Effects of Inflammatory Factor Expression Regulated by 12/15 Lipoxygenase on Obesity-Related Nephropathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Experimental Animals, Groups, and Administration
2.3. Sample Collection
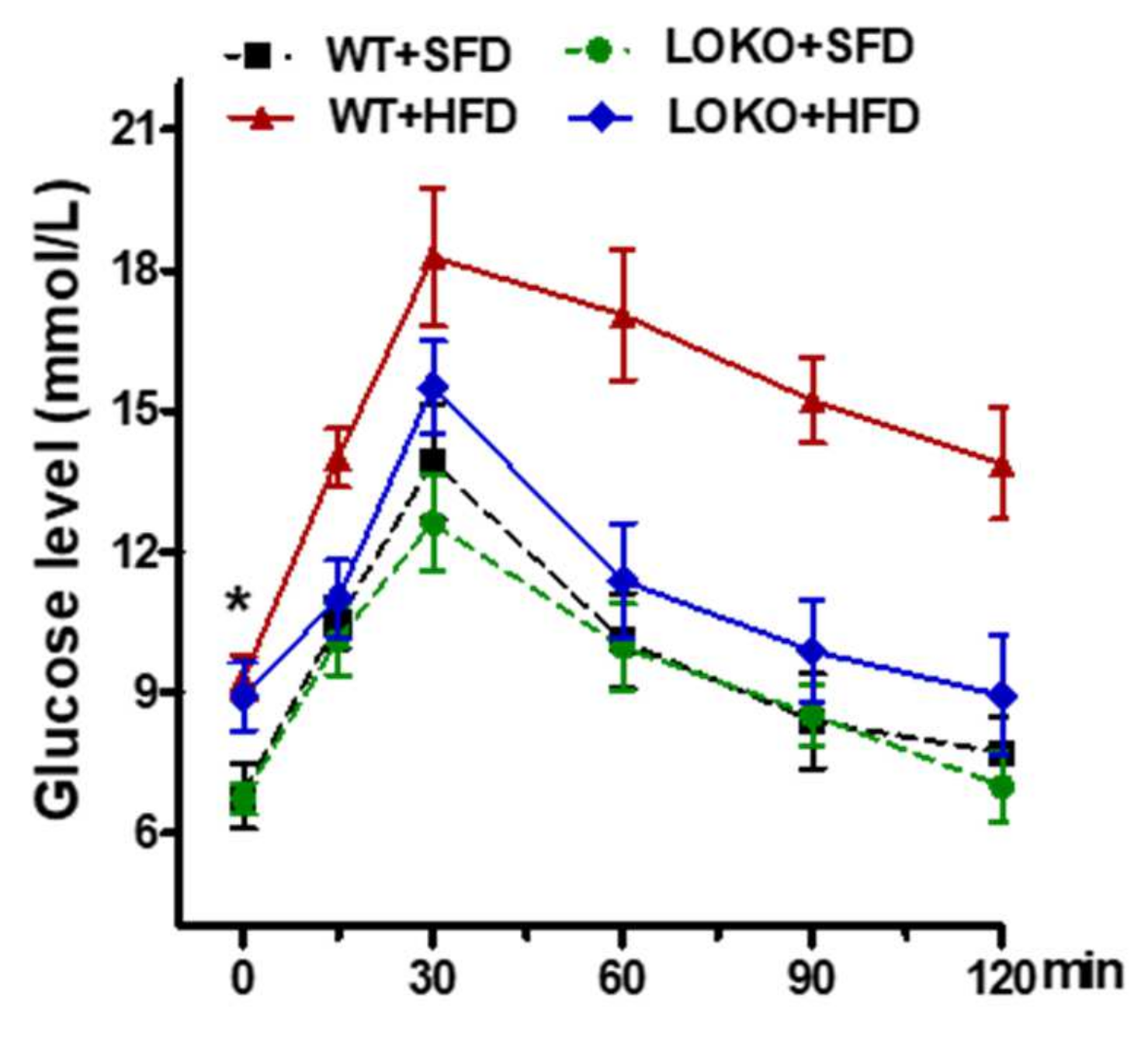
2.4. Glucose Tolerance Test (GTT)
2.5. Western Blotting Analysis
2.6. Chromatin Immunoprecipitation (ChIP Assay)
2.7. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
2.8. Histologic Staining and Morphometric Analyses
2.9. Statistical Analyses
3. Results
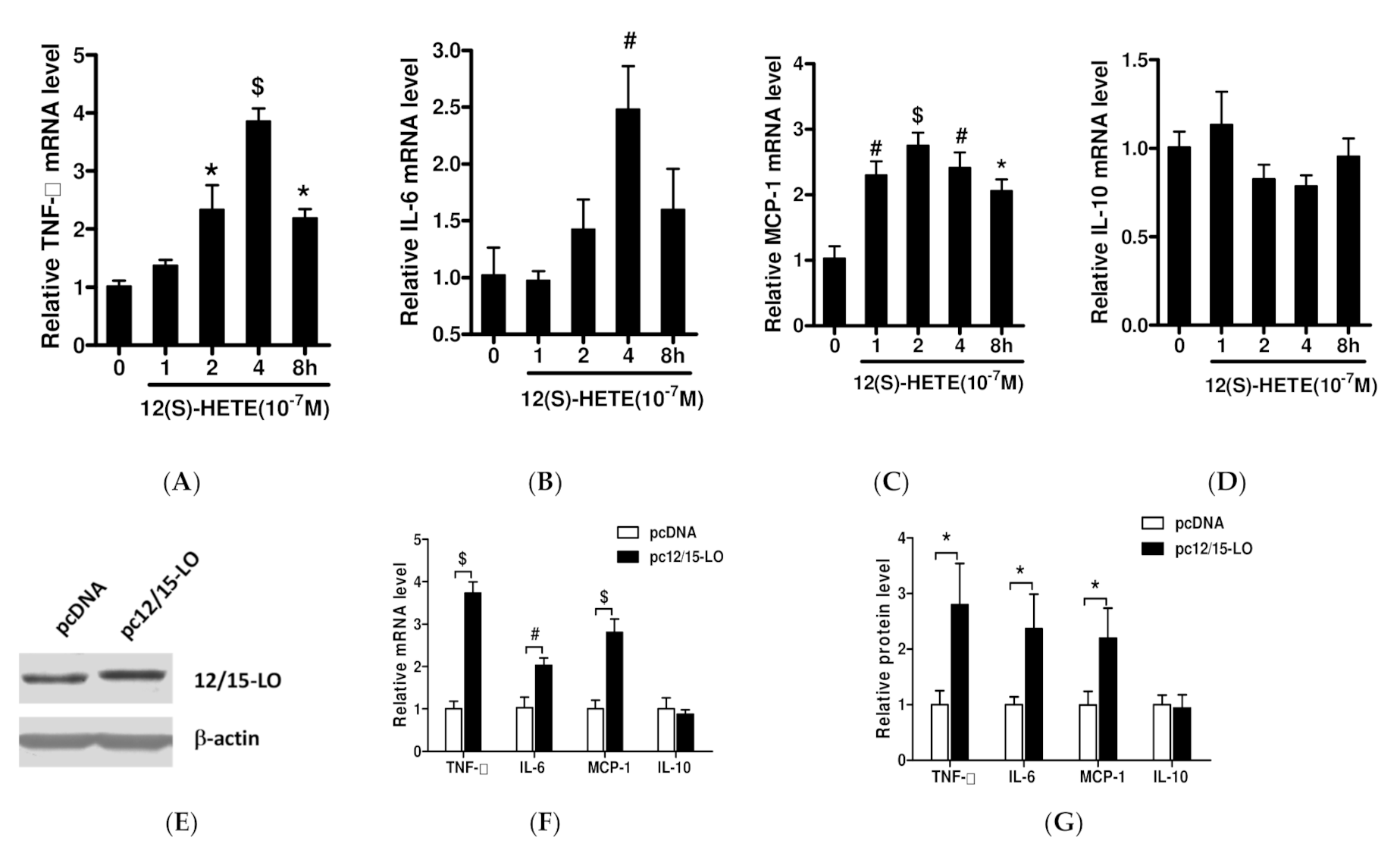
3.1. Effects of 12/15-LO and Its Metabolite 12(S)-HETE on the Expression of Adipose Inflammatory Factors
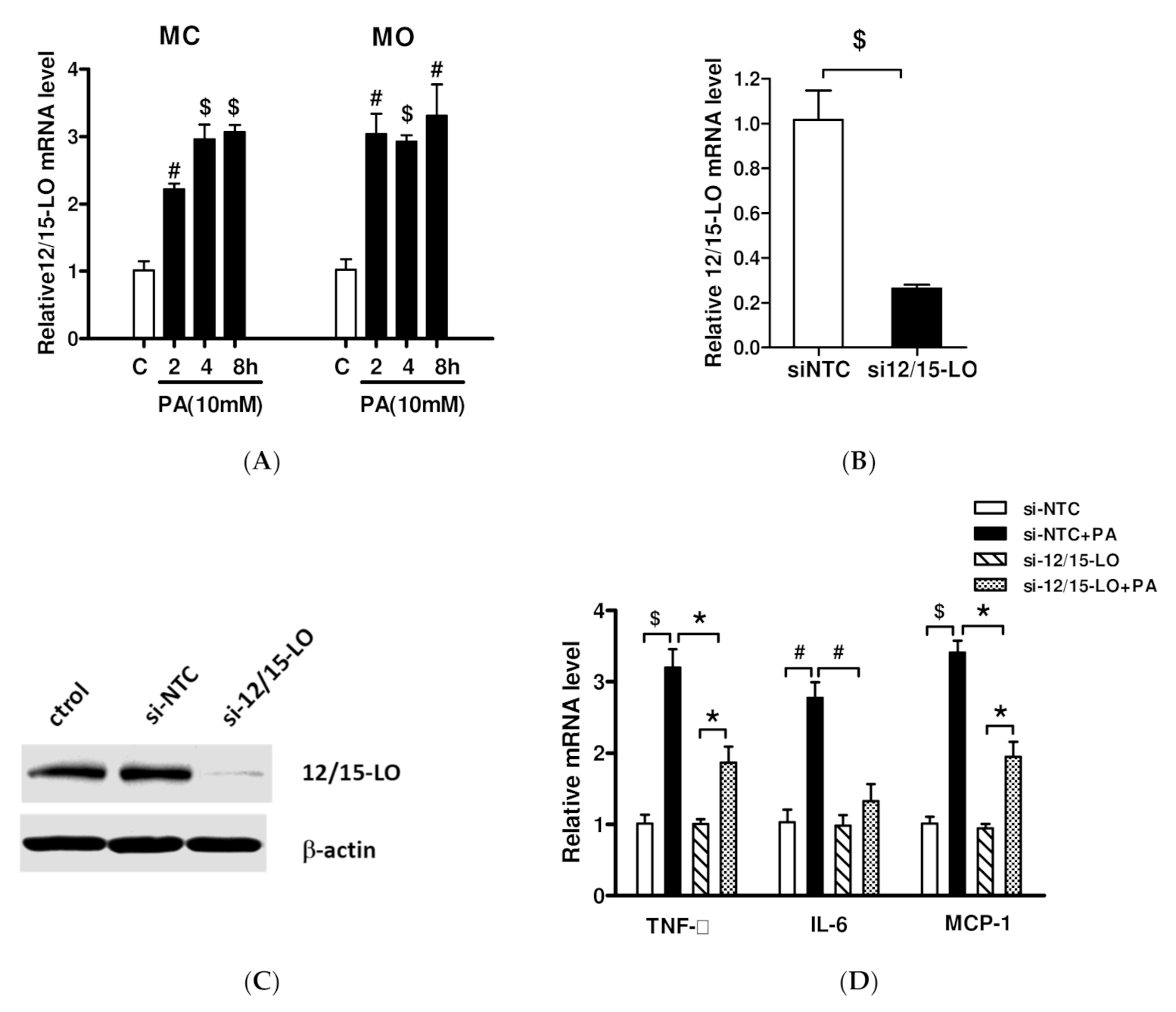
3.2. Effect of siRNA Inhibition of 12/15-LO Expression on Palmitic Acid-Regulated Inflammatory Factor Expression
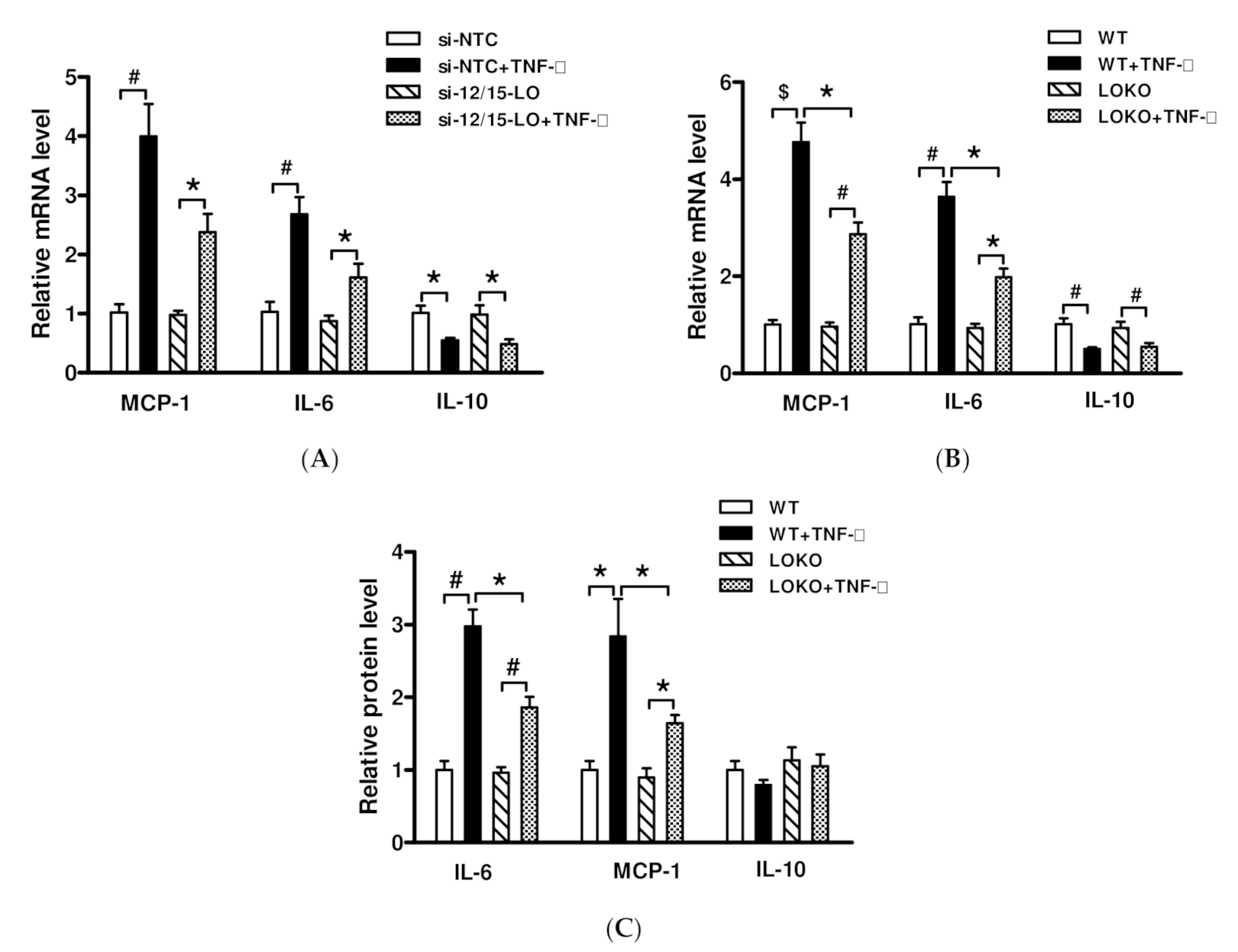
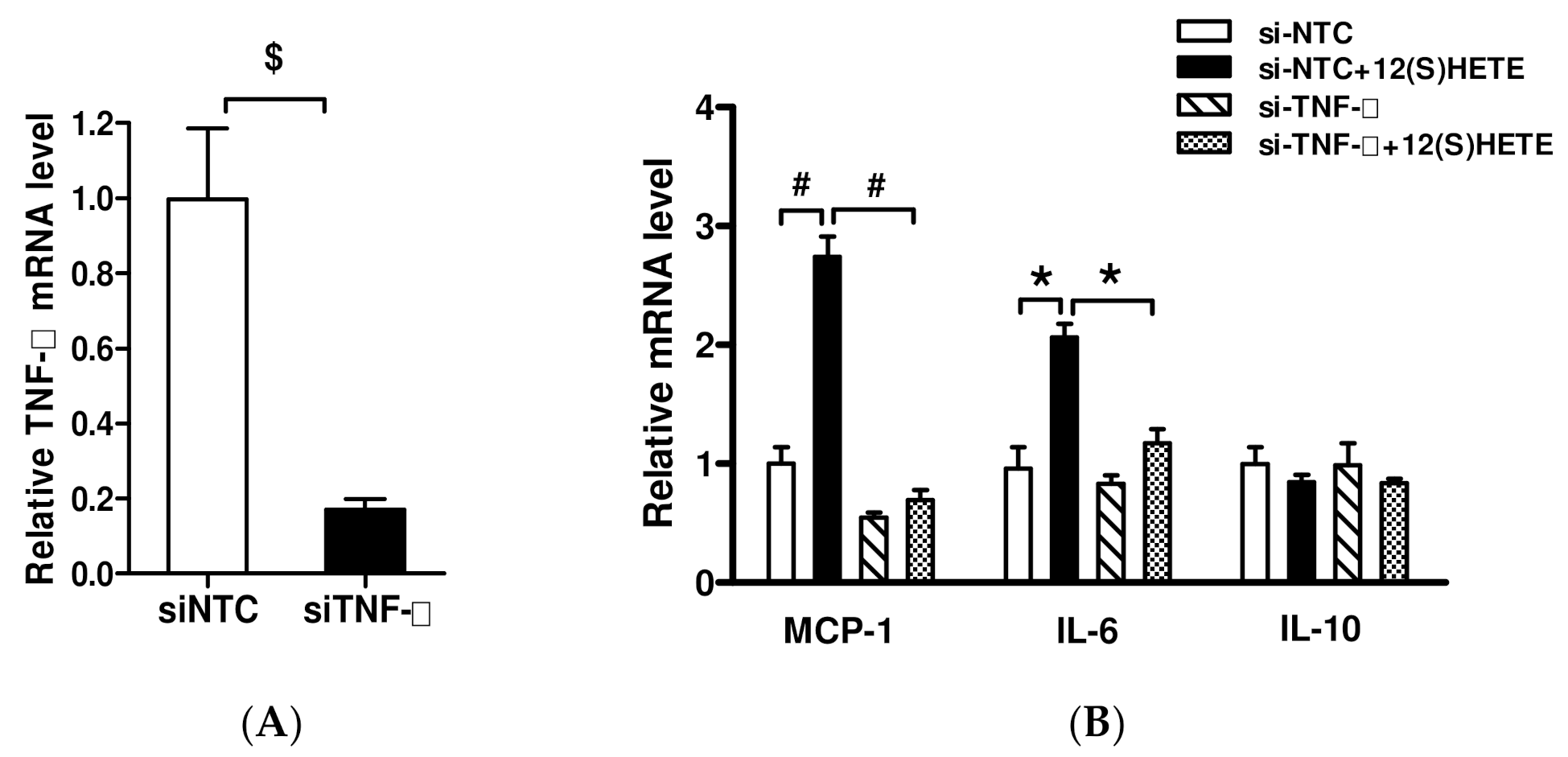
3.3. Effect of Inhibition of 12/15-LO Expression on TNF-α-Regulated Inflammatory Factor Expression
3.4. Effect of TNF-α Inhibition on 12/15-LO-Regulated Inflammatory Factors
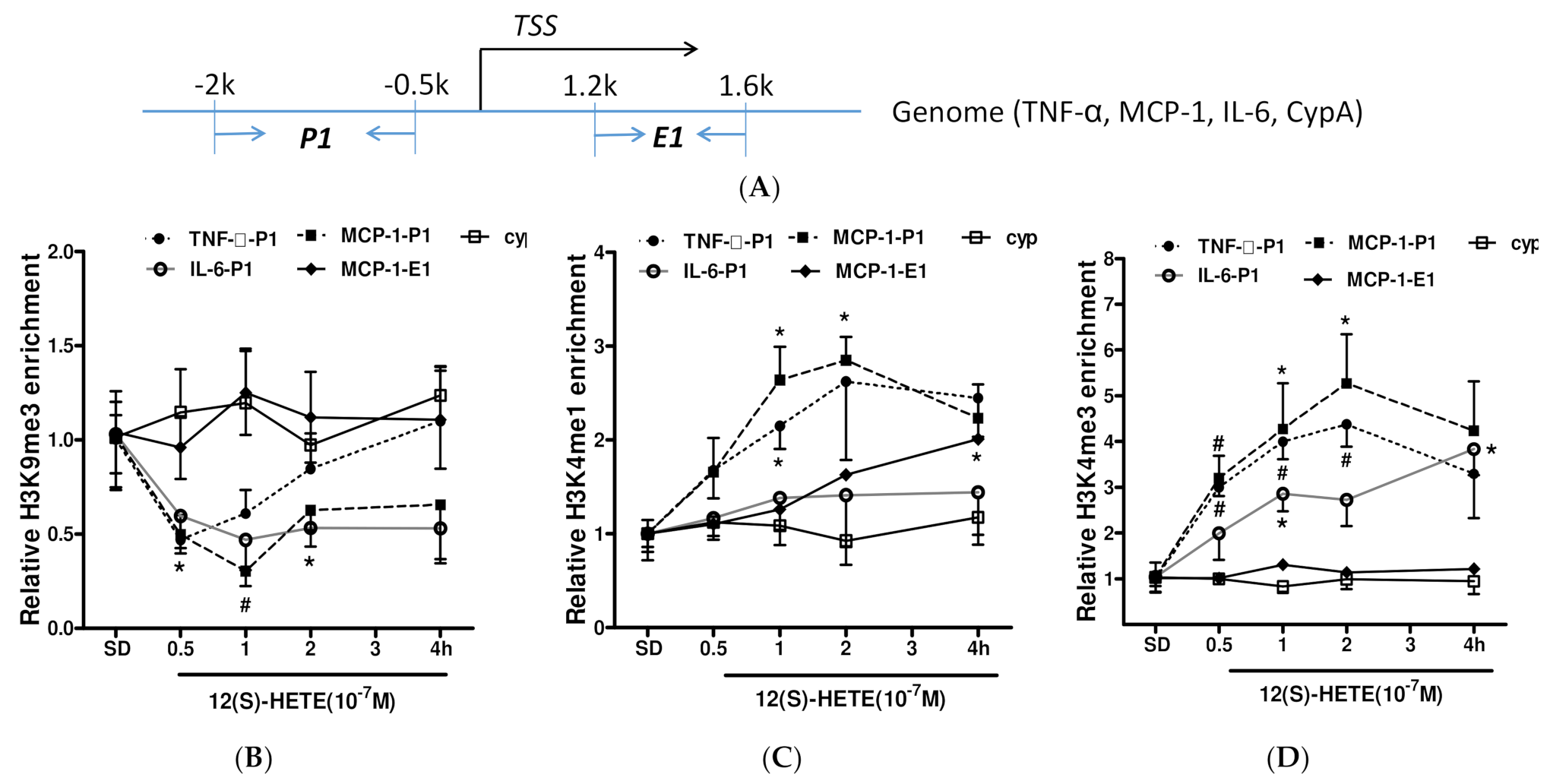
3.5. Histone Methylation Participates in the Regulation of Inflammatory Factor Expression by 12/15-LO Metabolites
3.6. Effects of 12/15-LO on Mouse Body Mass, Urinary Protein Content, and Other Indicators in Obese Mice
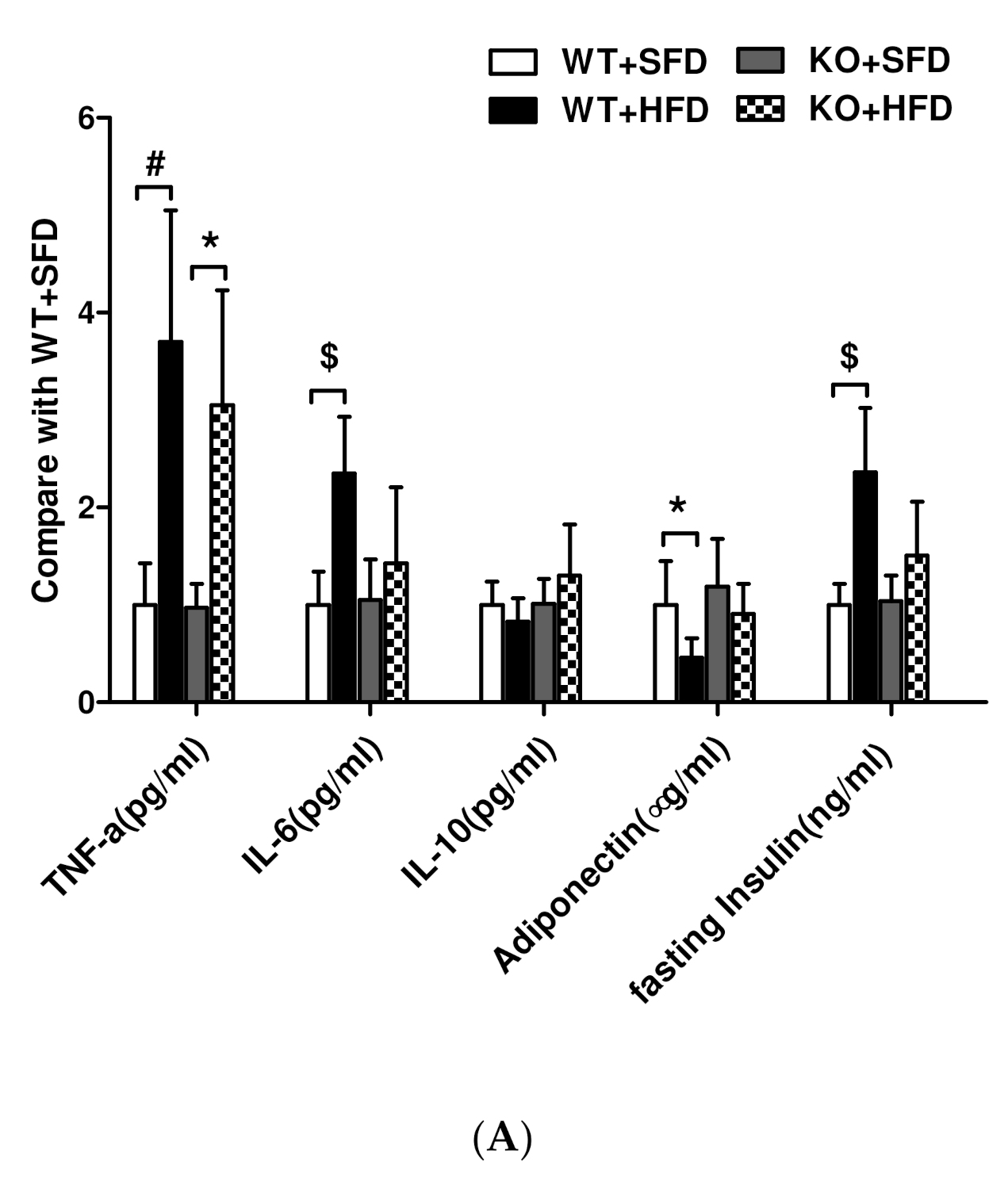
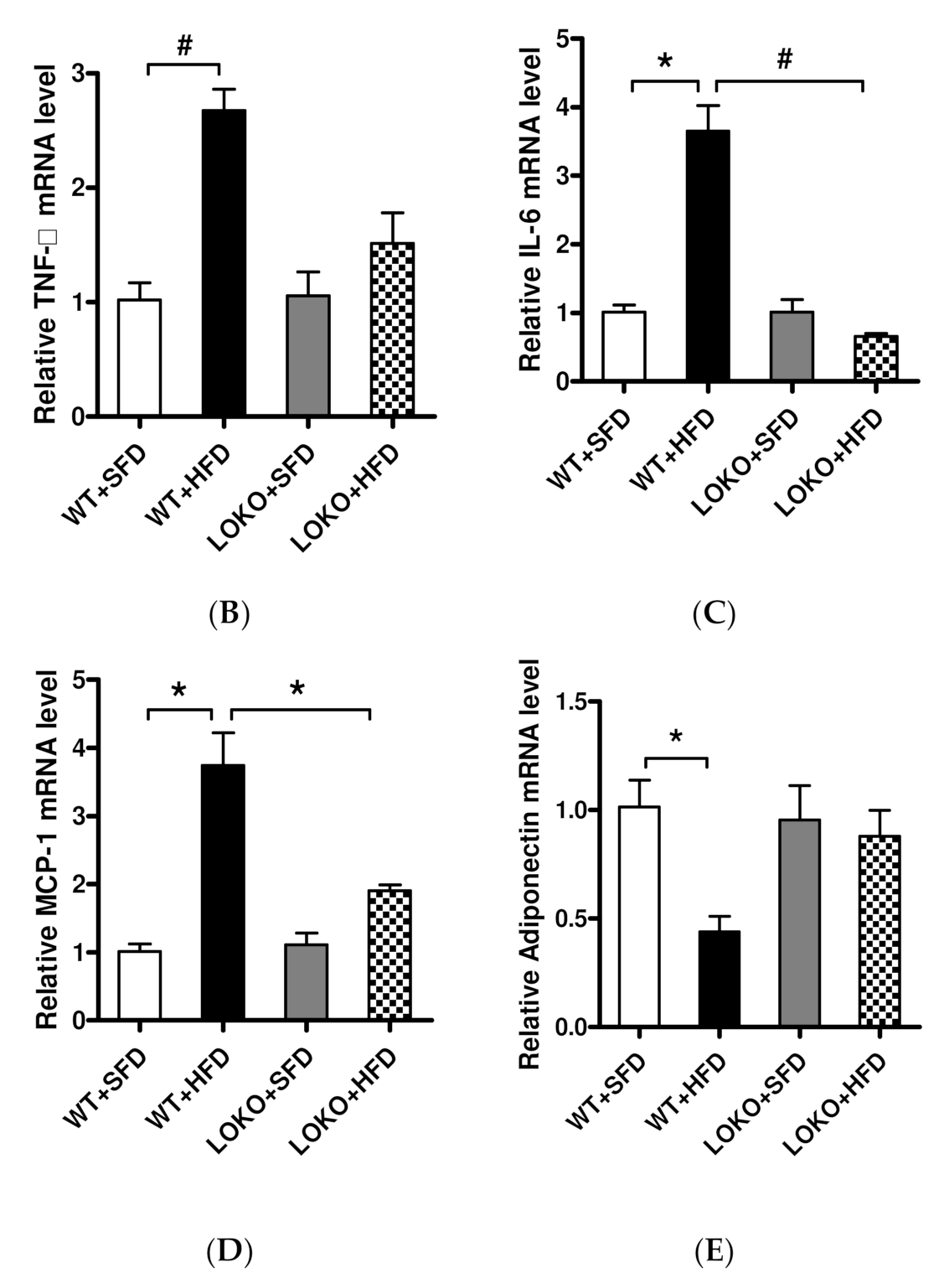
3.7. Expression of Lipid Inflammatory Cytokines and Adiponectin in Obese Mice
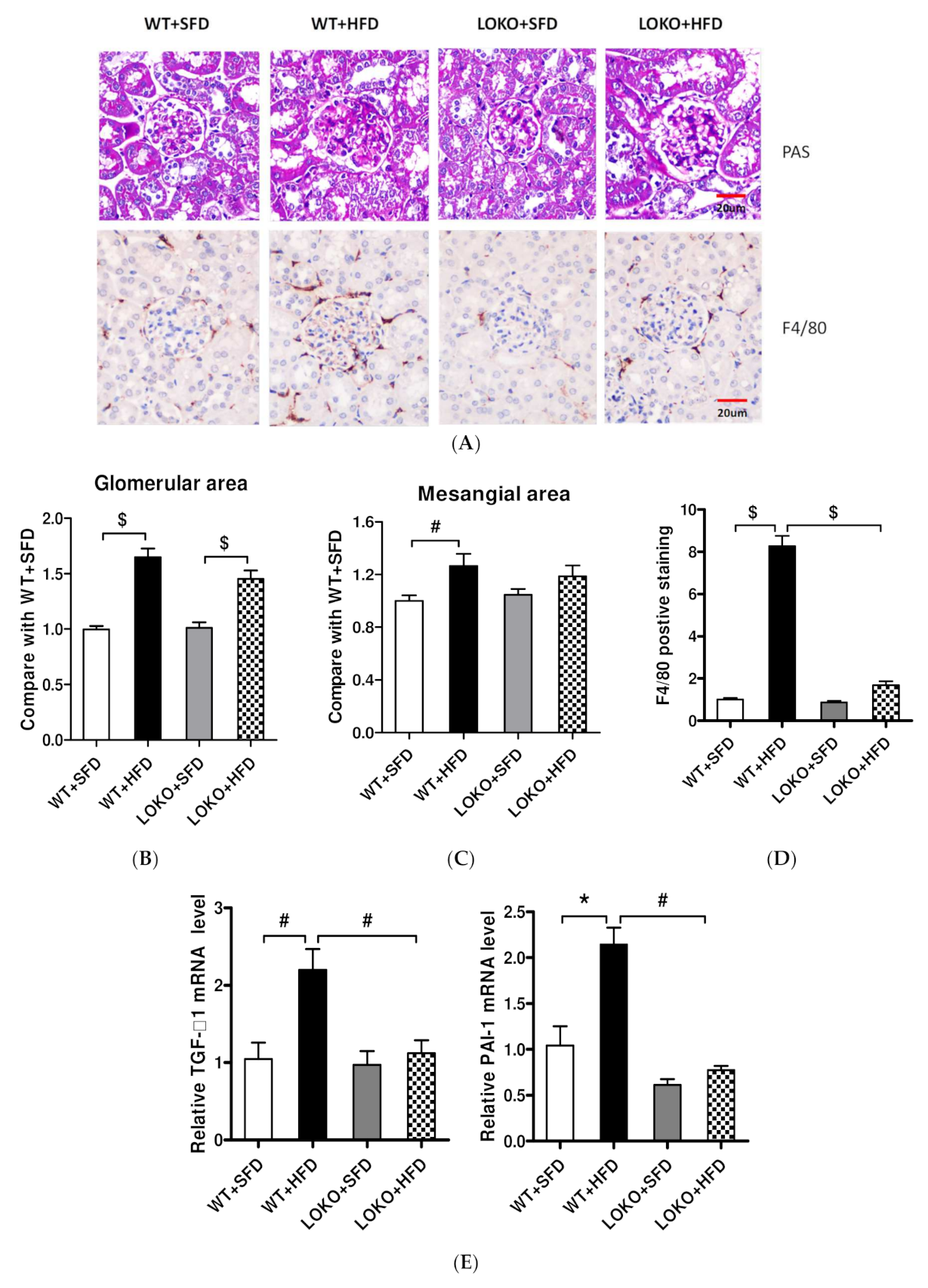
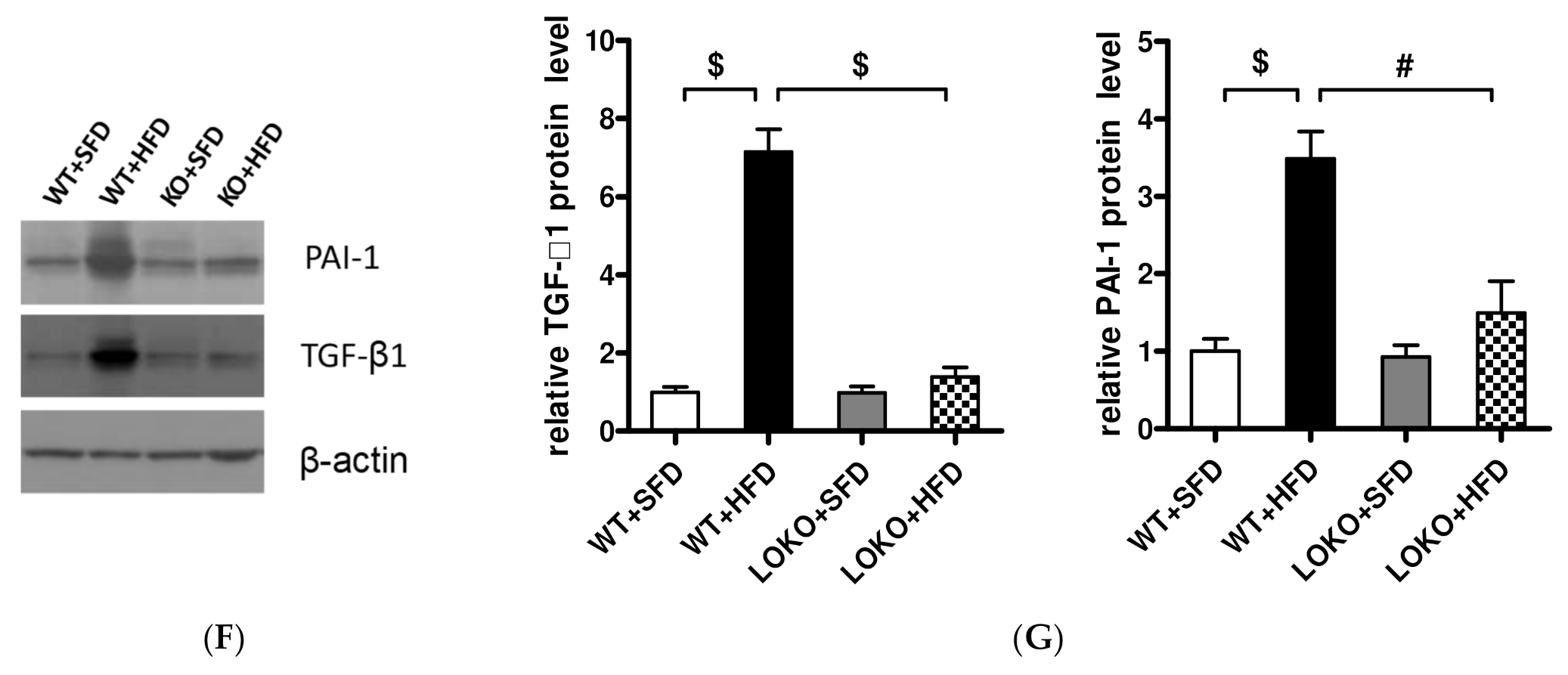
3.8. Pathological Renal Changes and Expression of Pathogenic Genes in Renal Tissues of Mice under Different Feeding Regimens
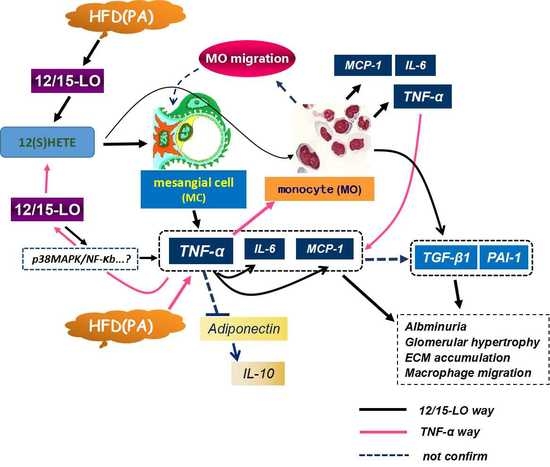
