Associations between Maternal Diet, Body Composition and Gut Microbial Ecology in Pregnancy
Abstract
:1. Introduction
2. Materials and Methods
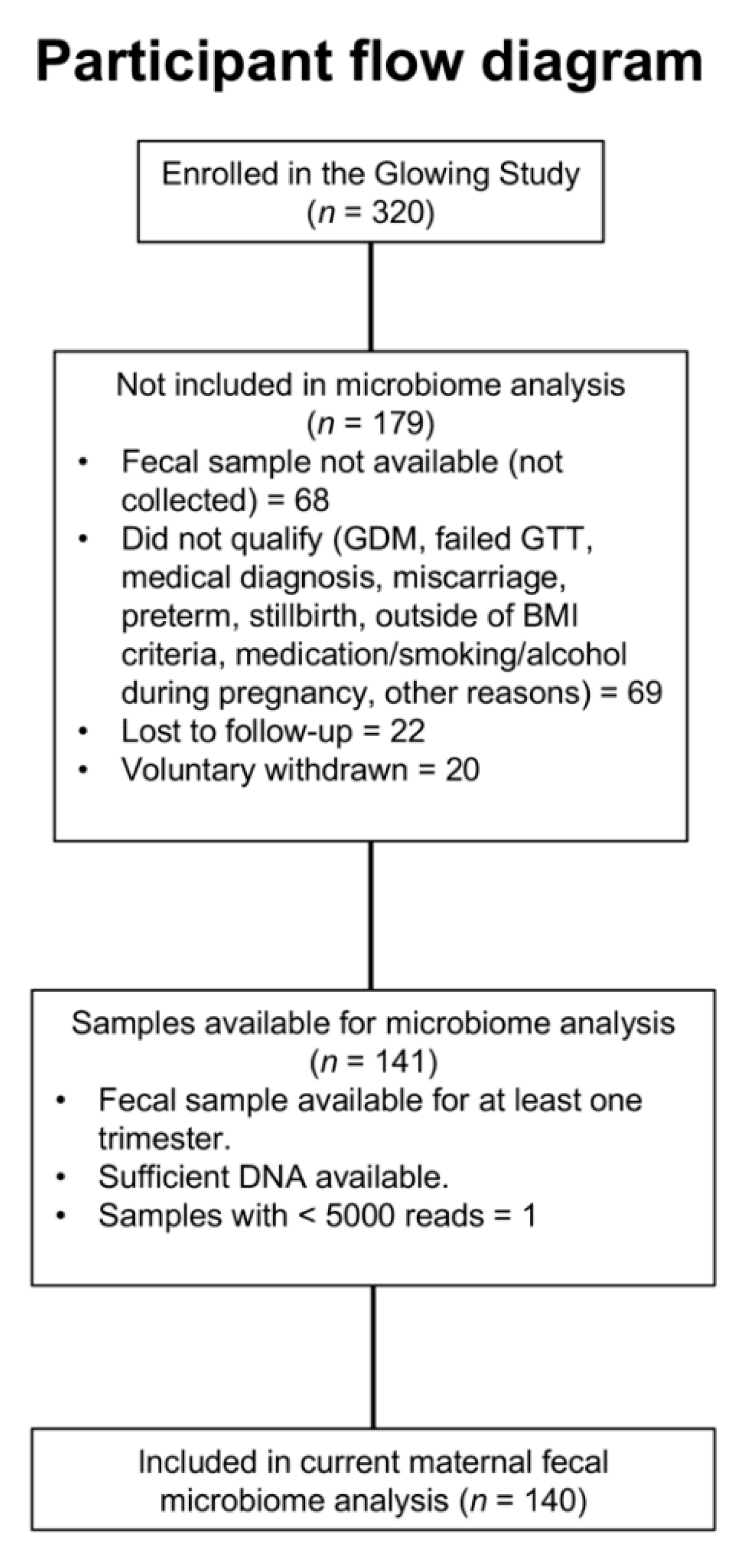
2.1. Study Design, Participants and Data Collection
2.2. Anthropometric and Body Composition Measurements
2.3. Dietary Analysis
2.4. Microbiome Analysis via 16S rRNA Amplicon Sequencing
2.5. Microbial Ecology Data Analysis
3. Results
3.1. Participant Characteristics
3.2. Alpha-Diversity across Pregnancy
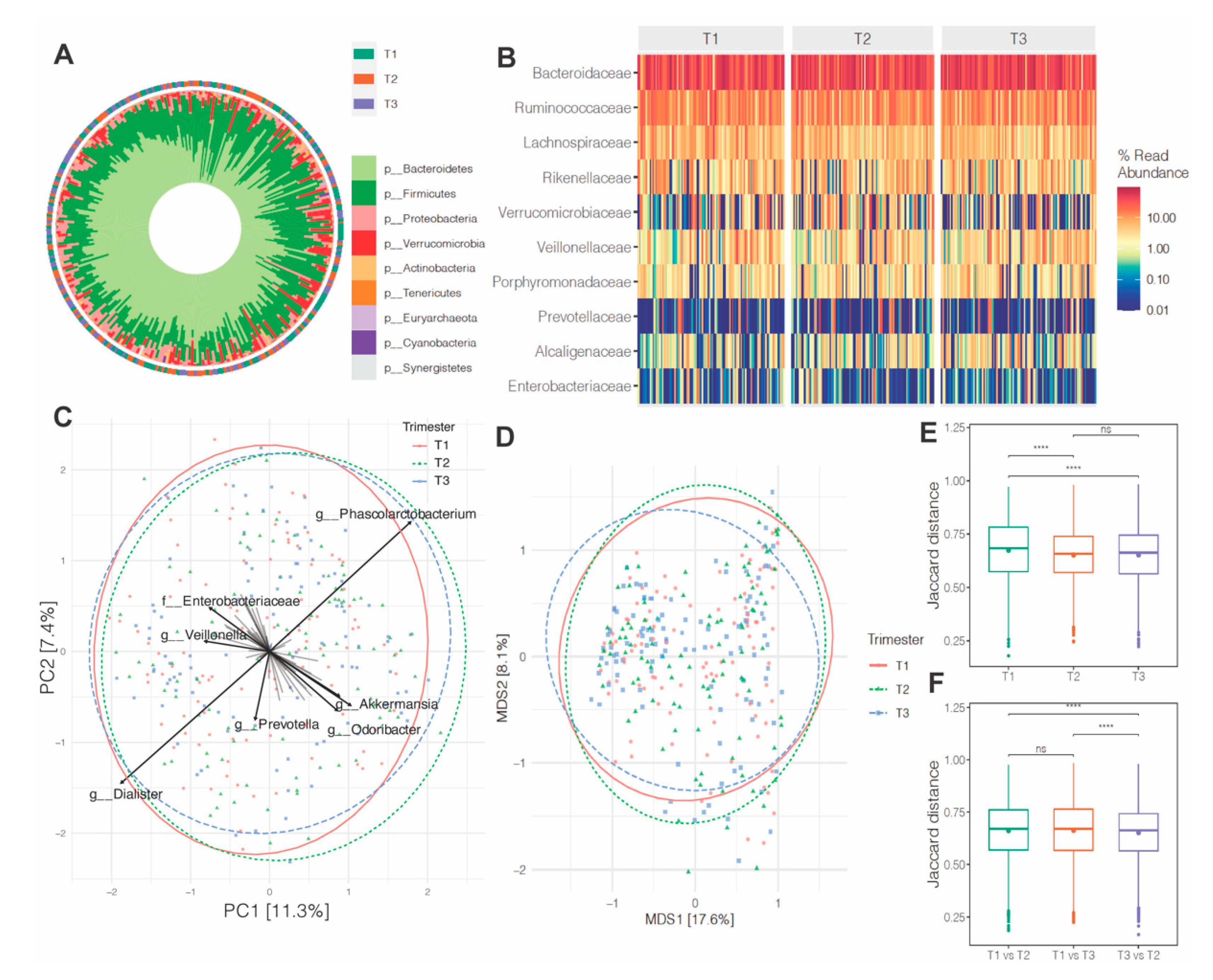
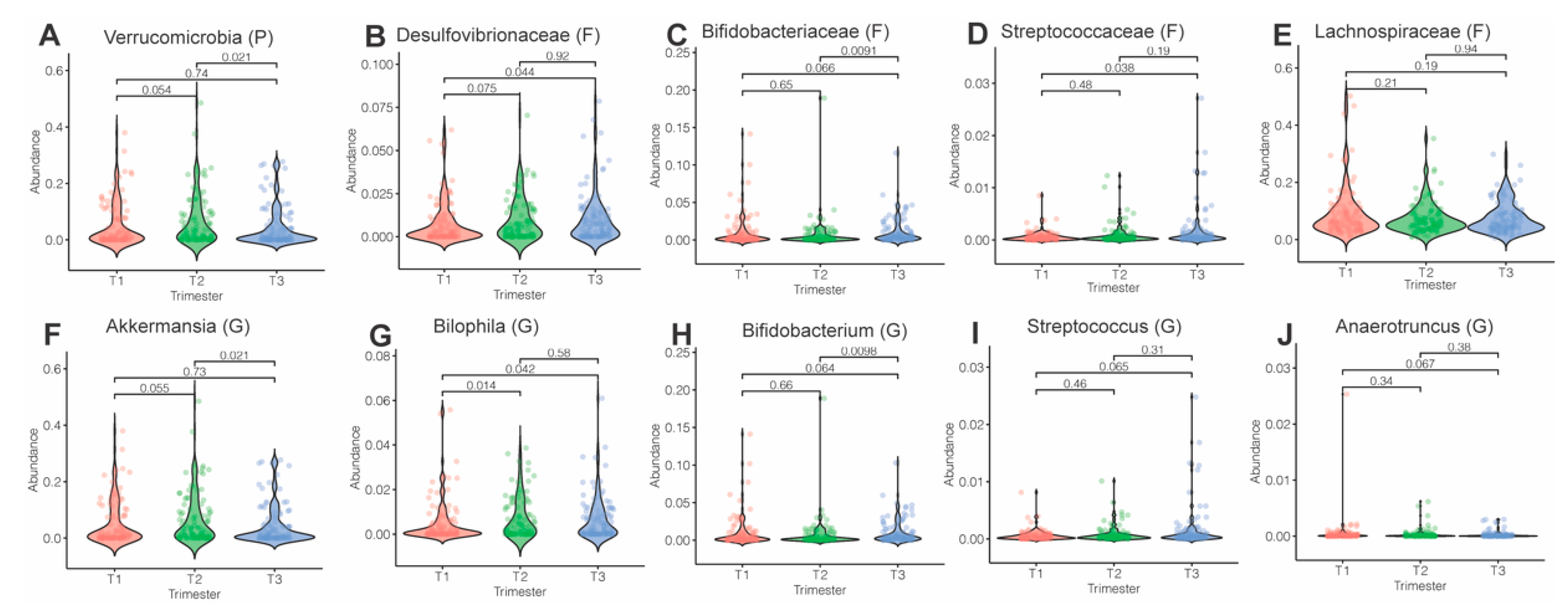
3.3. Microbiota Composition and Abundance across Pregnancy
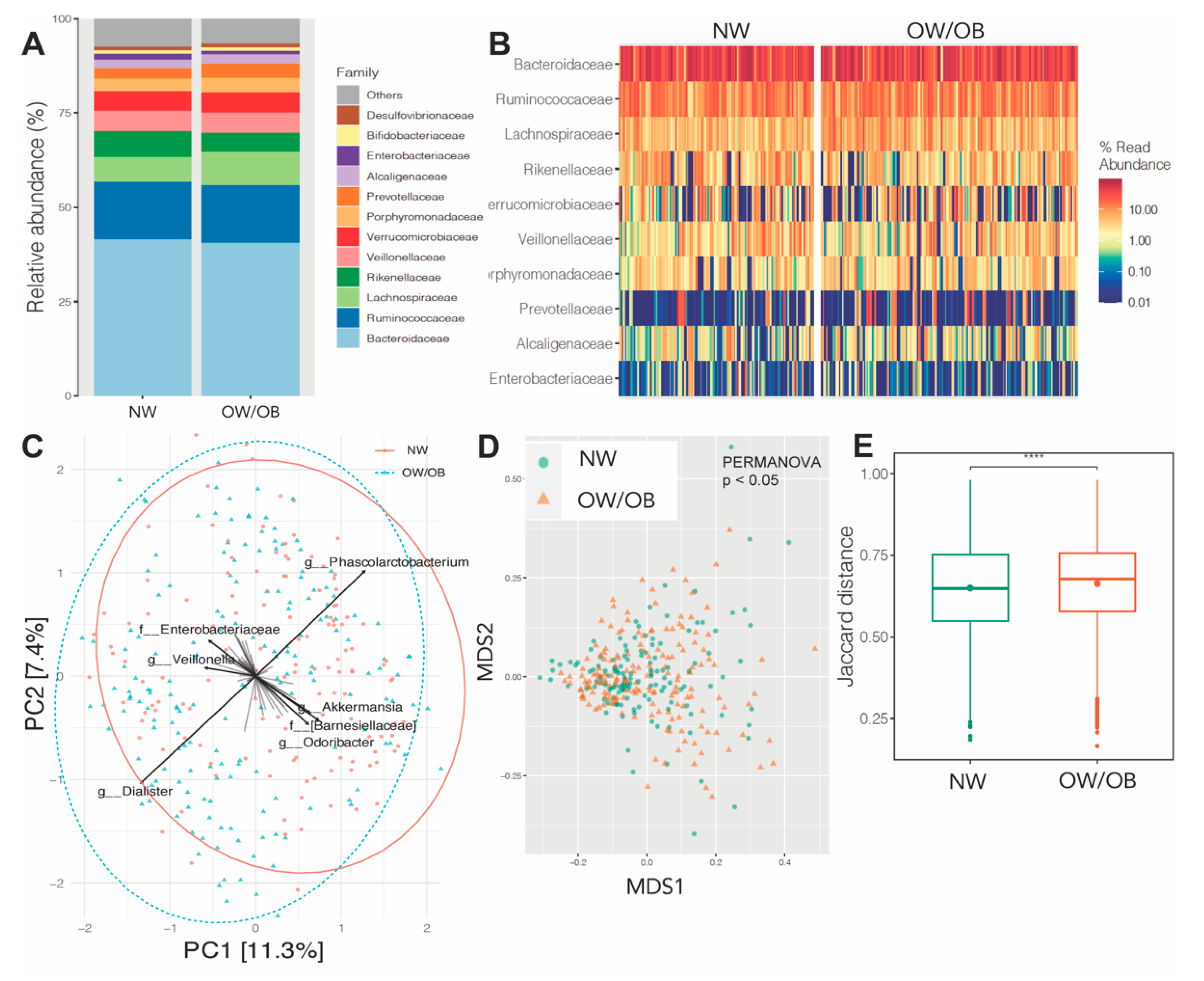
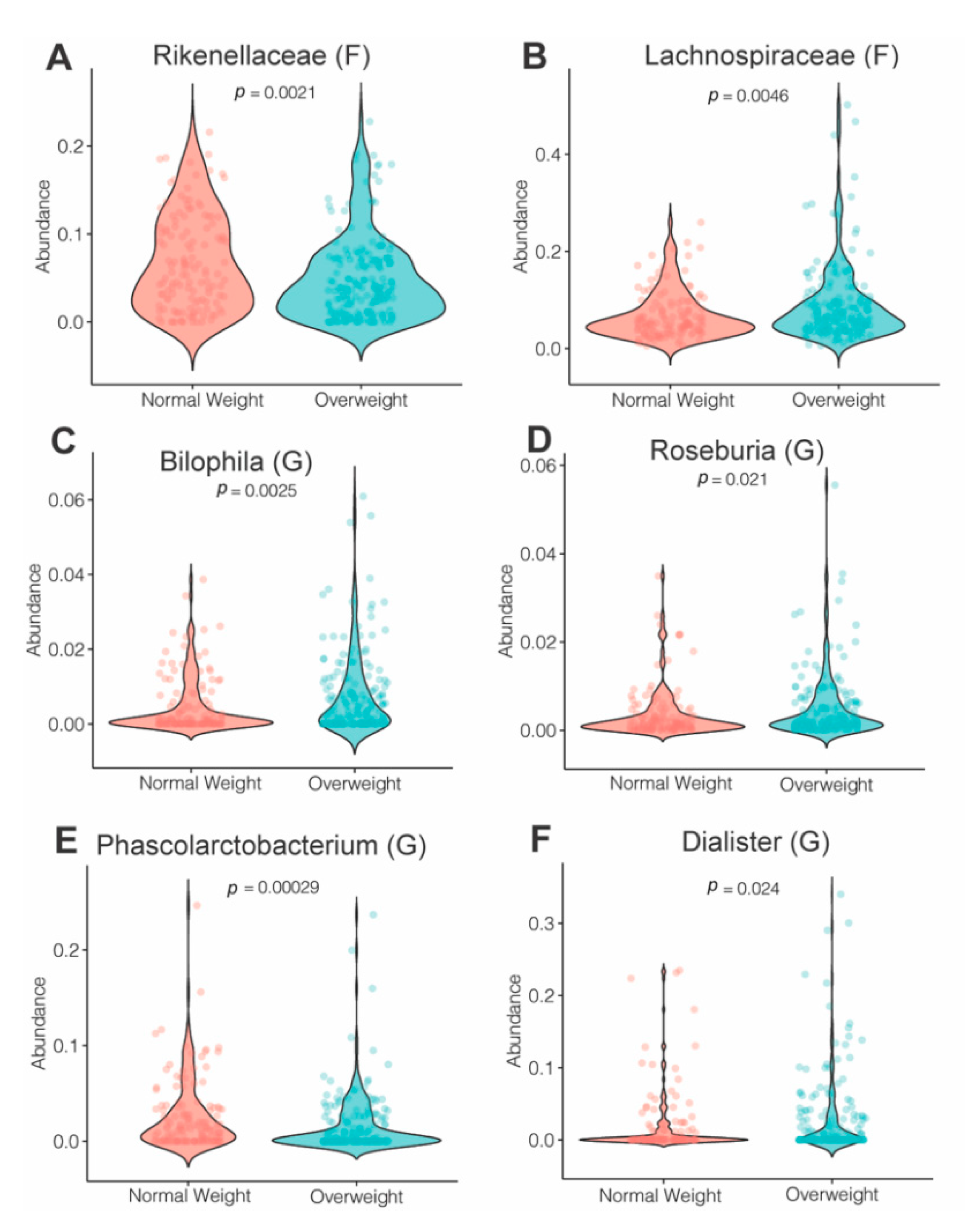
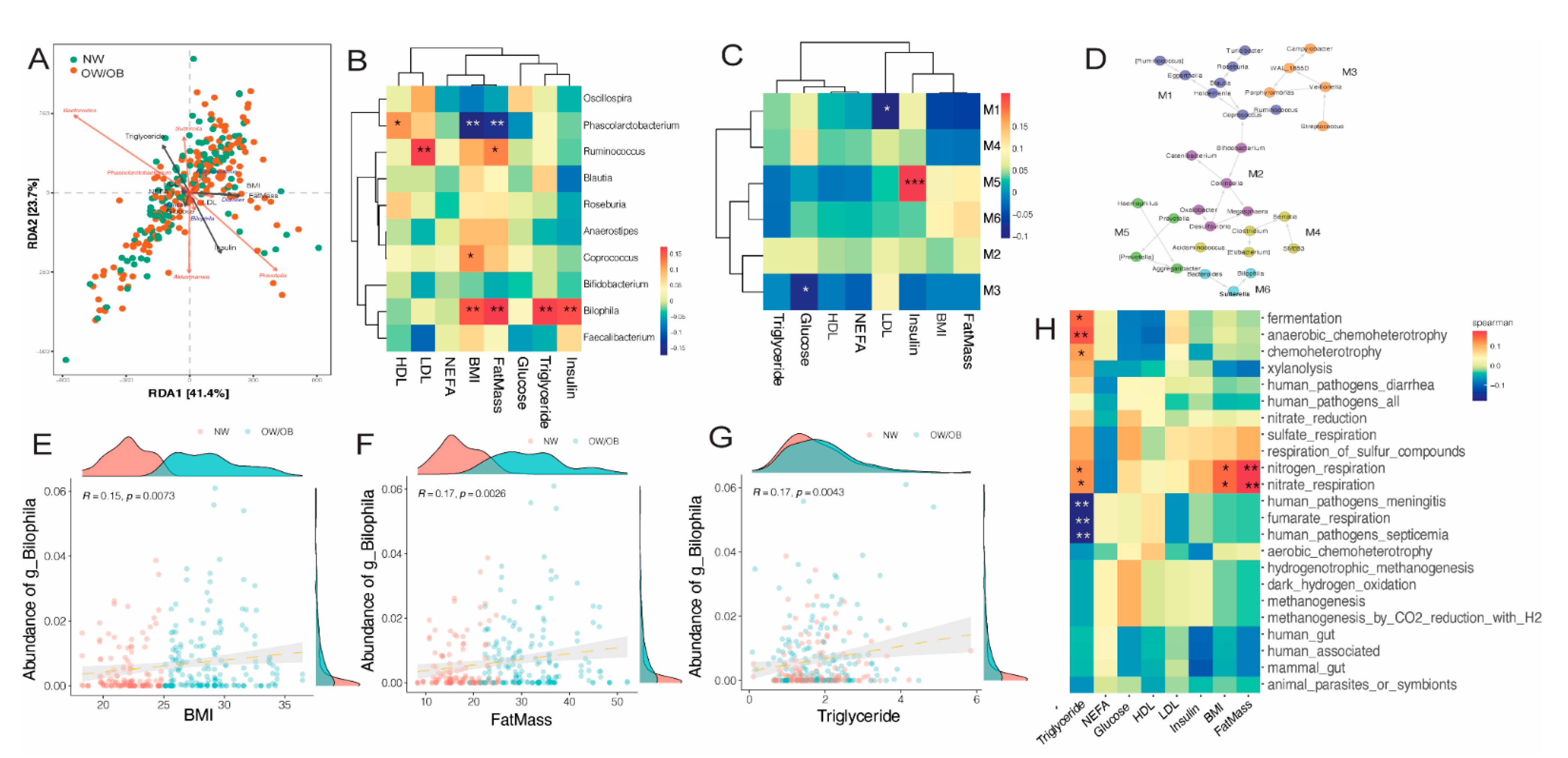
3.4. Effects of Maternal Obesity on Microbiota Composition and Abundance
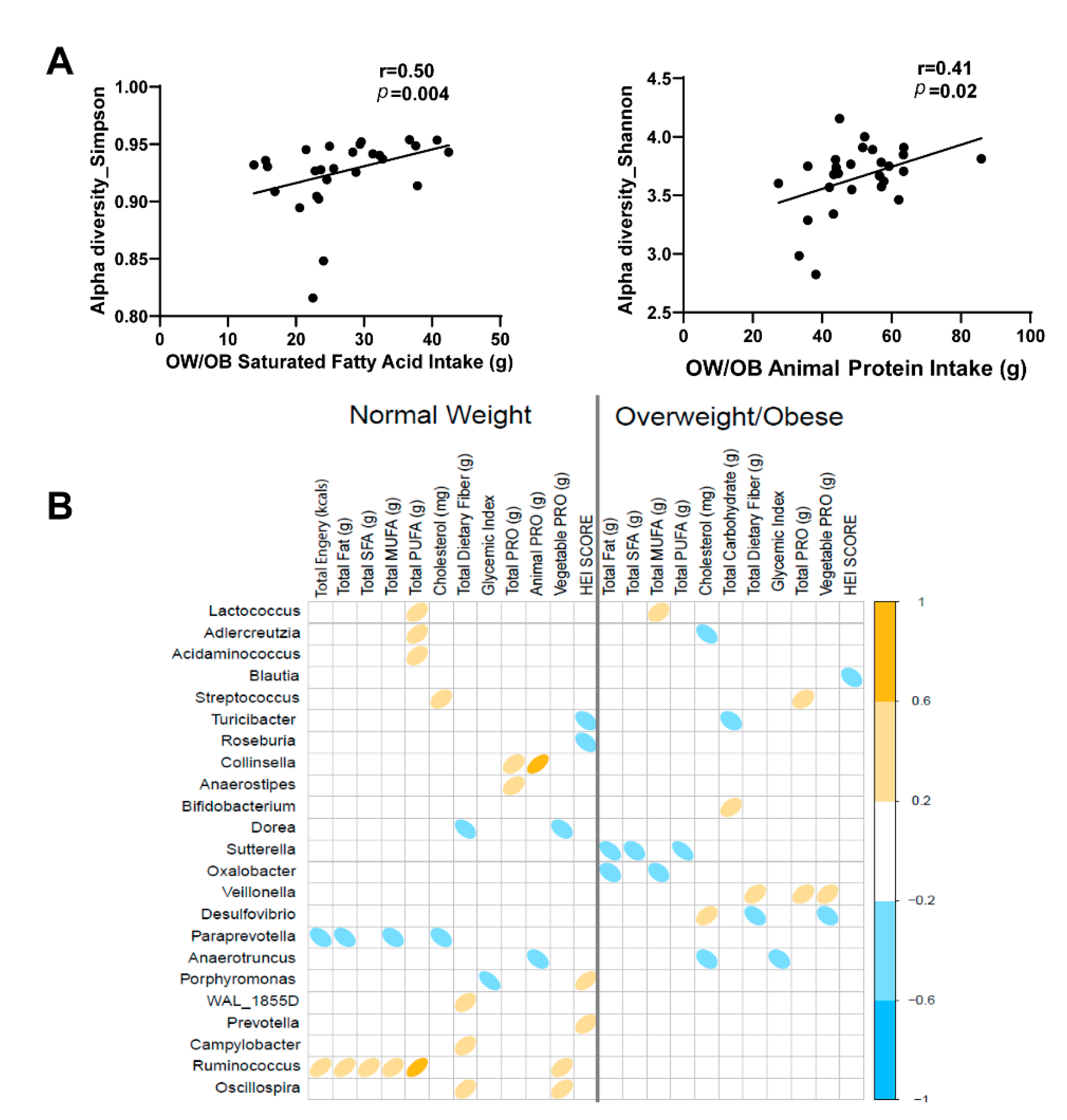
3.5. Impact of Diet on Maternal Gut Microbiome in Pregnancy
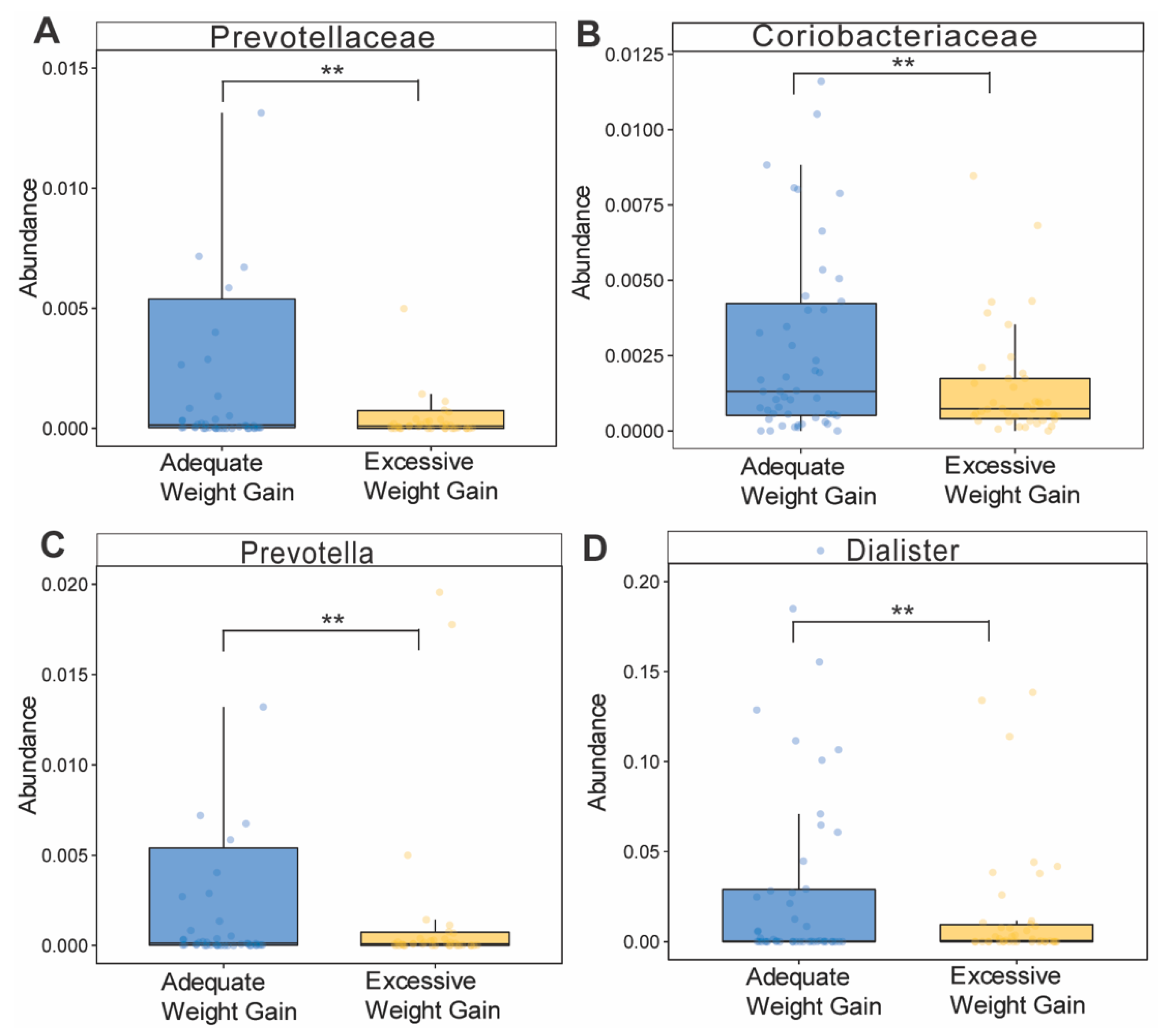
3.6. Effects of Gestational Weight Gain on Microbiota Composition and Abundance
3.7. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flegal, K.M.; Carroll, M.D.; Kit, B.K.; Ogden, C.L. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA 2012, 307, 491–497. [Google Scholar] [CrossRef] [Green Version]
- Aune, D.; Saugstad, O.D.; Henriksen, T.; Tonstad, S. Maternal body mass index and the risk of fetal death, stillbirth, and infant death: A systematic review and meta-analysis. JAMA 2014, 311, 1536–1546. [Google Scholar] [CrossRef] [Green Version]
