Protective Effect of Mitophagy Regulated by mTOR Signaling Pathway in Liver Fibrosis Associated with Selenium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Grouping and Intervention
2.2. Collection of Serum and Liver Samples
2.3. Selenium Concentration Detection
2.4. HE and Masson Stainings and TEM Observation
2.5. Metabolomics Analysis
2.5.1. Sample Preparation for Metabolomics Analysis
2.5.2. LC-MS/MS Metabolomics Analysis and Data Processing
2.5.3. Inter-Group Variation Analysis and Differential Metabolite Screening
2.5.4. Bioinformatics Analysis of Differential Metabolites
2.6. IHC Staining
2.7. Statistical Analysis
3. Results
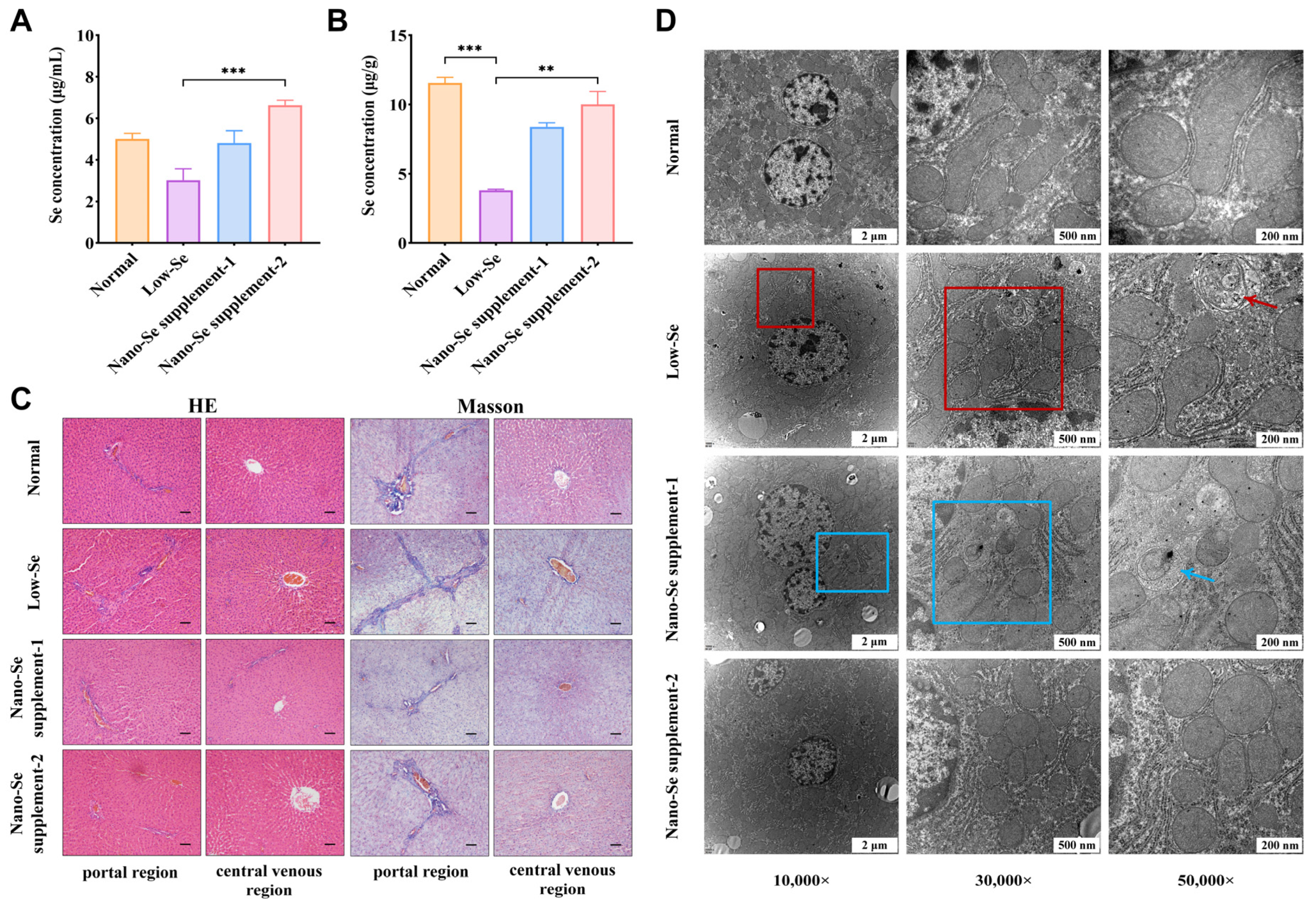
3.1. Selenium Concentrations of Serum and Liver Samples
3.2. HE and Masson Staining Results
3.3. Morphological Changes in the Ultrastructure of Liver
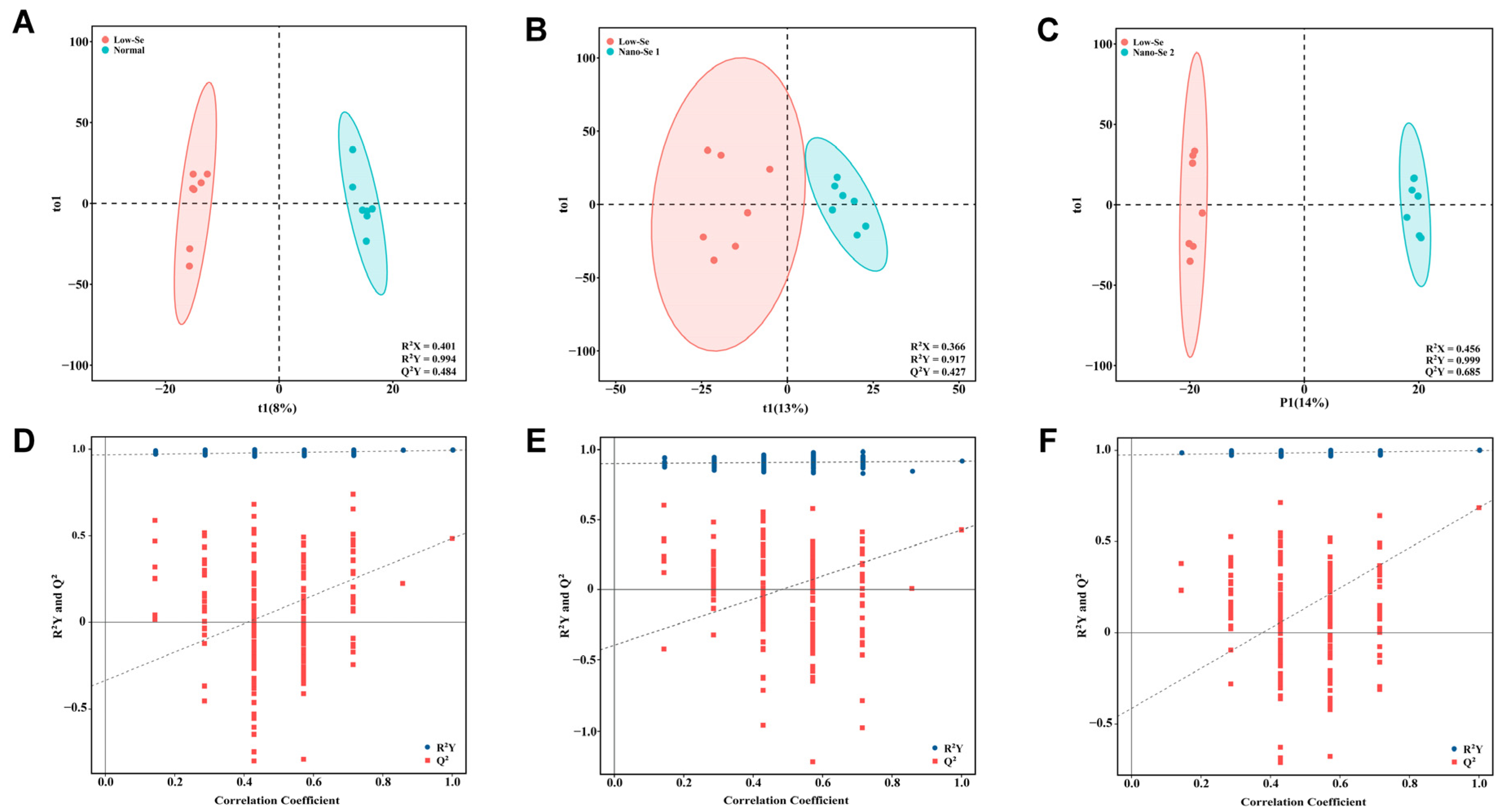
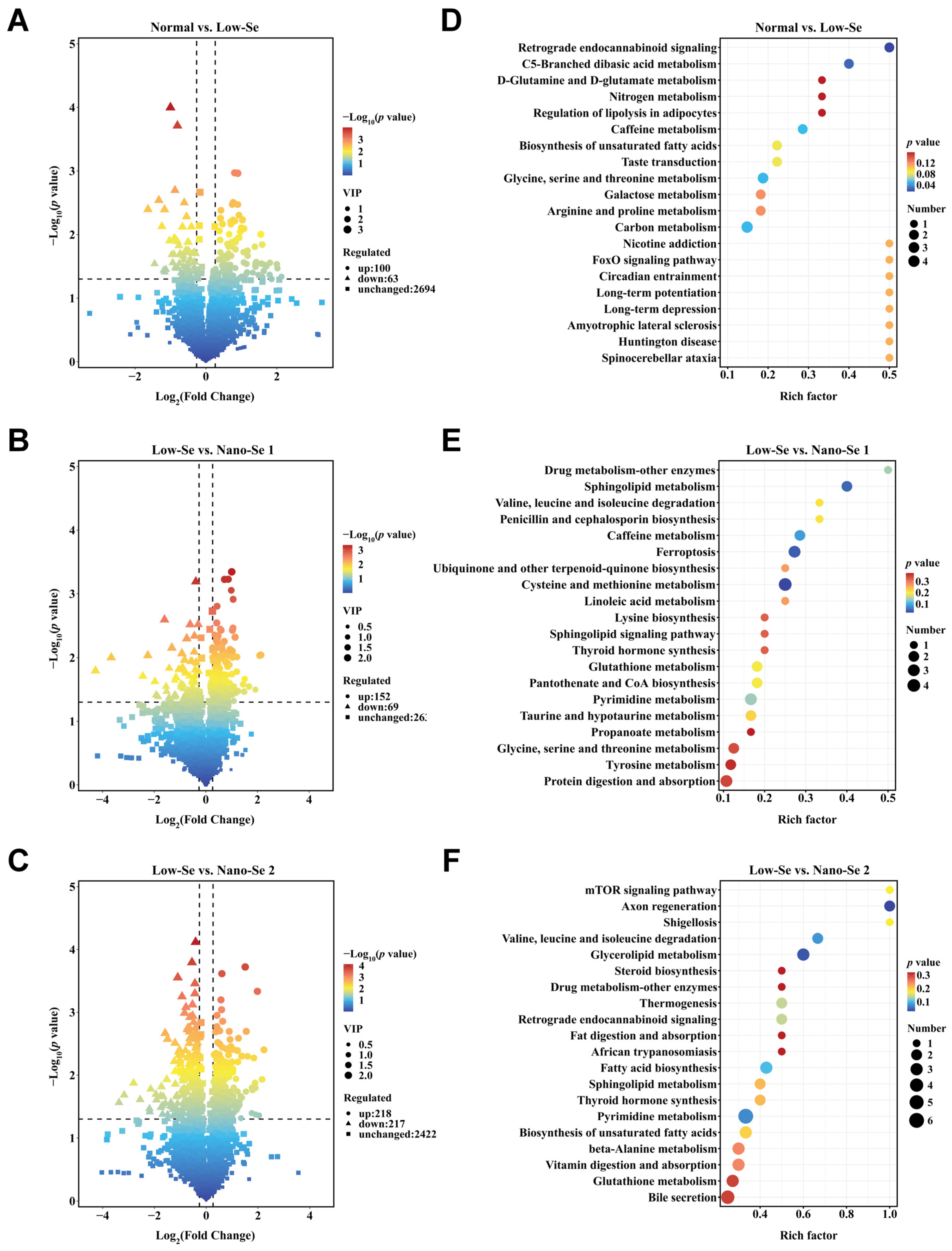
3.4. Metabolomics Analysis of Liver Tissue
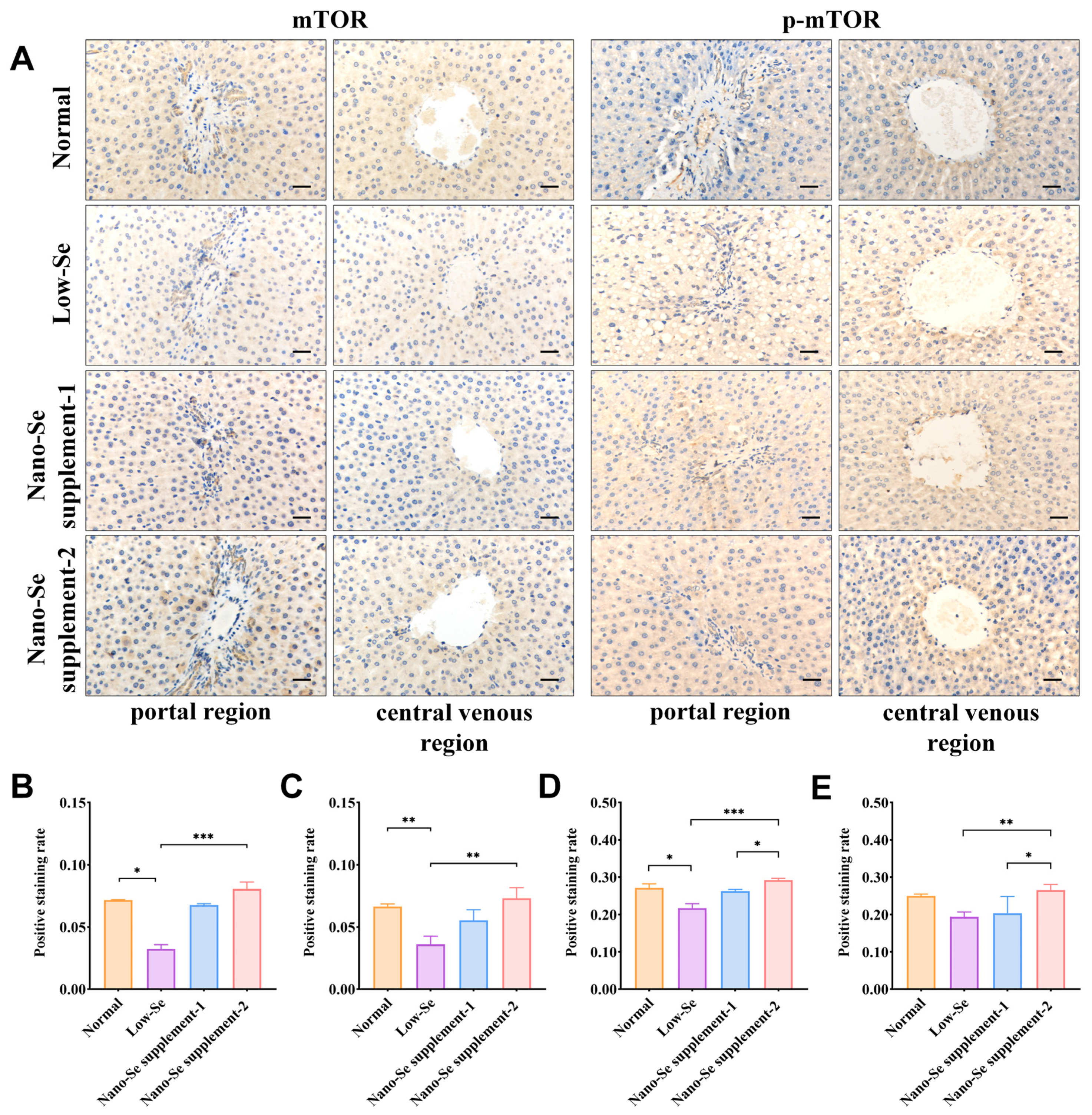
3.5. IHC Staining of mTOR, p-mTOR, ULK1, and p-ULK1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schomburg, L.; Hughes, D.J. The missing link? The potential role of selenium in the development of liver cancer and significance for the general population. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 707–709. [Google Scholar] [CrossRef] [PubMed]
- Motsenbocker, M.A.; Tappel, A.L. Effect of dietary selenium on plasma selenoprotein P, selenoprotein P1 and glutathione peroxidase in the rat. J. Nutr. 1984, 114, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.F.; Blotcky, A.J.; Jetton, M.M.; Hahn, H.K.; Burch, R.E. Serum levels of selenium, calcium, copper magnesium, manganese and zinc in various human diseases. J. Nutr. 1979, 109, 1432–1437. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Liang, H.; Yi, J.; Tan, W.; He, S.; Wang, S.; Li, F.; Wu, X.; Ma, J.; Shi, X.; et al. Long-term selenium-deficient diet induces liver damage by altering hepatocyte ultrastructure and MMP1/3 and TIMP1/3 expression in growing rats. Biol. Trace Elem. Res. 2017, 175, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Song, G.; Minuk, G.Y. Effects of hepatic stimulator substance, herbal medicine, selenium/vitamin E, and ciprofloxacin on cirrhosis in the rat. Gastroenterology 1996, 110, 1150–1155. [Google Scholar] [CrossRef]
- Drew, L. Liver cirrhosis: Scar wars. Nature 2018, 564, S74–S75. [Google Scholar] [CrossRef]
- Aydın, M.M.; Akçalı, K.C. Liver fibrosis. Turk. J. Gastroenterol. 2018, 29, 14–21. [Google Scholar] [CrossRef]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Toosi, A.E.K. Liver fibrosis: Causes and methods of assessment, a review. Rom. J. Intern. Med. 2015, 53, 304–314. [Google Scholar]
- Pickles, S.; Vigié, P.; Youle, R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Du, W.; Li, Y.; Shi, C.; Hu, N.; Ma, S.; Wang, W.; Ren, J. Effects of melatonin on fatty liver disease: The role of NR4A1/DNA-PKcs/p53 pathway, mitochondrial fission, and mitophagy. J. Pineal. Res. 2018, 64, 12450–12469. [Google Scholar] [CrossRef]
- Um, J.H.; Yun, J. Emerging role of mitophagy in human diseases and physiology. BMB Rep. 2017, 50, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Turkseven, S.; Bolognesi, M.; Brocca, A.; Pesce, P.; Angeli, P.; Di Pascoli, M. Mitochondria-targeted antioxidant mitoquinone attenuates liver inflammation and fibrosis in cirrhotic rats. Am. J. Physiol. Gastrointest Liver Physiol. 2020, 318, G298–G304. [Google Scholar] [CrossRef]