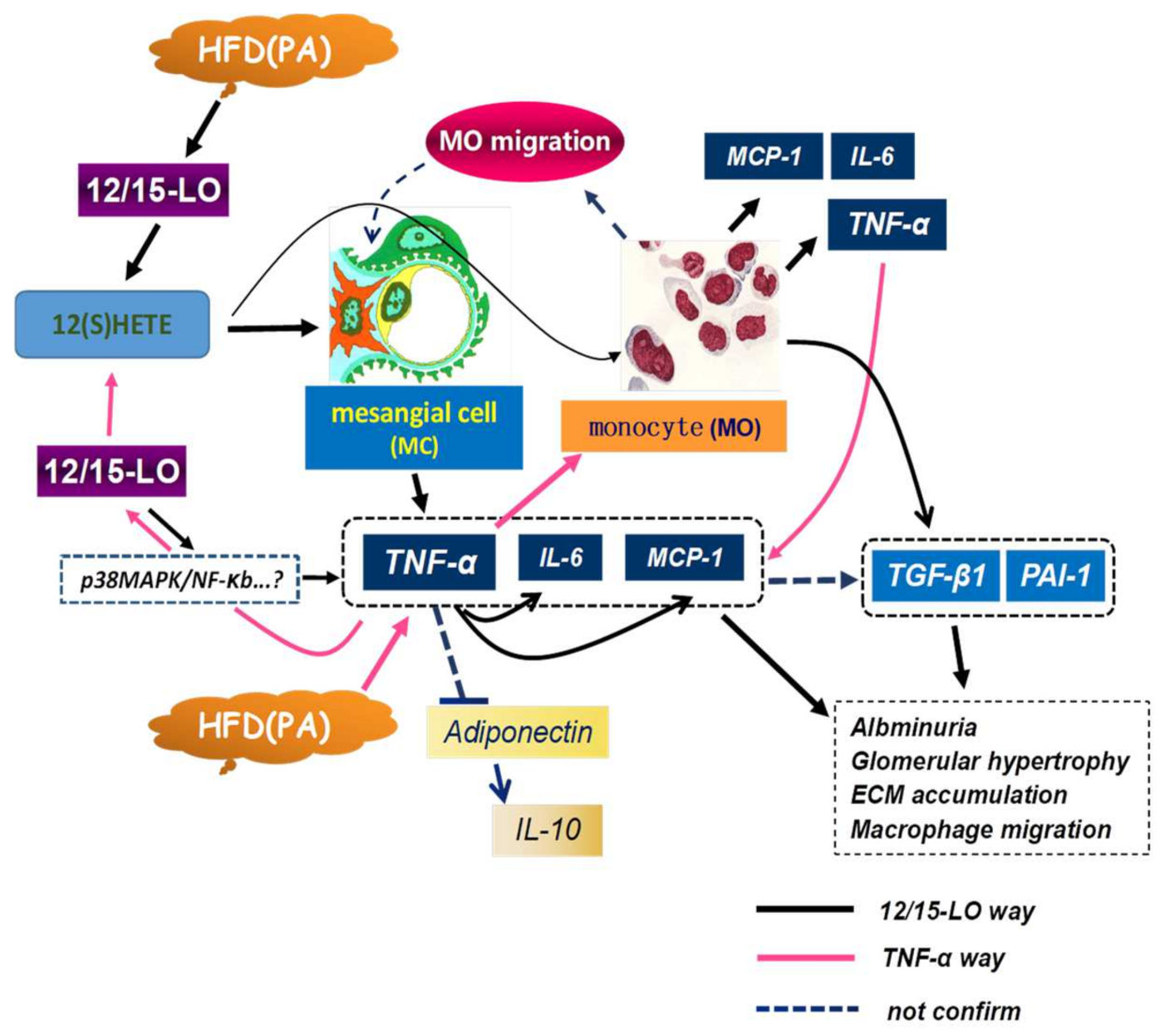
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajkovic, N.; Zamaklar, M.; Lalic, K.; Jotic, A.; Lukic, L.; Milicic, T.; Singh, S.; Stosic, L.; Lalic, N.M. Relationship between Obesity, Adipocytokines and Inflammatory Markers in Type 2 Diabetes: Relevance for Cardiovascular Risk Prevention. Int. J. Env. Res. Pub. He. 2014, 11, 4049–4065. [Google Scholar] [CrossRef] [PubMed]
- Stienstra, R.; van Diepen, J.A.; Tack, C.J.; Zaki, M.H.; van de Veerdonk, F.L.; Perera, D.; Neale, G.A.; Hooiveld, G.J.; Hijmans, A.; Vroegrijk, I.; et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 15324–15329. [Google Scholar] [CrossRef] [PubMed]
- Cao, H. Adipocytokines in obesity and metabolic disease. J. Endocrinol. 2014, 220, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Sikaris, K.A. The Clinical Biochemistry of Obesity-More Than Skin Deep. Heart Lung Circ. 2007, 16, S45–S50. [Google Scholar] [CrossRef]
- Decleves, A.E.; Zolkipli, Z.; Satriano, J.; Wang, L.; Nakayama, T.; Rogac, M.; Le, T.P.; Nortier, J.L.; Farquhar, M.G.; Naviaux, R.K.; et al. Regulation of lipid accumulation by AMP-activated kinase in high fat diet-induced kidney injury. Kidney. Int. 2014, 85, 611–623. [Google Scholar] [CrossRef]
- Yeung, J.; Holinstat, M. 12-lipoxygenase: A potential target for novel anti-platelet therapeutics. Cardiovasc. Hematol. Agents. Med. Chem. 2011, 9, 154–164. [Google Scholar] [CrossRef]
- Lopategi, A.; Lopez-Vicario, C.; Alcaraz-Quiles, J.; Garcia-Alonso, V.; Rius, B.; Titos, E.; Claria, J. Role of bioactive lipid mediators in obese adipose tissue inflammation and endocrine dysfunction. Mol. Cell. Endocrinol. 2016, 419, 44–59. [Google Scholar] [CrossRef]
