Milk Exosomes Transfer Oligosaccharides into Macrophages to Modulate Immunity and Attenuate Adherent-Invasive E. coli (AIEC) Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Milk Samples and Subjects
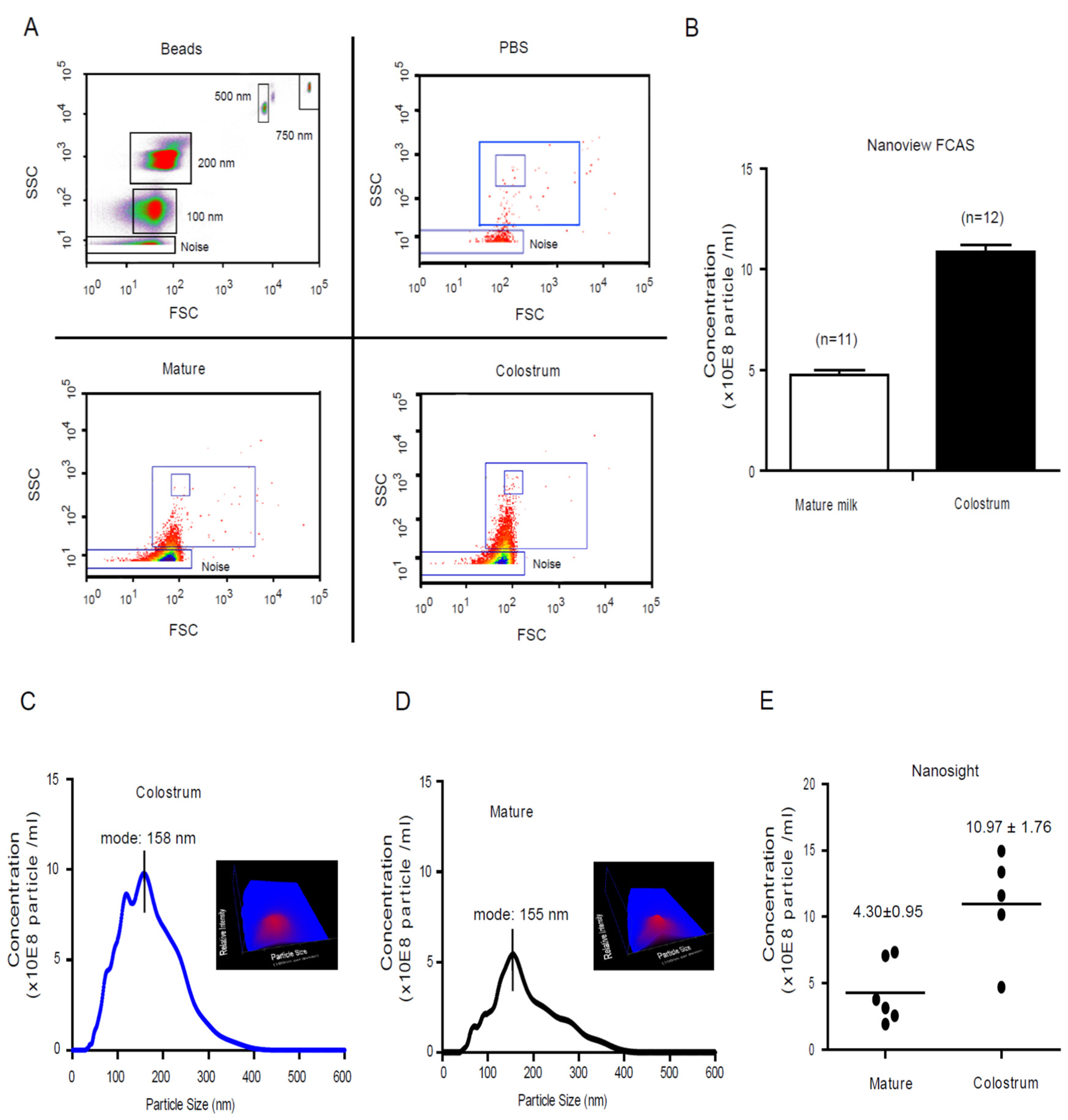
2.2. Nanosight Tracking Analysis on Human Milk Microparticles
2.3. Dynamic Analysis Human Milk Microparticle Population
2.4. Isolation of Exosome-like Vesicles by Differential Ultracentrifugation
2.5. Profile of Milk Exosome Encapsulated Oligosaccharides (MECO) Determined by LC-MS
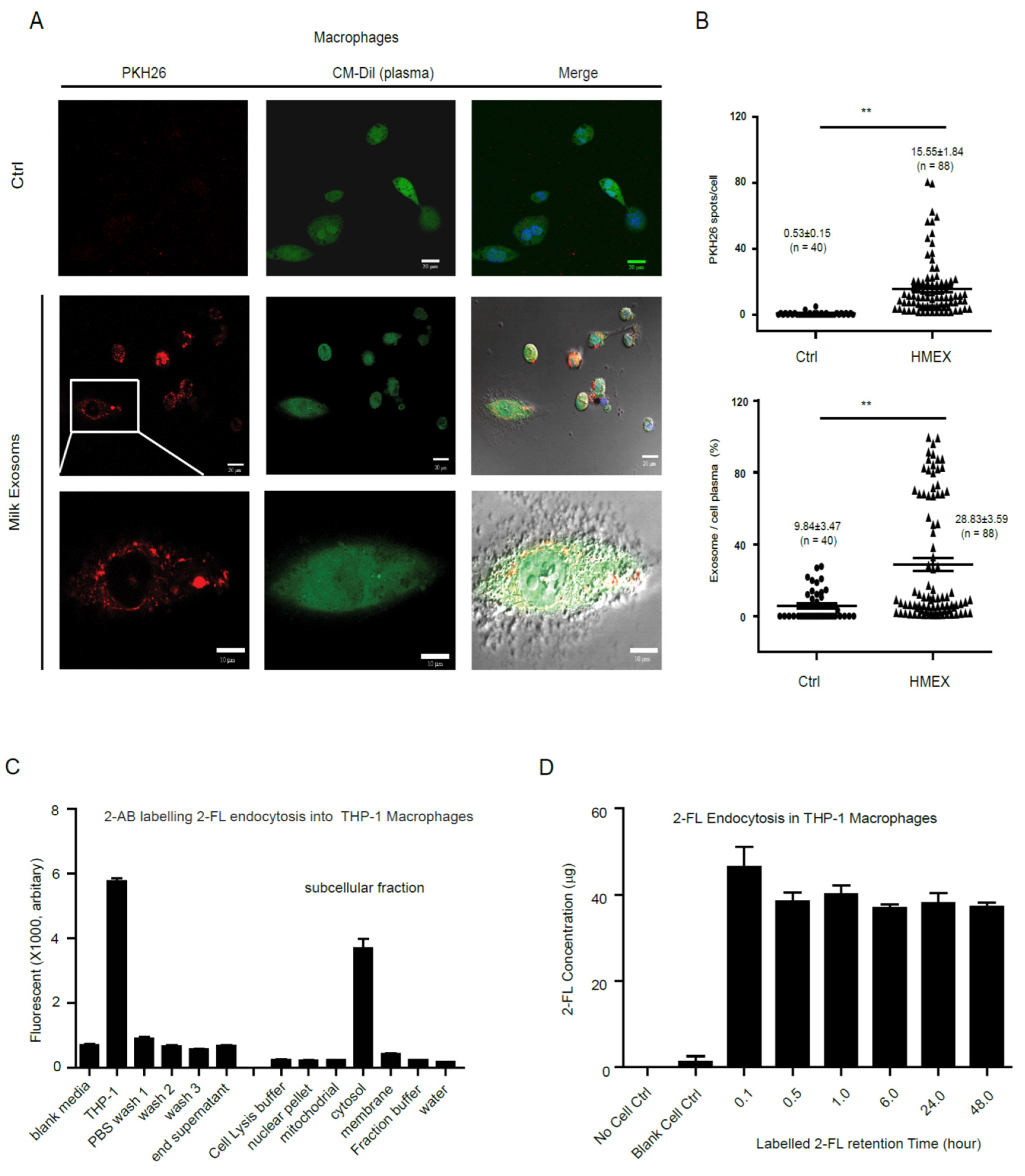