- Tian, Z.; Chen, Y.; Yao, N.; Hu, C.; Wu, Y.; Guo, D.; Liu, J.; Yang, Y.; Chen, T.; Zhao, Y.; et al. Role of mitophagy regulation by ROS in hepatic stellate cells during acute liver failure. Am. J. Physiol. Gastrointest Liver Physiol. 2018, 315, G374–G384. [Google Scholar] [CrossRef]
- Chao, X.; Ding, W.X. Role and mechanisms of autophagy in alcohol-induced liver injury. Adv. Pharmacol. 2019, 85, 109–131. [Google Scholar]
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, G.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010, 465, 942–946. [Google Scholar] [CrossRef]
- Ganley, I.G.; Lam, D.H.; Wang, J.R.; Ding, X.J.; Chen, S.; Jiang, X.J. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [Green Version]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Guo, X. A Chondroitin Sulfate A Nano-Se Micelle Particle and Its Preparation Method and Application. China, CN107260668B, 28 June 2020. [Google Scholar]
- Ishak, K.; Baptista, A.; Bianchi, L.; Callea, F.; De Groote, J.; Gudat, F.; Denk, H.; Desmet, V.; Korb, G.; MacSween, R.N. Histological grading and staging of chronic hepatitis. J. Hepatol. 1995, 22, 696–699. [Google Scholar] [CrossRef]
- Burk, R.F.; Hill, K.E. Regulation of Se metabolism and transport. Annu. Rev. Nutr. 2015, 7, 109–134. [Google Scholar] [CrossRef]
- Zhou, J.C.; Zheng, S.; Mo, J.; Liang, X.; Xu, Y.; Zhang, H.; Gong, C.; Liu, X.L.; Lei, X.G. Dietary selenium deficiency or excess reduces sperm quality and testicular mRNA abundance of nuclear glutathione peroxidase 4 in rats. J. Nutr. 2017, 147, 1947–1953. [Google Scholar] [CrossRef]