- Xu, H.Z.; Cheng, Y.L.; Wang, W.N.; Wu, H.; Zhang, Y.Y.; Zang, C.S.; Xu, Z.G. 12-Lipoxygenase Inhibition on Microalbuminuria in Type-1 and Type-2 Diabetes Is Associated with Changes of Glomerular Angiotensin II Type 1 Receptor Related to Insulin Resistance. Int. J. Mol. Sci. 2016, 17, 684. [Google Scholar] [CrossRef]
- Dobrian, A.D.; Morris, M.A.; Taylor-Fishwick, D.A.; Holman, T.R.; Imai, Y.; Mirmira, R.G.; Nadler, J.L. Role of the 12-lipoxygenase pathway in diabetes pathogenesis and complications. Pharmacol. Ther. 2019, 195, 100–110. [Google Scholar] [CrossRef]
- Hernandez-Perez, M.; Chopra, G.; Fine, J.; Conteh, A.M.; Anderson, R.M.; Linnemann, A.K.; Benjamin, C.; Nelson, J.B.; Benninger, K.S.; Nadler, J.L.; et al. Inhibition of 12/15-Lipoxygenase Protects Against beta-Cell Oxidative Stress and Glycemic Deterioration in Mouse Models of Type 1 Diabetes. Diabetes 2017, 66, 2875–2887. [Google Scholar] [CrossRef]
- Tersey, S.A.; Maier, B.; Nishiki, Y.; Maganti, A.V.; Nadler, J.L.; Mirmira, R.G. 12-lipoxygenase promotes obesity-induced oxidative stress in pancreatic islets. Mol. Cell. Biol. 2014, 34, 3735–3745. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.G.; Yuan, H.; Lanting, L.L.; Li, S.L.; Wang, M.; Shanmugam, N.; Kato, M.; Adler, S.G.; Reddy, M.A.; Natarajan, R. Products of 12/15-Lipoxygenase Upregulate the Angiotensin II Receptor. J. Am. Soc. Nephrol. 2008, 19, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Reddy, M.A.; Deshpande, S.; Jia, Y.; Park, J.T.; Lanting, L.L.; Jin, W.; Kato, M.; Xu, Z.G.; Das, S.; et al. Epigenetic Histone Modifications Involved in Profibrotic Gene Regulation by 12/15-Lipoxygenase and Its Oxidized Lipid Products in Diabetic Nephropathy. Antioxid. Redox Signal. 