Impact of Lipid Genetic Risk Score and Saturated Fatty Acid Intake on Central Obesity in an Asian Indian Population
Abstract
1. Introduction
2. Methods
2.1. Study Participants
2.2. Anthropometric and Biochemical Measurements
2.3. Dietary Assessment
2.4. SNP Selection and Genotyping
2.5. Construction of GRS
2.6. Statistical Analysis
3. Results
3.1. Characteristics of the Study Participants
3.2. Association of GRS with Lipid and Obesity-Related Traits
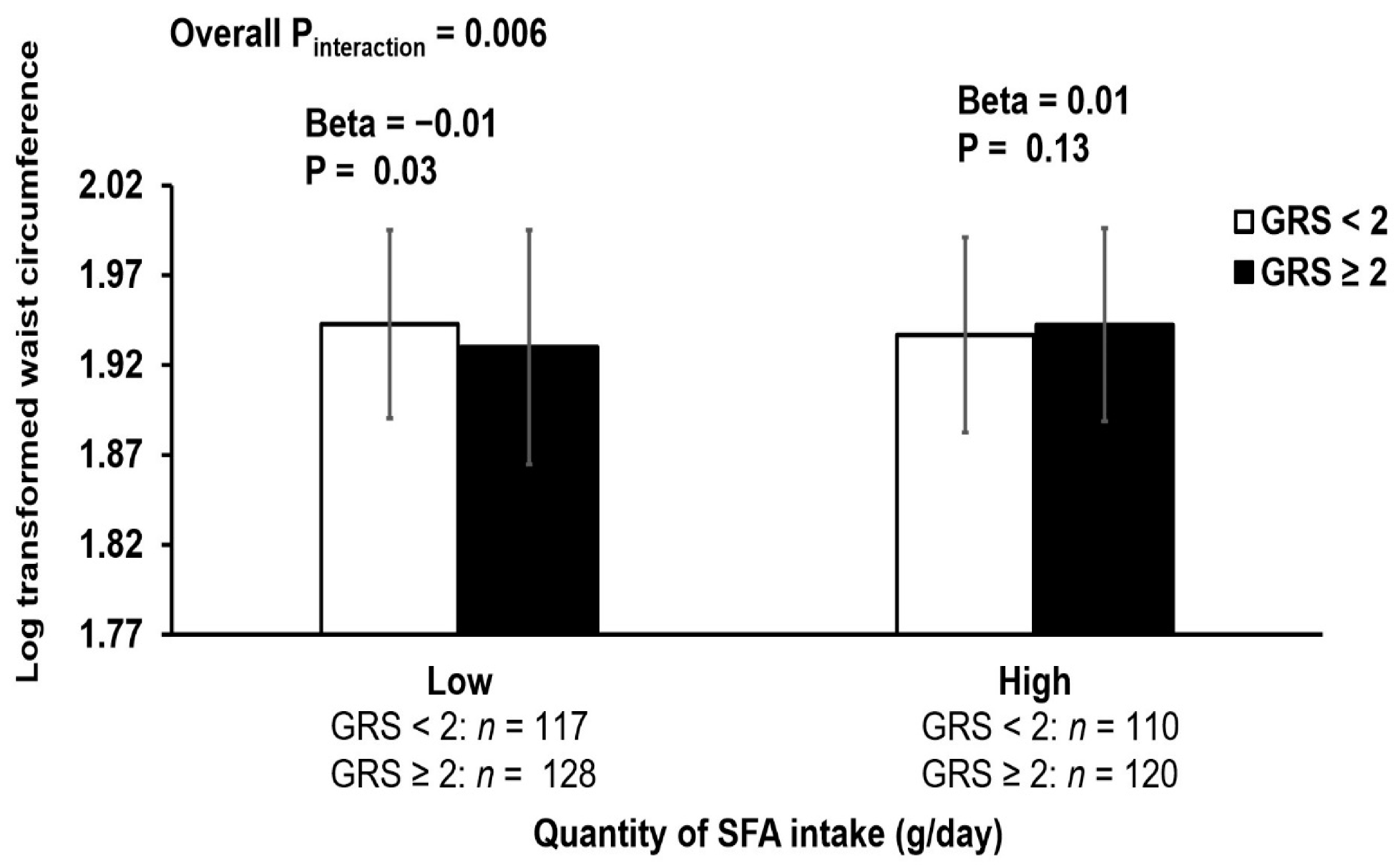
3.3. Interaction of GRS with Dietary Factors on Lipid and Obesity Related Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vimaleswaran, K.S.; Bodhini, D.; Lakshmipriya, N.; Ramya, K.; Anjana, R.M.; Sudha, V.; Lovegrove, J.A.; Kinra, S.; Mohan, V.; Radha, V. Interaction between FTO gene variants and lifestyle factors on metabolic traits in an Asian Indian population. Nutr. Metab. 2016, 13, 39. [Google Scholar] [CrossRef]
- Mohan, V.; Deepa, R. Adipocytokines and the expanding ‘Asian Indian Phenotype’. J. Assoc. Physicians India 2006, 54, 685–686. [Google Scholar] [PubMed]
- Gujral, U.P.; Mohan, V.; Pradeepa, R.; Deepa, M.; Anjana, R.M.; Mehta, N.K.; Gregg, E.W.; Narayan, K. Ethnic Variations in Diabetes and Prediabetes Prevalence and the roles of Insulin Resistance and β-cell Function: The CARRS and NHANES Studies. J. Clin. Transl. Endocrinol. 2016, 4, 19–27. [Google Scholar] [CrossRef]
- Chen, G.-C.; Arthur, R.; Iyengar, N.M.; Kamensky, V.; Xue, X.; Wassertheil-Smoller, S.; Allison, M.A.; Shadyab, A.H.; Wild, R.A.; Sun, Y.; et al. Association between regional body fat and cardiovascular disease risk among postmenopausal women with normal body mass index. Eur. Heart J. 2019, 40, 2849–2855. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Schick, F.; Häring, H.-U. Causes, Characteristics, and Consequences of Metabolically Unhealthy Normal Weight in Humans. Cell Metab. 2017, 26, 292–300. [Google Scholar] [CrossRef]
- Blüher, M. Metabolically Healthy Obesity. Endocr. Rev. 2020, 41, bnaa004. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, S.A.; Connell, J.M. The link between abdominal obesity, metabolic syndrome and cardiovascular disease. Nutr. Metab. Cardiovasc. Dis. 2007, 17, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Powell-Wiley, T.M.; Poirier, P.; Burke, L.E.; Després, J.P.; Gordon-Larsen, P.; Lavie, C.J.; Lear, S.A.; Ndumele, C.E.; Neeland, I.J.; Sanders, P.; et al. Obesity and Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation 2021, 143, e984–e1010. [Google Scholar] [CrossRef]
- Sahakyan, K.R.; Somers, V.K.; Rodriguez-Escudero, J.P.; Hodge, D.O.; Carter, R.E.; Sochor, O.; Coutinho, T.; Jensen, M.D.; Roger, V.L.; Singh, P.; et al. Normal-Weight Central Obesity: Implications for Total and Cardiovascular Mortality. Ann. Intern. Med. 2015, 163, 827–835. [Google Scholar] [CrossRef]
- Weinstock, P.H.; Levak-Frank, S.; Hudgins, L.C.; Radner, H.; Friedman, J.M.; Zechner, R.; Breslow, J.L. Lipoprotein lipase controls fatty acid entry into adipose tissue, but fat mass is preserved by endogenous synthesis in mice deficient in adipose tissue lipoprotein lipase. Proc. Natl. Acad. Sci. USA 1997, 94, 10261–10266. [Google Scholar] [CrossRef]
- Wang, H.; Eckel, R.H. Lipoprotein lipase: From gene to obesity. Am. J. Physiol.-Endocrinol. Metab. 2009, 297, E271–E288. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tucker, K.L.; Smith, C.E.; Lee, Y.C.; Huang, T.; Richardson, K.; Parnell, L.D.; Lai, C.Q.; Young, K.L.; Justice, A.E.; et al. Lipoprotein lipase variants interact with polyunsaturated fatty acids for obesity traits in women: Replication in two populations. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 1323–1329. [Google Scholar] [CrossRef]