- Neri, C.; Edlow, A.G. Effects of Maternal Obesity on Fetal Programming: Molecular Approaches. Cold Spring Harb. Perspect. Med. 2015, 6, a026591. [Google Scholar] [CrossRef] [Green Version]
- Stanislawski, M.A.; Dabelea, D.; Wagner, B.D.; Sontag, M.K.; Lozupone, C.A.; Eggesbo, M. Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants. Microbiome 2017, 5, 113. [Google Scholar] [CrossRef]
- Perez, P.F.; Dore, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. Bacterial imprinting of the neonatal immune system: Lessons from maternal cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef]
- Walker, R.W.; Clemente, J.C.; Peter, I.; Loos, R.J.F. The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatric Obes. 2017, 12, 3–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poston, L. Maternal obesity, gestational weight gain and diet as determinants of offspring long term health. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 627–639. [Google Scholar] [CrossRef]
- Shapiro, A.L.; Kaar, J.L.; Crume, T.L.; Starling, A.P.; Siega-Riz, A.M.; Ringham, B.M.; Glueck, D.H.; Norris, J.M.; Barbour, L.A.; Friedman, J.E.; et al. Maternal diet quality in pregnancy and neonatal adiposity: The Healthy Start Study. Int. J. Obes. 2016, 40, 1056–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Jumpertz, R.; Le, D.S.; Turnbaugh, P.J.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011, 94, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [Green Version]