2016, 24, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Khodabandeloo, H.; Gorgani-Firuzjaee, S.; Panahi, S.; Meshkani, R. Molecular and cellular mechanisms linking inflammation to insulin resistance and β-cell dysfunction. Transl. Res. 2016, 167, 228–256. [Google Scholar] [CrossRef]
- Chakrabarti, S.K.; Wen, Y.; Dobrian, A.D.; Cole, B.K.; Ma, Q.; Pei, H.; Williams, M.D.; Bevard, M.H.; Vandenhoff, G.E.; Keller, S.R.; et al. Evidence for activation of inflammatory lipoxygenase pathways in visceral adipose tissue of obese Zucker rats. Am. J. Physiol. Endocrinol. Metab. 2011, 300, 175–187. [Google Scholar] [CrossRef]
- Faulkner, J.; Pye, C.; Al-Shabrawey, M.; Elmarakby, A.A. Inhibition of 12/15-Lipoxygenase Reduces Renal Inflammation and Injury in Streptozotocin-Induced Diabetic Mice. J. Diabetes. Metab. 2015, 6, 555. [Google Scholar]
- Ma, K.; Nunemaker, C.S.; Wu, R.; Chakrabarti, S.K.; Taylor-Fishwick, D.A.; Nadler, J.L. 12-Lipoxygenase Products Reduce Insulin Secretion and {beta}-Cell Viability in Human Islets. J. Clin. Endocrinol. Metab. 2010, 95, 887–893. [Google Scholar] [CrossRef]
- Sethi, J.K.; Hotamisligil, G.S. Metabolic Messengers: Tumour necrosis factor. Nat. Metab. 2021, 3, 1302–1312. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J. Role of obesity-induced inflammation in the development of insulin resistance and type 2 diabetes: History of the research and remaining questions. Ann. Pediatr. Endocrinol. Metab. 2021, 26, 1–13. [Google Scholar] [CrossRef]
- Taylor, E.B. The complex role of adipokines in obesity, inflammation, and autoimmunity. Clin. Sci. 2021, 135, 731–752. [Google Scholar] [CrossRef]