- Gao, X.J.; Tang, B.; Liang, H.H.; Yi, L.; Wei, Z.G. Selenium deficiency induced an inflammatory response by the HSP60-TLR2-MAPKs signalling pathway in the liver of carp. Fish Shellfish Immunol. 2019, 4, 688–694. [Google Scholar] [CrossRef]
- Yao, L.L.; Du, Q.; Yao, H.D.; Chen, X.; Zhang, Z.W.; Xu, S.W. Roles of oxidative stress and endoplasmic reticulum stress in selenium deficiency-induced apoptosis in chicken liver. Biometals 2015, 28, 255–265. [Google Scholar] [CrossRef]
- Hu, X.; Chandler, J.D.; Orr, M.L.; Hao, L.; Liu, K.; Uppal, K.; Go, Y.M.; Jones, D.P. Selenium supplement alters hepatic energy and fatty acid metabolism in mice. J. Nutr. 2018, 148, 675–684. [Google Scholar] [CrossRef]
- Hu, X.; Chandler, J.D.; Fernandes, J.; Orr, M.L.; Hao, L.; Uppal, K.; Neujahr, D.C.; Jones, D.P.; Go, Y.M. Selenium supplement prevents metabolic and transcriptomic responses to cadmium in mouse lung. Biochim. Biophys. Acta. Gen. Subj. 2018, 11, 2417–2426. [Google Scholar] [CrossRef]
- Zhao, Z.P.; Barcus, M.; Kim, J.; Lum, K.L.; Mills, C.; Lei, X.G. High dietary selenium intake alters lipid metabolism and protein synthesis in liver and muscle of pigs. J. Nutr. 2016, 146, 1625–1633. [Google Scholar] [CrossRef]
- Liu, L.; Geng, X.; Cai, Y.; Copple, B.; Yoshinaga, M.; Shen, J.; Nebert, D.W.; Wang, H.; Liu, Z. Hepatic ZIP8 deficiency is associated with disrupted selenium homeostasis, liver pathology, and tumor formation. Am. J. Physiol. Gastrointest Liver Physiol. 2018, 315, G569–G579. [Google Scholar] [CrossRef]
- Kolachi, N.F.; Kazi, T.G.; Afridi, H.I.; Kazi, N.G.; Khan, S. Investigation of essential trace and toxic elements in biological samples (blood, serum and scalp hair) of liver cirrhotic/cancer female patients before and after mineral supplement. Clin. Nutr. 2012, 31, 967–973. [Google Scholar] [CrossRef]
