; Sniffen, C.J.; Crooker, B.A.; Knowlton, P.H. Effects of Nonstructural Carbohydrate, Urea and Soluble Protein Levels on Microbial Protein Synthesis in Continuous Culture of Rumen Contents. J. Anim. Sci. 1978, 47, 944–956. [Google Scholar] [CrossRef]
- Sinclair, L.A.; Garnsworthy, P.C.; Newbold, J.R.; Buttery, P.J. Effects of synchronizing the rate of dietary energy and nitrogen release in diets with a similar carbohydrate composition on rumen fermentation and microbial protein synthesis in sheep. J. Agric. Sci. 1995, 124, 463–472. [Google Scholar] [CrossRef]
- Hume, I.D.; Moir, R.J.; Somers, M. Synthesis of microbial protein in the rumen. I. Influence of the level of nitrogen intake. Aust. J. Agric. Res. 1970, 21, 283–296. [Google Scholar] [CrossRef]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Wiser, A.H.; Press, M.O.; Langford, K.W.; Liachko, I.; Snelling, T.J.; Dewhurst, R.J.; Walker, A.W.; et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 2018, 9, 870. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Wu, J.-J.; Xie, Y.-Y.; Zhu, S.-L.; Zhong, Y.-F.; Liu, J.-X.; Sun, H.-Z. Investigation of fiber utilization in the rumen of dairy cows based on metagenome-assembled genomes and single-cell RNA sequencing. Microbiome 2022, 10, 11. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Sun, H.-Z.; Wu, X.-H.; Liu, J.-X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Li, M.; Zhong, H.; Li, M.; Zheng, N.; Wang, J. Contribution of Ruminal Bacteriome to the Individual Variation of Nitrogen Utilization Efficiency of Dairy Cows. Front. Microbiol. 2022, 13, 815225. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; McKain, N.; Broderick, G.A.; Rode, L.M.; Walker, N.D.; Newbold, C.J.; Kopecny, J. Peptidases of the rumen bacterium, Prevotella ruminicola. Anaerobe 1997, 3, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Yu, Z. Genomic Insights into the Distribution of Peptidases and Proteolytic Capacity among Prevotella and Paraprevotella Species. Microbiol. Spectr. 2022, 10, e02185-21. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, S.; Betancur, C.; Mesa, H.; Isaza, G.; Jovel, J. Lower methane emissions were associated with higher abundance of ruminal Prevotella in a cohort of Colombian buffalos. BMC Microbiol. 2020, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Arntzen, M.O.; Varnai, A.; Mackie, R.I.; Eijsink, V.G.H.; Pope, P.B. Outer membrane vesicles from Fibrobacter succinogenes S85 contain an array of carbohydrate-active enzymes with versatile polysaccharide-degrading capacity. Environ. Microbiol. 2017, 19, 2701–2714. [Google Scholar] [CrossRef]
- Fondevila, M.; Dehority, B. Interactions between Fibrobacter succinogenes, Prevotella ruminicola, and Ruminococcus flavefaciens in the Digestion of Cellulose from Forages. J. Anim. Sci. 1996, 74, 678–684. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Wang, Y.; Li, S.; Cao, Z.; Ji, S.; He, Y.; Zhang, H. Effect of Dietary Forage to Concentrate Ratios on Dynamic Profile Changes and Interactions of Ruminal Microbiota and Metabolites in Holstein Heifers. Front. Microbiol. 2017, 8, 2206. [Google Scholar] [CrossRef]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef]
- Polyorach, S.; Wanapat, M.; Cherdthong, A. Influence of Yeast Fermented Cassava Chip Protein (YEFECAP) and Roughage to Concentrate Ratio on Ruminal Fermentation and Microorganisms Using In vitro Gas Production Technique. Asian-Australas. J. Anim. Sci. 2014, 27, 36–45. [Google Scholar] [CrossRef]