- Barter Philip, J.; Brewer, H.B.; Chapman, M.J.; Hennekens Charles, H.; Rader Daniel, J.; Tall Alan, R. Cholesteryl Ester Transfer Protein. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Barter, P.J. The causes and consequences of low levels of high density lipoproteins in patients with diabetes. Diabetes Metab. J. 2011, 35, 101–106. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, S.J.; Feskens, E.J.; Bos, M.B.; Hoelen, D.W.; Heijligenberg, R.; Bromhaar, M.G.; de Groot, L.C.; de Vries, J.H.; Müller, M.; Afman, L.A. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am. J. Clin. Nutr 2009, 90, 1656–1664. [Google Scholar] [CrossRef]
- Liu, X.; Garban, J.; Jones, P.J.; Vanden Heuvel, J.; Lamarche, B.; Jenkins, D.J.; Connelly, P.W.; Couture, P.; Pu, S.; Fleming, J.A.; et al. Diets Low in Saturated Fat with Different Unsaturated Fatty Acid Profiles Similarly Increase Serum-Mediated Cholesterol Efflux from THP-1 Macrophages in a Population with or at Risk for Metabolic Syndrome: The Canola Oil Multicenter Intervention Trial. J. Nutr. 2018, 148, 721–728. [Google Scholar] [CrossRef]
- Goto, T. A review of the studies on food-derived factors which regulate energy metabolism via the modulation of lipid-sensing nuclear receptors. Biosci. Biotechnol. Biochem. 2019, 83, 579–588. [Google Scholar] [CrossRef]
- Vimaleswaran, K.S.; Minihane, A.M.; Li, Y.; Gill, R.; Lovegrove, J.A.; Williams, C.M.; Jackson, K.G. The APOB insertion/deletion polymorphism (rs17240441) influences postprandial lipaemia in healthy adults. Nutr. Metab. 2015, 12, 7. [Google Scholar] [CrossRef]
- Ayyappa, K.A.; Shatwan, I.; Bodhini, D.; Bramwell, L.R.; Ramya, K.; Sudha, V.; Anjana, R.M.; Lovegrove, J.A.; Mohan, V.; Radha, V.; et al. High fat diet modifies the association of lipoprotein lipase gene polymorphism with high density lipoprotein cholesterol in an Asian Indian population. Nutr. Metab. 2017, 14, 8. [Google Scholar] [CrossRef]
- Shatwan, I.M.; Winther, K.H.; Ellahi, B.; Elwood, P.; Ben-Shlomo, Y.; Givens, I.; Rayman, M.P.; Lovegrove, J.A.; Vimaleswaran, K.S. Association of apolipoprotein E gene polymorphisms with blood lipids and their interaction with dietary factors. Lipids Health Dis. 2018, 17, 98. [Google Scholar] [CrossRef]
- Khushdeep, B.; Prasad, G.; Giri, A.K.; Kauser, Y.; Upadhyay, M.; Basu, A.; Tandon, N.; Bharadwaj, D. Genome-wide association study of blood lipids in Indians confirms universality of established variants. J. Hum. Genet. 2019, 64, 573–587. [Google Scholar]
- Adeyemo, A.; Bentley, A.R.; Meilleur, K.G.; Doumatey, A.P.; Chen, G.; Zhou, J.; Shriner, D.; Huang, H.; Herbert, A.; Gerry, N.P.; et al. Transferability and Fine Mapping of genome-wide associated loci for lipids in African Americans. BMC Med. Genet. 2012, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Kurano, M.; Tsukamoto, K.; Kamitsuji, S.; Kamatani, N.; Hara, M.; Ishikawa, T.; Kim, B.-J.; Moon, S.; Kim, Y.J.; Teramoto, T. Genome-wide association study of serum lipids confirms previously reported associations as well as new associations of common SNPs within PCSK7 gene with triglyceride. J. Hum. Genet. 2016, 61, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Zabaneh, D.; Balding, D.J. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS ONE 2010, 5, e11961. [Google Scholar] [CrossRef]
- Kathiresan, S.; Melander, O.; Guiducci, C.; Surti, A.; Burtt, N.P.; Rieder, M.J.; Cooper, G.M.; Roos, C.; Voight, B.F.; Havulinna, A.S.; et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008, 40, 189–197. [Google Scholar] [CrossRef]
- Lettre, G.; Palmer, C.D.; Young, T.; Ejebe, K.G.; Allayee, H.; Benjamin, E.J.; Bennett, F.; Bowden, D.W.; Chakravarti, A.; Dreisbach, A.; et al. Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project. PLOS Genet. 2011, 7, e1001300. [Google Scholar] [CrossRef]
- Waterworth, D.M.; Ricketts, S.L.; Song, K.; Chen, L.; Zhao, J.H.; Ripatti, S.; Aulchenko, Y.S.; Zhang, W.; Yuan, X.; Lim, N.; et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arter. Thromb. Vasc. Biol. 2010, 30, 2264–2276. [Google Scholar] [CrossRef]