- Burk, R.F.; Hill, K.E.; Motley, A.K.; Byrne, D.W.; Norsworthy, B.K. Selenium deficiency occurs in some patients with moderate-to-severe cirrhosis and can be corrected by administration of selenate but not selenomethionine: A randomized controlled trial. Am. J. Clin. Nutr. 2015, 102, 1126–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prystupa, A.; Kiciński, P.; Luchowska-Kocot, D.; Błażewicz, A.; Niedziałek, J.; Mizerski, G.; Jojczuk, M.; Ochal, A.; Sak, J.J.; Załuska, W. Association between serum selenium concentrations and levels of proinflammatory and profibrotic cytokines-interleukin-6 and growth differentiation factor-15, in patients with alcoholic liver cirrhosis. Int. J. Environ. Res. Public Health 2017, 14, 437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Guo, Y.F.; Qiu, C.W.; Deng, G.Z.; Guo, M.Y. Protective Action of Se-Supplement Against Acute Alcoholism Is Regulated by Selenoprotein P (SelP) in the Liver. Biol. Trace. Elem. Res. 2017, 175, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Hesketh, J.; Huang, D.; Gan, F.; Hao, S.; Tang, S.; Guo, Y.; Huang, K. Protective effects of selenium-glutathione-enriched probiotics on CCl4-induced liver fibrosis. J. Nutr. Biochem. 2018, 58, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Mickiewicz, B.; Villemaire, M.L.; Sandercock, L.E.; Jirik, F.R.; Vogel, H.J. Metabolic changes associated with selenium deficiency in mice. Biometals 2014, 27, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Berntssen, M.; Sundal, T.K.; Olsvik, P.A.; Amlund, H.; Rasinger, J.D.; Sele, V.; Hamre, K.; Hillestad, M.; Buttle, L.; Ørnsrud, R. Sensitivity and toxic mode of action of dietary organic and inorganic selenium in Atlantic salmon (Salmo salar). Aquat. Toxicol. 2017, 11, 116–126. [Google Scholar] [CrossRef]