- Arora, T.; Sharma, R. Fermentation potential of the gut microbiome: Implications for energy homeostasis and weight management. Nutr. Rev. 2011, 69, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- Maher, S.E.; O’Brien, E.C.; Moore, R.L.; Byrne, D.F.; Geraghty, A.A.; Saldova, R.; Murphy, E.F.; Van Sinderen, D.; Cotter, P.D.; McAuliffe, F.M. The association between the maternal diet and the maternal and infant gut microbiome: A systematic review. Br. J. Nutr. 2020, 1–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Backhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef] [Green Version]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Guo, R.; Li, S.; Liang, F.; Tian, C.; Zhao, X.; Long, Y.; Liu, F.; Jiang, M.; Zhang, Y.; et al. Systematic analysis of gut microbiota in pregnant women and its correlations with individual heterogeneity. NPJ Biofilms Microbiomes 2020, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Diaz, E.C.; Cleves, M.A.; DiCarlo, M.; Sobik, S.R.; Ruebel, M.L.; Thakali, K.M.; Sims, C.R.; Dajani, N.K.; Krukowski, R.A.; Børsheim, E.; et al. Parental adiposity differentially associates with newborn body composition. Pediatric Obes. 2020, 15, e12596. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Wankhade, U.D.; Zhong, Y.; Lazarenko, O.P.; Chintapalli, S.V.; Piccolo, B.D.; Chen, J.R.; Shankar, K. Sex-Specific Changes in Gut Microbiome Composition following Blueberry Consumption in C57BL/6J Mice. Nutrients 2019, 11, 313. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rideout, J.R.; He, Y.; Navas-Molina, J.A.; Walters, W.A.; Ursell, L.K.; Gibbons, S.M.; Chase, J.; McDonald, D.; Gonzalez, A.; Robbins-Pianka, A.; et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2014, 2, e545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Barnett, D.J.; Arts, I.C.; Panders, J. microViz: An R package for microbiome data visualization and statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- Subrina, I.; Sanz, H.; Vila, J. Building Bivariate Tables: The compareGroups Package for R. J. Stat. Softw. 2014, 57, 6297. [Google Scholar]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-scale Meta-omics Studies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rasmussen, K.M.; Yaktine, A.L. (Eds.) Weight Gain during Pregnancy: Reexamining the Guidelines; National Academies Press: Washington, DC, USA, 2009. [Google Scholar]
- Smid, M.C.; Ricks, N.M.; Panzer, A.; McCoy, A.N.; Azcarate-Peril, M.A.; Keku, T.O.; Boggess, K.A. Maternal Gut Microbiome Biodiversity in Pregnancy. Am. J. Perinatol. 2018, 35, 24–30. [Google Scholar] [PubMed]
- Chung, S.Y.; Ravel, J.; Regan, M. Clinical Relevance of Gastrointestinal Microbiota during Pregnancy: A Primer for Nurses. Biol. Res. Nurs. 2018, 20, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Sze, M.A.; Schloss, P.D. Erratum for Sze and Schloss, “Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome”. mBio 2017, 8, e01018-16. [Google Scholar] [CrossRef] [Green Version]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Li, D.; He, Y.; Li, Y.; Yang, Z.; Zhao, X.; Liu, Y.; Wang, Y.; Sun, J.; Feng, X.; et al. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 2019, 9, 13424. [Google Scholar] [CrossRef] [Green Version]