- Oh, S.W.; Lee, J.E.; Shin, E.; Kwon, H.; Choe, E.K.; Choi, S.Y.; Rhee, H.; Choi, S.H. Genome-wide association study of metabolic syndrome in Korean populations. PLoS ONE 2020, 15, e0227357. [Google Scholar]
- Zhou, L.; He, M.; Mo, Z.; Wu, C.; Yang, H.; Yu, D.; Yang, X.; Zhang, X.; Wang, Y.; Sun, J.; et al. A Genome Wide Association Study Identifies Common Variants Associated with Lipid Levels in the Chinese Population. PLoS ONE 2013, 8, e82420. [Google Scholar] [CrossRef]
- Chasman, D.I.; Paré, G.; Zee, R.Y.; Parker, A.N.; Cook, N.R.; Buring, J.E.; Kwiatkowski, D.J.; Rose, L.M.; Smith, J.D.; Williams, P.T.; et al. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and Apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ. Cardiovasc. Genet. 2008, 1, 21–30. [Google Scholar] [CrossRef]
- Sabatti, C.; Service, S.K.; Hartikainen, A.-L.; Pouta, A.; Ripatti, S.; Brodsky, J.; Jones, C.G.; Zaitlen, N.A.; Varilo, T.; Kaakinen, M.; et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009, 41, 35–46. [Google Scholar] [CrossRef]
- Wuni, R.; Kuhnle GG, C.; Wynn-Jones, A.A.; Vimaleswaran, K.S. A Nutrigenetic Update on CETP Gene-Diet Interactions on Lipid-Related Outcomes. Curr. Atheroscler. Rep. 2022, 24, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Radha, V.; Vimaleswaran, K.S.; Ayyappa, K.A.; Mohan, V. Association of lipoprotein lipase gene polymorphisms with obesity and type 2 diabetes in an Asian Indian population. Int. J. Obes 2007, 31, 913–918. [Google Scholar] [CrossRef]
- Hou, H.; Ma, R.; Guo, H.; He, J.; Hu, Y.; Mu, L.; Yan, Y.; Ma, J.; Li, S.; Zhang, J.; et al. Association between Six CETP Polymorphisms and Metabolic Syndrome in Uyghur Adults from Xinjiang, China. Int. J. Environ. Res. Public Health 2017, 14, 653. [Google Scholar] [CrossRef] [PubMed]
- Alsulami, S.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Tasrif, N.; Yani, F.F.; Lovegrove, J.A.; Sudji, I.R.; Lipoeto, N.I.; Vimaleswaran, K.S. Interaction between the genetic risk score and dietary protein intake on cardiometabolic traits in Southeast Asian. Genes Nutr. 2020, 15, 19. [Google Scholar] [CrossRef]
- Alsulami, S.; Nyakotey, D.A.; Dudek, K.; Bawah, A.M.; Lovegrove, J.A.; Annan, R.A.; Ellahi, B.; Vimaleswaran, K.S. Interaction between Metabolic Genetic Risk Score and Dietary Fatty Acid Intake on Central Obesity in a Ghanaian Population. Nutrients 2020, 12, 1906. [Google Scholar] [CrossRef] [PubMed]
- Janssens, A.C.; Moonesinghe, R.; Yang, Q.; Steyerberg, E.W.; van Duijn, C.M.; Khoury, M.J. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet. Med. 2007, 9, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Deepa, M.; Pradeepa, R.; Rema, M.; Mohan, A.; Deepa, R.; Shanthirani, S.; Mohan, V. The Chennai Urban Rural Epidemiology Study (CURES)--study design and methodology (urban component) (CURES-I). J. Assoc. Physicians India 2003, 51, 863–870. [Google Scholar] [PubMed]
- Vimaleswaran, K.S. A nutrigenetics approach to study the impact of genetic and lifestyle factors on cardiometabolic traits in various ethnic groups: Findings from the GeNuIne Collaboration. Proc. Nutr. Soc. 2020, 79, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Alathari, B.E.; Bodhini, D.; Jayashri, R.; Lakshmipriya, N.; Shanthi Rani, C.S.; Sudha, V.; Lovegrove, J.A.; Anjana, R.M.; Mohan, V.; Radha, V.; et al. A Nutrigenetic Approach to Investigate the Relationship between Metabolic Traits and Vitamin D Status in an Asian Indian Population. Nutrients 2020, 12, 1357. [Google Scholar] [CrossRef]