- Fernandes, J.; Hu, X.; Ryan Smith, M.; Go, Y.M.; Jones, D.P. Selenium at the redox interface of the genome, metabolome and exposome. Free Radic. Biol. Med. 2018, 11, 215–227. [Google Scholar] [CrossRef]
- Alehagen, U.; Johansson, P.; Aaseth, J.; Alexander, J.; Surowiec, I.; Lundstedt-Enkel, K.; Lundstedt, T. Significant changes in metabolic profiles after intervention with selenium and coenzyme Q10 in an elderly population. Biomolecules 2019, 9, 553. [Google Scholar] [CrossRef] [Green Version]
- Ahangar, N.; Naderi, M.; Noroozi, A.; Ghasemi, M.; Zamani, E.; Shaki, F. Zinc deficiency and oxidative stress involved in valproic acid induced hepatotoxicity: Protection by zinc and selenium supplementation. Biol. Trace Elem. Res. 2017, 179, 102–109. [Google Scholar] [CrossRef]
- Zhai, Q.; Xiao, Y.; Li, P.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Varied doses and chemical forms of selenium supplementation differentially affect mouse intestinal physiology. Food Funct. 2019, 10, 5398–5412. [Google Scholar] [CrossRef]
- Cuello, S.; Ramos, S.; Mateos, R.; Martín, M.A.; Madrid, Y.; Cámara, C.; Bravo, L.; Goya, L. Selenium methylselenocysteine protects human hepatoma HepG2 cells against oxidative stress induced by tert-butyl hydroperoxide. Anal. Bioanal. Chem. 2007, 389, 2167–2178. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.Z.; Zhang, T.; Lin, H.J.; Chang, Y.; Xing, J. Role of hydrogen sulfide on autophagy in liver injuries induced by selenium deficiency in chickens. Biol. Trace Elem. Res. 2017, 175, 194–203. [Google Scholar]