- Kume, S.; Uzu, T.; Araki, S.; Sugimoto, T.; Isshiki, K.; Chin-Kanasaki, M.; Sakaguchi, M.; Kubota, N.; Terauchi, Y.; Kadowaki, T.; et al. Role of altered renal lipid metabolism in the development of renal injury induced by a high-fat diet. J. Am. Soc. Nephrol. 2007, 18, 2715–2723. [Google Scholar] [CrossRef] [PubMed]
- Roubicek, T.; Bartlova, M.; Krajickova, J.; Haluzikova, D.; Mraz, M.; Lacinova, Z.; Kudla, M.; Teplan, V.; Haluzik, M. Increased production of proinflammatory cytokines in adipose tissue of patients with end-stage renal disease. Nutrition 2009, 25, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Ziyadeh, F.N.; Wolf, G. Pathogenesis of the podocytopathy and rpoteinuria in daibetic glomerulopathy. Curr. Diabetetes Rev. 2008, 4, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Tersey, S.A.; Bolanis, E.; Holman, T.R.; Maloney, D.J.; Nadler, J.L.; Mirmira, R.G. Minireview: 12-Lipoxygenase and Islet β-Cell Dysfunction in Diabetes. Mol. Endocrinol. 2015, 29, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Lanting, L.L.; Xu, Z.G.; Li, S.L.; Swiderski, P.; Putta, S.; Jonnalagadda, M.; Kato, M.; Natarajan, R. Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. Am. J. Physiol. Renal. Physiol. 2008, 295, 605–617. [Google Scholar] [CrossRef]
- Kato, M.; Natarajan, R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat. Rev. Nephrol. 2019, 15, 327–345. [Google Scholar] [CrossRef]
- Rosen, E.D.; Kaestner, K.H.; Natarajan, R.; Patti, M.E.; Sallari, R.; Sander, M.; Susztak, K. Epigenetics and Epigenomics: Implications for Diabetes and Obesity. Diabetes 2018, 67, 1923–1931. [Google Scholar] [CrossRef]
- Leung, A.; Trac, C.; Du, J.; Natarajan, R.; Schones, D.E. Persistent Chromatin Modifications Induced by High Fat Diet. J. Biol. Chem. 2016, 291, 10446–10455. [Google Scholar] [CrossRef]