- Muniz Pedrogo, D.A.; Jensen, M.D.; Van Dyke, C.T.; Murray, J.A.; Woods, J.A.; Chen, J.; Kashyap, P.C.; Nehra, V. Gut Microbial Carbohydrate Metabolism Hinders Weight Loss in Overweight Adults Undergoing Lifestyle Intervention With a Volumetric Diet. Mayo Clin. Proc. 2018, 93, 1104–1110. [Google Scholar] [CrossRef]
- Naderpoor, N.; Mousa, A.; Gomez-Arango, L.F.; Barrett, H.L.; Dekker Nitert, M.; de Courten, B. Faecal Microbiota Are Related to Insulin Sensitivity and Secretion in Overweight or Obese Adults. J. Clin. Med. 2019, 8, 452. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Long, W.; Hao, B.; Ding, D.; Ma, X.; Zhao, L.; Pang, X. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog. 2017, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Natividad, J.M.; Lamas, B.; Pham, H.P.; Michel, M.L.; Rainteau, D.; Bridonneau, C.; da Costa, G.; van Hylckama Vlieg, J.; Sovran, B.; Chamignon, C.; et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018, 9, 2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, Y.Y.; Ha, C.W.; Hoffmann, J.M.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Huang, F.; Zhao, A.; Lei, S.; Zhang, Y.; Xie, G.; Chen, T.; Qu, C.; Rajani, C.; Dong, B.; et al. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017, 15, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Button, J.E.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Li, G.H.; Huang, S.J.; Li, X.; Liu, X.S.; Du, Q.L. Response of gut microbiota to serum metabolome changes in intrahepatic cholestasis of pregnant patients. World J. Gastroenterol. 2020, 26, 7338–7351. [Google Scholar] [CrossRef]