- Liu, R.H.; Jia, T.T.; Cui, Y.; Lin, H.J.; Li, S. The protective effect of selenium on the chicken pancreas against cadmium toxicity via alleviating oxidative stress and autophagy. Biol. Trace Elem. Res. 2018, 184, 240–246. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, J.; Ge, J.; Wang, L.L.; Li, N.; Sun, X.T.; Cao, H.B.; Li, J.L. Selenium triggers Nrf2-mediated protection against cadmium-induced chicken hepatocyte autophagy and apoptosis. Toxicol. Vitr. 2017, 10, 349–356. [Google Scholar] [CrossRef]
- Song, G.L.; Chen, C.; Wu, Q.Y.; Zhang, Z.H.; Zheng, R.; Chen, Y.; Jia, S.Z.; Ni, J.Z. Selenium-enriched yeast inhibited β-amyloid production and modulated autophagy in a triple transgenic mouse model of Alzheimer’s disease. Metallomics 2018, 10, 1107–1115. [Google Scholar] [CrossRef]
- Gao, J.; Nie, W.; Wang, F.L.; Guo, Y.M. Maternal selenium supplementation enhanced skeletal muscle development through increasing protein synthesis and SelW mRNA levels of their offspring. Biol. Trace Elem. Res. 2018, 186, 238–248. [Google Scholar] [CrossRef]
- Wang, L.; Yuan, D.; Zheng, J.; Wu, X.; Wang, J.; Liu, X.; He, Y.; Zhang, C.; Liu, C.; Wang, T.; et al. Chikusetsu saponin IVa attenuates isoprenaline-induced myocardial fibrosis in mice through activation autophagy mediated by AMPK/mTOR/ULK1 signaling. Phytomedicine 2019, 5, 152764–152771. [Google Scholar] [CrossRef]
- Hossain, K.F.B.; Rahman, M.M.; Sikder, M.T.; Hosokawa, T.; Saito, T.; Kurasaki, M. Selenium modulates inorganic mercury induced cytotoxicity and intrinsic apoptosis in PC12 cells. Ecotoxicol. Environ. Saf. 2021, 207, 111262. [Google Scholar] [CrossRef]