- Pin, C.; Wanapat, M.; Wachirapakorn, C.; Rowlinson, P. Effect of Synchronizing Starch Sources and Protein (NPN) in the Rumen on Feed Intake, Rumen Microbial Fermentation, Nutrient Utilization and Performance of Lactating Dairy Cows. Asian-Australas. J. Anim. Sci. 2004, 17, 1400–1410. [Google Scholar] [CrossRef]
- Newbold, C.; McKain, N.; Wallace, J. The role of protozoa in ruminal peptide metabolism. In Biochemistry and Molecular Biology of “Anaerobic” Protozoa; Lloyd, D., Coombs, G., Paget, T.A., Eds.; Harwood Academic Press: London, UK, 1989; pp. 42–55. [Google Scholar]
- Akkada, A.; Howard, B. The biochemistry of rumen protozoa. 5. The nitrogen metabolism of Entodinium. Biochem. J. 1962, 82, 313–320. [Google Scholar] [CrossRef][Green Version]
- Cook, A.R. Urease activity in the rumen of sheep and the isolation of ureolytic bacteria. J. Gen. Microbiol. 1976, 92, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, B.C.; Coombs, G.H.; Williams, A.G. Proteinase activity in rumen ciliate protozoa. J. Gen. Microbiol. 1988, 134, 2605–2614. [Google Scholar] [CrossRef] [PubMed]
- Mickdam, E.; Khiaosa-Ard, R.; Metzler-Zebeli, B.U.; Klevenhusen, F.; Chizzola, R.; Zebeli, Q. Rumen microbial abundance and fermentation profile during severe subacute ruminal acidosis and its modulation by plant derived alkaloids in vitro. Anaerobe 2016, 39, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Khafipour, E.; Krause, D.; Plaizier, J. Alfalfa pellet-induced subacute ruminal acidosis in dairy cows increases bacterial endotoxin in the rumen without causing inflammation. J. Dairy Sci. 2009, 92, 1712–1724. [Google Scholar] [CrossRef]
- Wang, M.Z.; Wang, H.R.; Yu, L.H. Effects of NDF Content on Protozoal Community and Grazing Rate in Rumen. J. Anim. Vet. Adv. 2009, 8, 1746–1752. [Google Scholar]



| Item | Treatments | |||
|---|---|---|---|---|
| S20 | S30 | S40 | S50 | |
| Ingredient, % | ||||
| Rice straw | 50 | 50 | 50 | 50 |
| Corn | 35 | 34 | 33 | 33.4 |
| Soybean meal | 5 | 6.5 | 6.9 | 1 |
| Wheat bran | 4 | 6.45 | 9.25 | 14 |
| Corn protein meal | 6 | 2.65 | - | - |
| Urea | - | 0.4 | 0.85 | 1.6 |
| Total | 100 | 100 | 100 | 100 |
| Nutritive level, g/kg | ||||
| DM | 888.75 | 887.53 | 886.63 | 886.86 |
| CP | 137.91 | 137.22 | 137.89 | 138.93 |
| SP (% of CP) | 20.43 | 30.12 | 39.95 | 49.50 |
| EE | 43.00 | 43.10 | 43.26 | 42.08 |
| Ash | 65.75 | 66.27 | 66.85 | 65.86 |
| NDF | 350.23 | 358.3 | 366.62 | 380.67 |
| ADF | 199.88 | 198.86 | 199.06 | 205.46 |
| Ca | 1.02 | 1.02 | 1.03 | 0.98 |
| P | 2.44 | 2.47 | 2.53 | 2.58 |
| Sampling Time/h | Treatment | SEM | p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| S20 | S30 | S40 | S50 | S | T | S × T | ||
| 2 | 6.77 | 6.78 | 6.74 | 6.76 | 0.004 | <0.001 | 0.408 | <0.001 |
| 4 | 6.67 | 6.63 | 6.68 | 6.65 | ||||
| 8 | 6.55 | 6.54 | 6.56 | 6.52 | ||||
| 12 | 6.32 | 6.33 | 6.35 | 6.46 | ||||
| 24 | 5.90 | 5.94 | 6.00 | 6.02 | ||||
| Sampling Time/h | Treatment | SEM | p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| S20 | S30 | S40 | S50 | S | T | S × T | ||
| 2 | 18.65 | 17.52 | 17.21 | 19.24 | 0.257 | 0.002 | 0.002 | 0.037 |
| 4 | 11.79 b | 12.17 b | 12.66 b | 20.96 a | ||||
| 8 | 11.82 | 10.93 | 12.65 | 13.78 | ||||
| 12 | 11.67 c | 12.26 c | 14.48 b | 19.16 a | ||||
| 24 | 15.61 c | 15.85 c | 16.20 bc | 20.45 a | ||||
| Sampling Time/h | Treatment | SEM | p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| S20 | S30 | S40 | S50 | S | T | S × T | ||
| 2 | 1.40 | 1.37 | 1.36 | 1.26 | 0.043 | <0.001 | 0.258 | 0.884 |
| 4 | 1.39 | 1.23 | 1.46 | 1.75 | ||||