- Surendran, S.; Jayashri, R.; Drysdale, L.; Bodhini, D.; Lakshmipriya, N.; Shanthi Rani, C.S.; Sudha, V.; Lovegrove, J.A.; Anjana, R.M.; Mohan, V.; et al. Evidence for the association between FTO gene variants and vitamin B12 concentrations in an Asian Indian population. Genes Nutr. 2019, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Vimaleswaran, K.S. Gene–nutrient interactions on metabolic diseases: Findings from the GeNuIne Collaboration. Nutr. Bull. 2017, 42, 80–86. [Google Scholar] [CrossRef]
- Bodhini, D.; Gaal, S.; Shatwan, I.; Ramya, K.; Ellahi, B.; Surendran, S.; Sudha, V.; Anjana, M.R.; Mohan, V.; Lovegrove, J.A.; et al. Interaction between TCF7L2 polymorphism and dietary fat intake on high density lipoprotein cholesterol. PLoS ONE 2017, 12, e0188382. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Regional Office for the Western, P. In The Asia-Pacific Perspective: Redefining Obesity and Its Treatment; Health Communications Australia: Sydney, Australia, 2000. [Google Scholar]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem 1972, 18, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Sudha, V.; Radhika, G.; Sathya, R.M.; Ganesan, A.; Mohan, V. Reproducibility and validity of an interviewer-administered semi-quantitative food frequency questionnaire to assess dietary intake of urban adults in southern India. Int. J. Food Sci. Nutr. 2006, 57, 481–493. [Google Scholar] [CrossRef]
- Prakash, J.; Mittal, B.; Srivastava, A.; Awasthi, S.; Srivastava, N. The Association of a Rare Variant of -93, -53 Promoter Gene Polymorphisms of Lipoprotein Lipase gene with Obesity and Insulin Resistance. Oman Med. J. 2018, 33, 401–408. [Google Scholar] [CrossRef]
- Cho, Y.S.; Go, M.J.; Han, H.R.; Cha, S.H.; Kim, H.T.; Min, H.; Shin, H.D.; Park, C.; Han, B.G.; Cho, N.H.; et al. Association of lipoprotein lipase (LPL) single nucleotide polymorphisms with type 2 diabetes mellitus. Exp. Mol. Med. 2008, 40, 523–532. [Google Scholar] [CrossRef]
- Moghadasi, M.; Kelishadi, R.; Marateb, H.R.; Haghjooy Javanmard, S.; Mansourian, M.; Heshmat, R.; Esmaeil Motlagh, M. Logic Regression Analysis of Gene Polymorphisms and HDL Levels in a Nationally Representative Sample of Iranian Adolescents: The CASPIAN-III Study. Int. J. Endocrinol. Metab. 2017, 15, e14037. [Google Scholar] [CrossRef]
- Spirin, V.; Schmidt, S.; Pertsemlidis, A.; Cooper, R.S.; Cohen, J.C.; Sunyaev, S.R. Common single-nucleotide polymorphisms act in concert to affect plasma levels of high-density lipoprotein cholesterol. Am. J. Hum. Genet. 2007, 81, 1298–1303. [Google Scholar] [CrossRef][Green Version]
- Pradeepa, R.; Anjana, R.M.; Joshi, S.R.; Bhansali, A.; Deepa, M.; Joshi, P.P.; Dhandania, V.K.; Madhu, S.V.; Rao, P.V.; Geetha, L. Prevalence of generalized & abdominal obesity in urban & rural India-the ICMR-INDIAB Study (Phase-I)[ICMR-INDIAB-3]. Indian J. Med. Res. 2015, 142, 139. [Google Scholar]
- Mohan, V.; Deepa, R. Obesity and abdominal obesity in Asian Indians. Indian J. Med. Res. 2006, 123, 593–596. [Google Scholar] [PubMed]
- Shrivastava, U.; Misra, A.; Mohan, V.; Unnikrishnan, R.; Bachani, D. Obesity, Diabetes and Cardiovascular Diseases in India: Public Health Challenges. Curr. Diabetes Rev. 2017, 13, 65–80. [Google Scholar] [CrossRef]
- Gokulakrishnan, K.; Amutha, A.; Ranjani, H.; Bibin, S.Y.; Balakumar, M.; Pandey, G.K.; Anjana, R.M.; Ali, M.K.; Narayan, K.M.V.; Mohan, V. Relationship of adipokines and proinflammatory cytokines among asian indians with obesity and youth onset type 2 diabetes. Endocr. Pract. 2015, 21, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Lakshmipriya, N.; Gayathri, R.; Shanmugam, S.; Srinivasan, R.; Krishnaswamy, K.; Jeevan, R.; Unnikrishnan, R.; Anjana, R.; Sudha, V.; Mohan, V. Dietary fatty-acid profile of south Indian adults and its association with type 2 diabetes––CURES 151. J. Diabetol. 2020, 11, 13–24. [Google Scholar]
- World Health Organisation. Healthy Diet. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/healthy-diet (accessed on 6 January 2022).