- Li, L.; Hai, J.; Li, Z.; Zhang, Y.; Peng, H.; Li, K.; Weng, X. Resveratrol modulates autophagy and NF-κB activity in a murine model for treating non-alcoholic fatty liver disease. Food Chem. Toxicol. 2014, 6, 166–173. [Google Scholar] [CrossRef]

| Group | Distance (μm) | Ishak Score |
|---|---|---|
| Normal | 235.68 (208.98, 272.95) ***, ### | 0 (0, 0) *** |
| Low-selenium | 158.64 (130.11, 182.84) | 2 (1, 4) |
| Nano-selenium supplement-1 | 187.27 (167.16, 213.64) *** | 1 (0, 2.75) |
| Nano-selenium supplement-2 | 224.09 (190.45, 266.36) ***, # | 1 (0, 1) *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, L.; Guo, Z.; Liu, H.; Liu, J.; Lin, X.; Deng, H.; Liu, X.; Zhao, Y.; Xiao, X.; Lei, J.; et al. Protective Effect of Mitophagy Regulated by mTOR Signaling Pathway in Liver Fibrosis Associated with Selenium. Nutrients 2022, 14, 2410. https://doi.org/10.3390/nu14122410
Qiao L, Guo Z, Liu H, Liu J, Lin X, Deng H, Liu X, Zhao Y, Xiao X, Lei J, et al. Protective Effect of Mitophagy Regulated by mTOR Signaling Pathway in Liver Fibrosis Associated with Selenium. Nutrients. 2022; 14(12):2410. https://doi.org/10.3390/nu14122410
Chicago/Turabian StyleQiao, Lichun, Ziwei Guo, Haobiao Liu, Jiaxin Liu, Xue Lin, Huan Deng, Xuan Liu, Yan Zhao, Xiang Xiao, Jian Lei, and et al. 2022. "Protective Effect of Mitophagy Regulated by mTOR Signaling Pathway in Liver Fibrosis Associated with Selenium" Nutrients 14, no. 12: 2410. https://doi.org/10.3390/nu14122410
APA StyleQiao, L., Guo, Z., Liu, H., Liu, J., Lin, X., Deng, H., Liu, X., Zhao, Y., Xiao, X., Lei, J., & Han, J. (2022). Protective Effect of Mitophagy Regulated by mTOR Signaling Pathway in Liver Fibrosis Associated with Selenium. Nutrients, 14(12), 2410. https://doi.org/10.3390/nu14122410