| 8 | 1.67 | 1.74 | 1.67 | 1.72 | ||||
| 12 | 1.76 | 1.58 | 1.79 | 1.65 | ||||
| 24 | 2.15 | 1.76 | 1.84 | 2.21 | ||||
| Item | Treatment | SEM | p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| S20 | S30 | S40 | S50 | T | L | Q | ||
| TVFA, mM | 78.67 b | 91.69 a | 89.30 a | 80.04 b | 2.065 | 0.031 | 0.050 | 0.017 |
| Acetate, mM | 46.50 b | 53.88 a | 52.66 a | 47.58 b | 1.205 | 0.022 | 0.015 | 0.005 |
| Propionate, mM | 20.76 b | 24.56 a | 23.46 a | 21.19 b | 0.565 | 0.030 | 0.181 | 0.036 |
| Butyrate, mM | 9.36 | 10.58 | 10.39 | 9.88 | 0.259 | 0.253 | 0.149 | 0.067 |
| Isovalerate, mM | 0.84 | 0.98 | 0.95 | 0.85 | 0.023 | 0.712 | 0.650 | 0.369 |
| Valerate, mM | 0.69 b | 0.81 a | 0.78 ab | 0.70 b | 0.019 | 0.033 | 0.223 | 0.045 |
| Isobutyrate, mM | 0.53 | 0.59 | 0.55 | 0.53 | 0.011 | 0.411 | 0.333 | 0.182 |
| A/P | 2.24 | 2.19 | 2.25 | 2.24 | 0.009 | 0.108 | 0.373 | 0.187 |
| Phylum | Genus | Species | Treatment | SEM | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| S20 | S30 | S40 | S50 | T | L | Q | ||||
| Firmicutes | Coprococcus | Rumen_bacterium | 4.23 | 2.99 | 6.88 | 2.99 | 0.852 | 0.359 | 0.984 | 0.446 |
| Bacteroidota | Rikenellaceae_RC9_gut_group | Bacteroidales_bacterium | 3.81 | 3.70 | 5.32 | 3.11 | 0.392 | 0.236 | 0.885 | 0.178 |
| Bacteroidota | Prevotella | Prevotella_ruminicola | 1.48 b | 1.83 a | 1.95 a | 1.50 b | 0.079 | 0.044 | 0.756 | 0.008 |
| Bacteroidota | Bacteroidales_RF16_group | Rumen_bacterium | 1.55 | 1.62 | 1.69 | 1.55 | 0.057 | 0.825 | 0.901 | 0.417 |
| Firmicutes | Selenomonas | Selenomonas_ruminantium | 1.11 | 1.01 | 0.79 | 1.21 | 0.078 | 0.281 | 0.919 | 0.110 |
| Proteobacteria | Ruminobacter | Ruminobacter_amylophilus | 0.45 | 0.38 | 0.39 | 0.80 | 0.101 | 0.445 | 0.267 | 0.266 |
| Fibrobacterota | Fibrobacter | Fibrobacter_succinogenes | 0.51 ab | 0.72 a | 0.52 ab | 0.31 b | 0.054 | 0.034 | 0.038 | 0.031 |
| Firmicutes | Selenomonas | Rumen_bacterium | 0.55 | 0.43 | 0.39 | 0.50 | 0.030 | 0.185 | 0.438 | 0.048 |
| Bacteroidota | Rikenellaceae_RC9_gut_group | Rumen_bacterium | 0.25 | 0.28 | 0.38 | 0.31 | 0.021 | 0.165 | 0.135 | 0.237 |
| Firmicutes | Christensenellaceae_R-7_group | Bacterium_AC2043 | 0.12 a | 0.07 ab | 0.03 b | 0.13 a | 0.014 | 0.006 | 0.961 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Wei, W.; Yang, S.; Huang, Z.; Li, C.; Yu, X.; Qi, R.; Liu, W.; Loor, J.J.; Wang, M.; et al. Regulation of Dietary Protein Solubility Improves Ruminal Nitrogen Metabolism In Vitro: Role of Bacteria–Protozoa Interactions. Nutrients 2022, 14, 2972. https://doi.org/10.3390/nu14142972
Zhang Z, Wei W, Yang S, Huang Z, Li C, Yu X, Qi R, Liu W, Loor JJ, Wang M, et al. Regulation of Dietary Protein Solubility Improves Ruminal Nitrogen Metabolism In Vitro: Role of Bacteria–Protozoa Interactions. Nutrients. 2022; 14(14):2972. https://doi.org/10.3390/nu14142972
Chicago/Turabian StyleZhang, Zhenbin, Wenjun Wei, Sihan Yang, Zeliang Huang, Chuang Li, Xiang Yu, Ruxin Qi, Wujun Liu, Juan J. Loor, Mengzhi Wang, and et al. 2022. "Regulation of Dietary Protein Solubility Improves Ruminal Nitrogen Metabolism In Vitro: Role of Bacteria–Protozoa Interactions" Nutrients 14, no. 14: 2972. https://doi.org/10.3390/nu14142972
APA StyleZhang, Z., Wei, W., Yang, S., Huang, Z., Li, C., Yu, X., Qi, R., Liu, W., Loor, J. J., Wang, M., & Zhang, X. (2022). Regulation of Dietary Protein Solubility Improves Ruminal Nitrogen Metabolism In Vitro: Role of Bacteria–Protozoa Interactions. Nutrients, 14(14), 2972. https://doi.org/10.3390/nu14142972