- Noriega, B.S.; Sanchez-Gonzalez, M.A.; Salyakina, D.; Coffman, J. Understanding the Impact of Omega-3 Rich Diet on the Gut Microbiota. Case Rep. Med. 2016, 2016, 3089303. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Mantrana, I.; Selma-Royo, M.; Gonzalez, S.; Parra-Llorca, A.; Martinez-Costa, C.; Collado, M.C. Distinct maternal microbiota clusters are associated with diet during pregnancy: Impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes 2020, 11, 962–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astbury, S.; Atallah, E.; Vijay, A.; Aithal, G.P.; Grove, J.I.; Valdes, A.M. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 2020, 11, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Liu, Y.Y.; Wan, X.Z.; Huang, Z.R.; Liu, B.; Zhao, C. Regulatory Efficacy of the Polyunsaturated Fatty Acids from Microalgae Spirulina platensis on Lipid Metabolism and Gut Microbiota in High-Fat Diet Rats. Int. J. Mol. Sci. 2018, 19, 3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santacruz, A.; Collado, M.C.; Garcia-Valdes, L.; Segura, M.T.; Martin-Lagos, J.A.; Anjos, T.; Marti-Romero, M.; Lopez, R.M.; Florido, J.; Campoy, C.; et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 2010, 104, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furet, J.P.; Kong, L.C.; Tap, J.; Poitou, C.; Basdevant, A.; Bouillot, J.L.; Mariat, D.; Corthier, G.; Dore, J.; Henegar, C.; et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: Links with metabolic and low-grade inflammation markers. Diabetes 2010, 59, 3049–3057. [Google Scholar] [CrossRef] [Green Version]
| All | Normal Weight (BMI <25 kg/m2) | Overweight/Obese (BMI ≥ 25 kg/m2) | p-value (NW vs. OW/OB) | |
|---|---|---|---|---|
| N (%) | 140 | 60 (42.9) | 80 (57.1) | |
| Age at enrollment (years) | 29.46 ± 0.32 | 29.45 ± 0.51 | 29.47 ± 0.41 | 0.970 |
| Race | ||||
| Caucasian | 122 (87.1) | 52 (86.7) | 70 (87.5) | |
| Non-Caucasian | 18 (12.9) | 8 (13.3) | 10 (12.5) | |
| Body Mass Index (kg/m2) | 25.91 ± 0.34 | 22.14 ± 0.22 | 28.73 ± 0.31 | <0.0001 |
| Fat Mass (%) | 25.47 ± 0.79 | 17.32 ± 0.57 | 31.58 ± 0.80 | <0.0001 |
| Gestational Weight Gain (kg) | 12.01 ± 0.38 | 12.81 ± 0.38 | 11.41 ± 0.58 | 0.047 |
| IOM Weight Gain Category | <0.0001 | |||
| Inadequate | 19 (13.7) | 10 (16.9) | 9 (11.3) | |
| Adequate | 61 (43.9) | 36 (61.0) | 25 (31.3) | |
| Excessive | 59 (42.4) | 13 (22.0) | 46 (57.5) |
| Pregnancy Stage | T1 | T2 | T3 | p-Value Overall | p.T1 vs. T2 | p.T1 vs. T3 | p.T2 vs. T3 |
|---|---|---|---|---|---|---|---|
| n | n = 101 | n = 98 | n = 107 | ||||
| Chao1 | 40.1 (5.46) | 40.9 (5.92) | 40.4 (5.55) | 0.641 | 0.615 | 0.915 | 0.844 |
| Shannon | 2.02 (0.35) | 2.00 (0.37) | 1.96 (0.39) | 0.61 | 0.938 | 0.59 | 0.805 |
| Inverse Simpson | 4.64 (1.96) | 4.55 (1.87) | 4.34 (1.78) | 0.498 | 0.929 | 0.481 | 0.718 |