- Bomsztyk, K.; Denisenko, O. Epigenetic alterations in acute kidney injury. Semin. Nephrol. 2013, 33, 327–340. [Google Scholar] [CrossRef]
- Zager, R.A.; Johnson, A.C. Renal ischemia-reperfusion injury upregulates histone-modifying enzyme systems and alters histone expression at proinflammatory/profibrotic genes. Am. J. Physiol. Renal. Physiol. 2009, 296, 1032–1041. [Google Scholar] [CrossRef][Green Version]
- Miao, F.; Chen, Z.; Genuth, S.; Paterson, A.; Zhang, L.X.; Wu, X.W.; Li, S.M.; Cleary, P.; Riggs, A.; Harlan, D.M.; et al. Evaluating the Role of Epigenetic Histone Modifications in the Metabolic Memory of Type 1 Diabetes. Diabetes 2014, 63, 1748–1762. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.C.; Liu, N.; Ma, F.Z.; Sun, W.X.; Wu, H.; Xu, Z.G.; Yuan, H. Role of histone modification in 12-lipoxygenase-associated p21 gene regulation. Mol. Med. Rep. 2016, 14, 3978–3984. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Villeneuve, L.M.; Reddy, M.A.; Lanting, L.L.; Wang, M.; Meng, L.; Natarajan, R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc. Natl. Acad. Sci. USA 2008, 105, 9047–9052. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Reddy, M.A.; Miao, F.; Shanmugam, N.; Yee, J.K.; Hawkins, D.; Ren, B.; Natarajan, R. Role of the Histone H3 Lysine 4 Methyltransferase, SET7/9, in the Regulation of NF-κB-dependent Inflammatory Genes: Relevance to diabetes and inflammation. J. Biol. Chem. 2008, 283, 26771–26781. [Google Scholar] [CrossRef] [PubMed]
| Primers (5′→3′) | ||
|---|---|---|
| Gene (Species) | Sense | Anti-Sense |
| cDNA primers | ||
| 12/15-LO (R/M) | CGCTGGCACTCTGTTTGAAGCG | TGGATGGCTATGGGCAAGA |
| TNF-α (M) | TGTTGCCTCCTCTTTTGCTT | TGGTCACCAAATCAGCGTTA |
| MCP-1 (M) | TTAAAAACCTGGATCGGAACC | GCATTAGCTTCAGATTTACGG |