- Misra, A.; Sharma, R.; Gulati, S.; Joshi, S.R.; Sharma, V.; Ghafoorunissa Ibrahim, A.; Joshi, S.; Laxmaiah, A.; Kurpad, A.; Raj, R.K.; et al. Consensus dietary guidelines for healthy living and prevention of obesity, the metabolic syndrome, diabetes, and related disorders in Asian Indians. Diabetes Technol. 2011, 13, 683–694. [Google Scholar] [CrossRef]
- Moleres, A.; Milagro, F.I.; Marcos, A.; González Zorzano, E.; Campoy, C.; Garagorri, J.M.; Azcona-Sanjulian, M.C.; Martínez, J.A.; Marti, A. Common variants in genes related to lipid and energy metabolism are associated with weight loss after an intervention in overweight/obese adolescents. Nutr. Hosp. 2014, 30, 75–83. [Google Scholar]
- Coram, M.A.; Duan, Q.; Hoffmann, T.J.; Thornton, T.; Knowles, J.W.; Johnson, N.A.; Ochs-Balcom, H.M.; Donlon, T.A.; Martin, L.W.; Eaton, C.B.; et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 2013, 92, 904–916. [Google Scholar] [CrossRef]
- Piers, L.S.; Walker, K.Z.; Stoney, R.M.; Soares, M.J.; O’Dea, K. The influence of the type of dietary fat on postprandial fat oxidation rates: Monounsaturated (olive oil) vs. saturated fat (cream). Int. J. Obes. Relat. Metab. Disord 2002, 26, 814–821. [Google Scholar] [CrossRef]
- Jones, P.J.; Jew, S.; AbuMweis, S. The effect of dietary oleic, linoleic, and linolenic acids on fat oxidation and energy expenditure in healthy men. Metabolism 2008, 57, 1198–1203. [Google Scholar] [CrossRef]
- Vargas-Alarcón, G.; Pérez-Méndez, O.; Posadas-Sánchez, R.; Peña-Duque, M.A.; Martínez-Ríos, M.A.; Delgadillo-Rodriguez, H.; Fragoso, J.M. The rs4783961 and rs708272 genetic variants of the CETP gene are associated with coronary artery disease, but not with restenosis after coronary stenting. Arch. Cardiol Mex. 2021. [Google Scholar]
- Frisdal, E.; Klerkx, A.H.; Le Goff, W.; Tanck, M.W.; Lagarde, J.P.; Jukema, J.W.; Kastelein, J.J.; Chapman, M.J.; Guerin, M. Functional interaction between -629C/A, -971G/A and -1337C/T polymorphisms in the CETP gene is a major determinant of promoter activity and plasma CETP concentration in the REGRESS Study. Hum. Mol. Genet. 2005, 14, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Chen, S.Y.; Liu, S.P.; Lin, J.M.; Lin, H.J.; Lei, Y.J.; Chung, Y.C.; Chen, Y.C.; Wang, Y.H.; Liao, W.L.; et al. Cholesteryl Ester Transfer Protein Genetic Variants Associated with Risk for Type 2 Diabetes and Diabetic Kidney Disease in Taiwanese Population. Genes 2019, 10, 782. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K.; Lettre, G.; Young, T.; Farlow, D.N.; Pirruccello, J.P.; Ejebe, K.G.; Keating, B.J.; Yang, Q.; Chen, M.H.; Lapchyk, N.; et al. Candidate gene association resource (CARe): Design, methods, and proof of concept. Circ. Cardiovasc. Genet. 2010, 3, 267–275. [Google Scholar] [CrossRef] [PubMed]