| Fisher | 4.38 (0.66) | 4.43 (0.71) | 4.37 (0.64) | 0.767 | 0.829 | 0.996 | 0.777 |
| NW | Overweight/Obese | p-Value | |
|---|---|---|---|
| Chao1 | 40.7 (5.23) | 40.5 (5.77) | 0.81 |
| Shannon | 2.00 (0.34) | 1.99 (0.38) | 0.766 |
| Inverse Simpson | 4.46 (1.82) | 4.58 (1.95) | 0.576 |
| Fisher | 4.38 (0.58) | 4.38 (0.71) | 0.949 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruebel, M.L.; Gilley, S.P.; Sims, C.R.; Zhong, Y.; Turner, D.; Chintapalli, S.V.; Piccolo, B.D.; Andres, A.; Shankar, K. Associations between Maternal Diet, Body Composition and Gut Microbial Ecology in Pregnancy. Nutrients 2021, 13, 3295. https://doi.org/10.3390/nu13093295
Ruebel ML, Gilley SP, Sims CR, Zhong Y, Turner D, Chintapalli SV, Piccolo BD, Andres A, Shankar K. Associations between Maternal Diet, Body Composition and Gut Microbial Ecology in Pregnancy. Nutrients. 2021; 13(9):3295. https://doi.org/10.3390/nu13093295
Chicago/Turabian StyleRuebel, Meghan L., Stephanie P. Gilley, Clark R. Sims, Ying Zhong, Donald Turner, Sree V. Chintapalli, Brian D. Piccolo, Aline Andres, and Kartik Shankar. 2021. "Associations between Maternal Diet, Body Composition and Gut Microbial Ecology in Pregnancy" Nutrients 13, no. 9: 3295. https://doi.org/10.3390/nu13093295
APA StyleRuebel, M. L., Gilley, S. P., Sims, C. R., Zhong, Y., Turner, D., Chintapalli, S. V., Piccolo, B. D., Andres, A., & Shankar, K. (2021). Associations between Maternal Diet, Body Composition and Gut Microbial Ecology in Pregnancy. Nutrients, 13(9), 3295. https://doi.org/10.3390/nu13093295