| IL-6 (M) | ACAAAGCCAGAGTCCTTC | ACCACAGTGAGGAATGTCCAC |
| IL-10 (M) | CCCAAGCTTATGCCTGGCTCAGCACTGC | CGCGGATCCTTAGCTTTTCATTTTGATC |
| IL-1β (M) | TTGACGGACC CCAAAAG ATG | AGAAGGTGCTCATG TCCTCA |
| IL-12 (M) | TCTGCAGAGAAGGTCACACT | ATGAAGAAGCTGGTGCTGTA |
| CXCL10 (M) | CTCATCCTGCTGGGTCTGAG | CCTATGGCCCTCATTCTCAC |
| β-actin (R/M) | CCCTGTATGCCTCTGGTCGT | CGGACGCAGCTCAGTAACAGTCCG |
| ChIP primers | ||
| IL-6 pro (M) | CGTTTATGATTCTTTCGATGCTAAACG | GTGGGCTCCAGAGCAGAATGAG |
| MCP-1 pro (M) | CACTAACTGAGGCCATGAACAGGTTAGTG | GCAAACCAGCACAAATGTAGCC |
| MCP-1 enh (M) | ATTTCCACGCTCTTATCCTACTCTG | TCACCATTGCAAAGTGAATTGG |
| TNF-α pro (M) | AACCGAGACAGAAGGTGCAG | TGTGCCAACAACTGCCTTTA |
| CypA pro (M) | GCAGGTAGGTCCTTGAGCTTGTC | CGCTAGAAGACCCTTCACCATAGCG |
| WT + SFD | WT + HFD | LOKO + SFD | LOKO + HFD | |
|---|---|---|---|---|
| Fasting glucose 1 wk (mg/dL) | 137 ± 22.0 | 128 ± 21.2 | 126 ± 22.1 | 127 ± 19.7 |
| Fasting glucose 8 wk (mg/dL) | 143 ± 26.5 | 189 ± 33.7 | 141 ± 26.3 | 177 ± 31.6 |
| Fasting glucose 14 wk (mg/dL) | 155 ± 29.7 | 243 ± 46.4 # | 144 ± 34.6 | 226 ± 40.1 † |
| Serum creatinine 14 wk (mg/dL) | 0.22 ± 0.02 | 0.31 ± 0.03 | 0.20 ± 0.01 | 0.28 ± 0.02 |
| Body weight 1 wk (g) | 20.57 ± 0.53 | 20.43± 1.27 | 23.00 ± 0.89 | 22.43 ± 1.62 |
| Body weight 8 wk (g) | 26.57 ± 0.98 | 34.57 ± 2.94 $ | 30.17 ± 1.17 | 35.86 ± 3.34 ‡ |
| Body weight 14 wk (g) | 29.57 ± 0.79 | 45.14 ± 2.91 $ | 31.83 ± 1.47 | 44.29 ± 4.72 £ |
| Kidney weight 14wk (g) | 0.44 ± 0.02 | 0.60 ± 0.02 $ | 0.48 ± 0.03 | 0.59 ± 0.03 £ |
| K/B (%) | 1.492 ± 0.049 | 1.340 ± 0.083 * | 1.517± 0.081 | 1.337 ± 0.16 † |
| K/T (g/cm) | 0.224 ± 0.010 | 0.316 ± 0.017 $ | 0.247 ± 0.012 | 0.310 ± 0.018 £ |