- de Luis, D.; Izaola, O.; Primo, D.; Gomez, E.; Lopez, J.J.; Ortola, A.; Aller, R. Association of a cholesteryl ester transfer protein variant (rs1800777) with fat mass, HDL cholesterol levels, and metabolic syndrome. Endocrinol. Diabetes Nutr. (Engl. Ed.) 2018, 65, 387–393. [Google Scholar]
- Povel, C.M.; Boer, J.M.A.; Imholz, S.; Dollé, M.E.T.; Feskens, E.J.M. Genetic variants in lipid metabolism are independently associated with multiple features of the metabolic syndrome. Lipids Health Dis. 2011, 10, 118. [Google Scholar] [CrossRef]
- Ebenbichler, C.F.; Laimer, M.; Kaser, S.; Ritsch, A.; Sandhofer, A.; Weiss, H.; Aigner, F.; Patsch, J.R. Relationship Between Cholesteryl Ester Transfer Protein and Atherogenic Lipoprotein Profile in Morbidly Obese Women. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Stadler, J.T.; Lackner, S.; Mörkl, S.; Trakaki, A.; Scharnagl, H.; Borenich, A.; Wonisch, W.; Mangge, H.; Zelzer, S.; Meier-Allard, N.; et al. Obesity Affects HDL Metabolism, Composition and Subclass Distribution. Biomedicines 2021, 9, 242. [Google Scholar] [CrossRef]
- Park, K.-H.; Yadav, D.; Kim, S.-J.; Kim, J.-R.; Cho, K.-H. Slim Body Weight Is Highly Associated With Enhanced Lipoprotein Functionality, Higher HDL-C, and Large HDL Particle Size in Young Women. Front. Endocrinol. 2018, 9, 406. [Google Scholar] [CrossRef]
- Salerno, A.G.; Silva, T.R.; Amaral ME, C.; Alberici, L.C.; Bonfleur, M.L.; Patrício, P.R.; Francesconi EP, M.S.; Grassi-Kassisse, D.M.; Vercesi, A.E.; Boschero, A.C.; et al. Overexpression of apolipoprotein CIII increases and CETP reverses diet-induced obesity in transgenic mice. Int. J. Obes. 2007, 31, 1586–1595. [Google Scholar] [CrossRef]
- de Souza, R.J.; Mente, A.; Maroleanu, A.; Cozma, A.I.; Ha, V.; Kishibe, T.; Uleryk, E.; Budylowski, P.; Schünemann, H.; Beyene, J.; et al. Intake of saturated and trans unsaturated fatty acids and risk of all cause mortality, cardiovascular disease, and type 2 diabetes: Systematic review and meta-analysis of observational studies. BMJ Br. Med. J. 2015, 351, h3978. [Google Scholar] [CrossRef]
- Ramsden, C.E.; Zamora, D.; Majchrzak-Hong, S.; Faurot, K.R.; Broste, S.K.; Frantz, R.P.; Davis, J.M.; Ringel, A.; Suchindran, C.M.; Hibbeln, J.R. Re-evaluation of the traditional diet-heart hypothesis: Analysis of recovered data from Minnesota Coronary Experiment (1968-1973). BMJ 2016, 353, i1246. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; van der Schouw, Y.T.; Soedamah-Muthu, S.S.; Spijkerman AM, W.; Sluijs, I. Intake of dietary saturated fatty acids and risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition-Netherlands cohort: Associations by types, sources of fatty acids and substitution by macronutrients. Eur. J. Nutr. 2019, 58, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Forouhi, N.G.; Koulman, A.; Sharp, S.J.; Imamura, F.; Kröger, J.; Schulze, M.B.; Crowe, F.L.; Huerta, J.M.; Guevara, M.; Beulens, J.W.; et al. Differences in the prospective association between individual plasma phospholipid saturated fatty acids and incident type 2 diabetes: The EPIC-InterAct case-cohort study. Lancet Diabetes Endocrinol. 2014, 2, 810–818. [Google Scholar] [CrossRef]
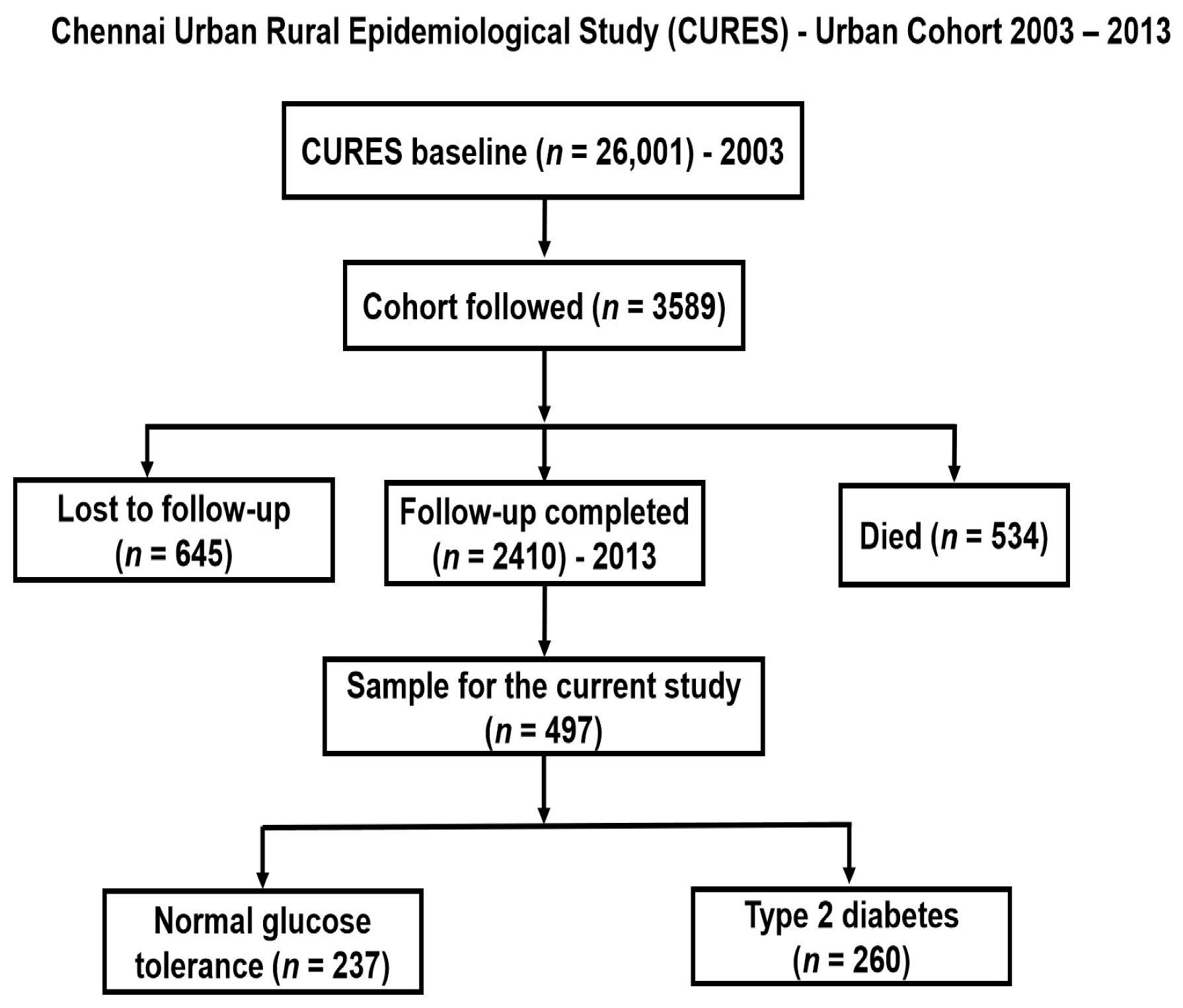
| All Participants (n = 497) | GRS < 2 (n = 239) | GRS ≥ 2 (n = 258) | p Value * | |
|---|---|---|---|---|
| Age (years) | 44 ± 10 | 45 ± 10 | 44 ± 9 | 0.34 |
| Sex [Men (%), Women (%)] | 225 (45), 272 (55) | 106 (47), 133 (49) | 119 (53), 139 (51) | 0.69 |
| BMI (kg/m2) | 24.6 ± 4.5 | 24.7 ± 4.7 | 24.4 ± 4.3 | 0.41 |
| WC (cm) | 87 ± 11 | 88 ± 12 | 87 ± 11 | 0.39 |
| WHR | 0.92 ± 0.08 | 0.92 ± 0.09 | 0.91 ± 0.08 | 0.57 |
| Obese cases (%) | 209 (42) | 109 (52) | 100 (48) | 0.12 |
| HDL (mg/dL) | 42 ± 10 | 42 ± 10 | 42 ± 10 | 0.79 |
| LDL (mg/dL) | 119 ± 32 | 118 ± 32 | 119 ± 32 | 0.81 |
| TG (mg/dL) | 165 ± 150 | 166 ± 120 | 164 ± 173 | 0.87 |
| Total cholesterol (mg/dL) | 191 ± 40 | 192 ± 42 | 190 ± 38 | 0.64 |
| Systolic BP (mmHg) | 122 ± 20 | 123 ± 22 | 120 ± 18 | 0.15 |
| Diastolic BP (mmHg) | 76 ± 11 | 76 ± 12 | 75 ± 11 | 0.60 |
| Fasting plasma glucose (mg/dL) | 126 ± 65 | 126 ± 64 | 127 ± 67 | 0.79 |
| Fasting serum insulin (μIU/mL) | 9 ± 6 | 9 ± 6 | 9 ± 7 | 0.89 |
| Insulin resistance | 3 ± 2 | 3 ± 2 | 2 ± 2 | 0.44 |
| HbA1c (%) | 7 ± 2 | 7 ± 2 | 7 ± 2 | 0.91 |
| Fat (g) | 67 ± 27 | 67 ± 26 | 67 ± 27 | 0.83 |
| Carbohydrate (g) | 410 ± 136 | 410 ± 134 | 411 ± 138 | 0.92 |
| Protein (g) | 72 ± 24 | 73 ± 24 | 72 ± 23 | 0.63 |
| Dietary fibre (g) | 32 ± 12 | 32 ± 12 | 32 ± 11 | 0.77 |
| Energy (kcal/day) | 2560 ± 822 | 2560 ± 809 | 2559 ± 834 | 0.99 |
| Total SFA (g) | 25 ± 11 | 25 ± 11 | 25 ± 11 | 0.91 |
| Total MUFA (g) | 20 ± 8 | 20 ± 8 | 21 ± 9 | 0.79 |
| Total PUFA (g) | 19 ± 9 | 18 ± 9 | 19 ± 10 | 0.77 |
| Plant protein (g/day) | 41 ± 14 | 40 ± 13 | 42 ± 14 | 0.23 |
| Animal protein (g/day) | 23 ± 13 | 23 ± 12 | 22 ± 13 | 0.75 |
| Smokers (%) | 88 (18) | 33 (38) | 55 (63) | 0.03 |
| Alcohol drinkers (%) | 123 (25) | 52 (42) | 71 (58) | 0.14 |
| T2D cases (%) | 260 (52) | 131 (50.4) | 129 (49.6) | 0.28 |
| Trait | GRS ∗ Fat (g) | GRS ∗ Carbohydrate (g) | GRS ∗ Protein (g) | GRS ∗ Dietary Fibre (g) |
|---|---|---|---|---|
| Beta Coefficient ± SE (Pinteraction) | Beta Coefficient ± SE (Pinteraction) | Beta Coefficient ± SE (Pinteraction) | Beta Coefficient ± SE (Pinteraction) | |
| BMI (kg/m2) | 0.05 ± 0.04 (0.21) a | 0.04 ± 0.05 (0.36) a | 0.04 ± 0.05 (0.35) a | −0.01 ± 0.04 (0.77) a |
| WC (cm) | 0.06 ± 0.03 (0.03) a | 0.05 ± 0.03 (0.18) a | 0.07 ± 0.04 (0.07) a | 0.00 ± 0.03 (0.93) a |
| Waist hip ratio | 0.01 ± 0.02 (0.52) b | 0.00 ± 0.02 (0.98) b | 0.01 ± 0.02 (0.58) b | −0.01 ± 0.02 (0.62) b |
| Common obesity | −1.76 ± 1.14 (0.12) a | 0.10 ± 0.08 (0.20) a | −2.52 ± 1.41 (0.08) a | −0.35 ± 1.26 (0.78) a |
| HDL (mg/dL) | −0.04 ± 0.05 (0.42) b | −0.07 ± 0.06 (0.23) b | −0.07 ± 0.06 (0.21) b | −0.04 ± 0.05 (0.47) b |
| LDL (mg/dL) | 0.02 ± 0.06 (0.82) b | 0.02 ± 0.08 (0.79) b | −0.01 ± 0.08 (0.90) b | −0.02 ± 0.07 (0.81) b |
| TG (mg/dL) | 0.10 ± 0.12 (0.39) b | −0.01 ± 0.15 (0.97) b | −0.02 ± 0.15 (0.89) b | 0.08 ± 0.13 (0.57) b |
| Total cholesterol (mg/dL) | 0.02 ± 0.04 (0.70) b | −0.00 ± 0.06 (0.98) b | −0.02 ± 0.06 (0.65) b | −0.00 ± 0.05 (0.98) b |
| Systolic BP (mmHg) | 0.03 ± 0.03 (0.35) b | 0.03 ± 0.04 (0.49) b | 0.03 ± 0.04 (0.48) b | 0.04 ± 0.03 (0.25) b |
| Diastolic BP (mmHg) | 0.02 ± 0.03 (0.50) b | 0.01 ± 0.04 (0.87) b | 0.03 ± 0.04 (0.51) b | 0.01 ± 0.04 (0.72) b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wuni, R.; Adela Nathania, E.; Ayyappa, A.K.; Lakshmipriya, N.; Ramya, K.; Gayathri, R.; Geetha, G.; Anjana, R.M.; Kuhnle, G.G.C.; Radha, V.; et al. Impact of Lipid Genetic Risk Score and Saturated Fatty Acid Intake on Central Obesity in an Asian Indian Population. Nutrients 2022, 14, 2713. https://doi.org/10.3390/nu14132713
Wuni R, Adela Nathania E, Ayyappa AK, Lakshmipriya N, Ramya K, Gayathri R, Geetha G, Anjana RM, Kuhnle GGC, Radha V, et al. Impact of Lipid Genetic Risk Score and Saturated Fatty Acid Intake on Central Obesity in an Asian Indian Population. Nutrients. 2022; 14(13):2713. https://doi.org/10.3390/nu14132713
Chicago/Turabian StyleWuni, Ramatu, Evelyn Adela Nathania, Ashok K. Ayyappa, Nagarajan Lakshmipriya, Kandaswamy Ramya, Rajagopal Gayathri, Gunasekaran Geetha, Ranjit Mohan Anjana, Gunter G. C. Kuhnle, Venkatesan Radha, and et al. 2022. "Impact of Lipid Genetic Risk Score and Saturated Fatty Acid Intake on Central Obesity in an Asian Indian Population" Nutrients 14, no. 13: 2713. https://doi.org/10.3390/nu14132713
APA StyleWuni, R., Adela Nathania, E., Ayyappa, A. K., Lakshmipriya, N., Ramya, K., Gayathri, R., Geetha, G., Anjana, R. M., Kuhnle, G. G. C., Radha, V., Mohan, V., Sudha, V., & Vimaleswaran, K. S. (2022). Impact of Lipid Genetic Risk Score and Saturated Fatty Acid Intake on Central Obesity in an Asian Indian Population. Nutrients, 14(13), 2713. https://doi.org/10.3390/nu14132713