| Urinary volume 1 wk (mL) | 2.67 ± 0.67 | 3.10 ± 0.48 | 2.85 ± 0.63 | 3.50 ± 0.56 |
| Proteinuria 1 wk (mg) | 21.39 ± 3.88 | 22.53 ± 4.89 | 24.17 ± 3.37 | 22.68 ± 4.22 |
| Urinary ACR 1 wk (μg/mg) | 35.19 ± 7.01 | 34.40 ± 10.29 | 33.40 ± 11.14 | 36.08 ± 7.18 |
| Urinary volume 8 wk (mL) | 3.59 ± 0.53 | 2.53 ± 0.56 $ | 4.40 ± 0.58 | 2.49 ± 0.38 † |
| Proteinuria 8 wk (mg) | 21.53 ± 4.10 | 45.53 ± 13.33 # | 24.16 ± 5.24 | 33.41 ± 7.63 † |
| Urinary ACR 8 wk (μg/mg) | 48.70 ± 10.38 | 85.66± 20.72 * | 56.32 ± 15.97 | 59.10 ± 12.61 |
| Urinary volume 14 wk (mL) | 2.86 ± 0.66 | 1.36 ± 0.59 # | 4.35 ± 1.31 | 1.47 ± 0.49 ‡ |
| Proteinuria 14 wk (mg) | 25.62 ± 6.33 | 42.82 ± 15.64 * | 24.86 ± 5.30 | 33.74 ±9.54 |
| Urinary ACR 14wk (μg/mg) | 46.63 ± 12.78 | 106.35 ± 22.78 * | 32.67 ± 7.41 | 72.51 ± 23.97 † |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Liu, Y.; Dong, D.; Yu, J.; Yuan, H. Effects of Inflammatory Factor Expression Regulated by 12/15 Lipoxygenase on Obesity-Related Nephropathy. Nutrients 2022, 14, 2743. https://doi.org/10.3390/nu14132743
Liu N, Liu Y, Dong D, Yu J, Yuan H. Effects of Inflammatory Factor Expression Regulated by 12/15 Lipoxygenase on Obesity-Related Nephropathy. Nutrients. 2022; 14(13):2743. https://doi.org/10.3390/nu14132743
Chicago/Turabian StyleLiu, Nian, Yang Liu, Dan Dong, Jinyu Yu, and Hang Yuan. 2022. "Effects of Inflammatory Factor Expression Regulated by 12/15 Lipoxygenase on Obesity-Related Nephropathy" Nutrients 14, no. 13: 2743. https://doi.org/10.3390/nu14132743
APA StyleLiu, N., Liu, Y., Dong, D., Yu, J., & Yuan, H. (2022). Effects of Inflammatory Factor Expression Regulated by 12/15 Lipoxygenase on Obesity-Related Nephropathy. Nutrients, 14(13), 2743. https://doi.org/10.3390/nu14132743