2.6. Colostrum Exosomes Labeled with PKH26
2.7. THP-1 Differentiated Macrophages Culture
2.8. Macrophages Uptake of Labeled Colostrum Exosomes
2.9. Milk Oligosaccharides 2′-FL Labeling
2.10. Internalization of Labeled 2′-FL into Macrophages and Subcellular Localization
2.11. Colostrum Exosome Encapsulated Oligosaccharides Treated THP-1 Derived Macrophages
2.12. Whole Transcriptome Expression Profiling of Macrophage
2.13. In Vivo Animals Experiments
2.14. Hematoxylin & Eosin Staining
2.15. Statistical Analysis
3. Results
3.1. Characterized the Dynamic Profile of Human Milk Exosomes
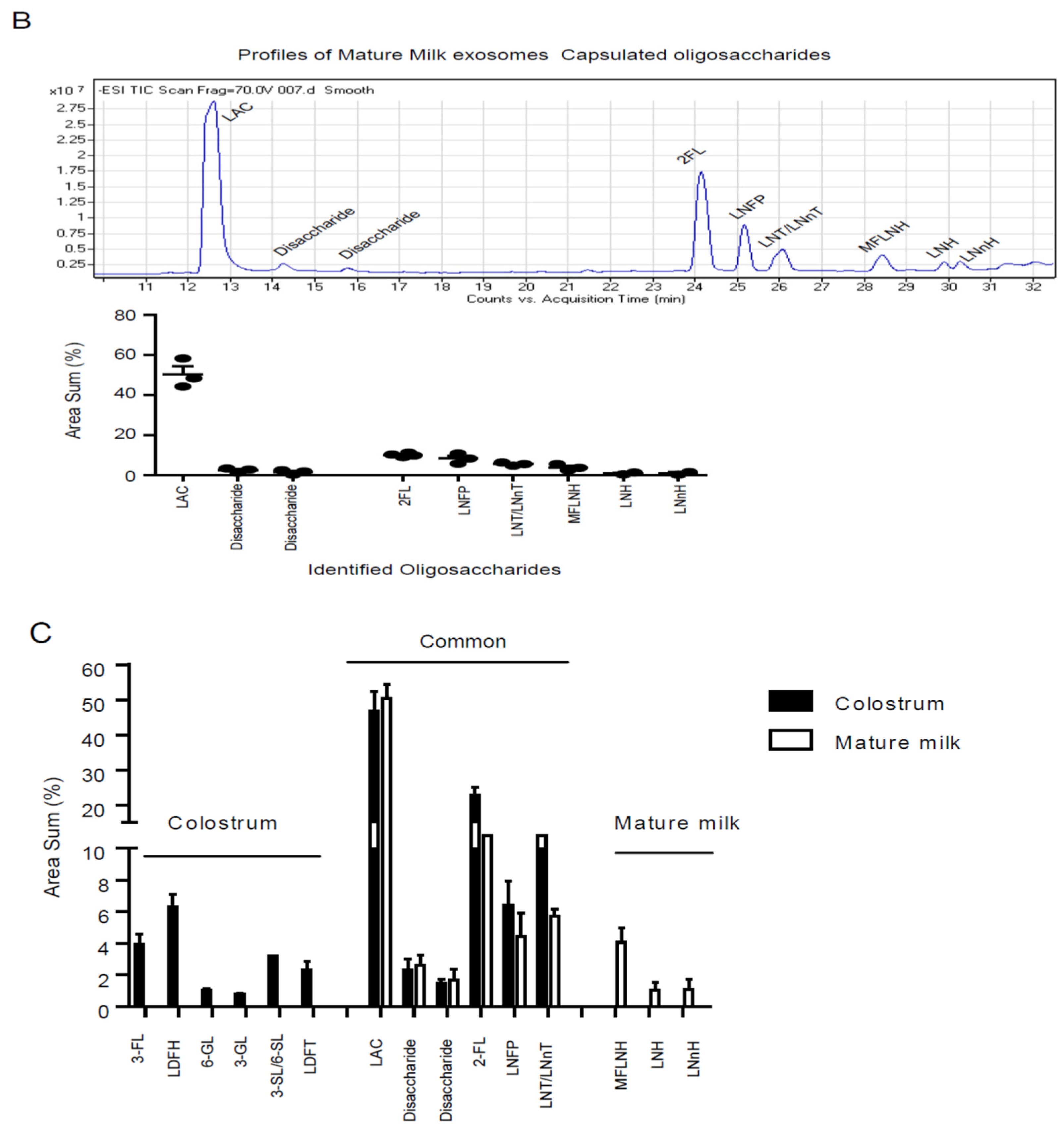
3.2. Characterization Profile of Human Milk Exosome Encapsulated Oligosaccharides by LC-MS
3.3. Colostrum Exosomes Capsulated Oligosaccharides Phagocytosis into Macrophages
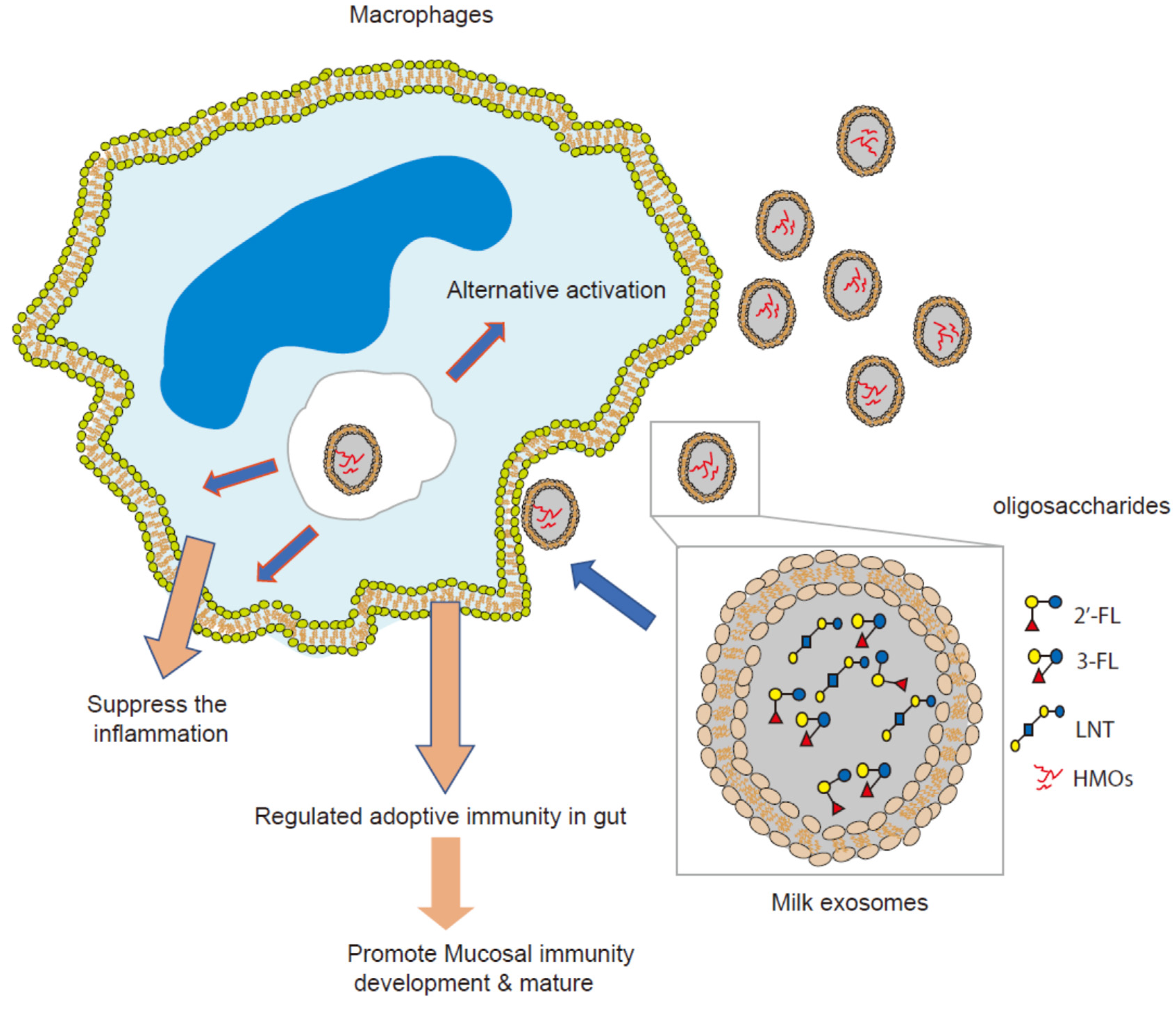
3.4. Colostrum Exosomes Capsulated Oligosaccharides (CECO) Modulate Macrophages Alternative Activation and Mucosal Immunity Development
3.5. Colostrum Exosome Enapsulated Oligosaccharides Attenuate AIEC Infection and Inflammation In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Newburg, D.S.; Ruiz-Palacios, G.M.; Morrow, A.L. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005, 25, 37–58. [Google Scholar] [CrossRef]
- He, Y.; Liu, S.; Leone, S.; Newburg, D.S. Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 2014, 7, 1326–1339. [Google Scholar] [CrossRef] [PubMed]
- Kurakevich, E.; Hennet, T.; Hausmann, M.; Rogler, G.; Borsig, L. Milk oligosaccharide sialyl (alpha2,3) lactose activates intestinal CD11c+ cells through TLR4. Proc. Natl. Acad. Sci. USA 2013, 110, 17444–17449. [Google Scholar] [CrossRef] [PubMed]
- Frederick, R. Mother’s milk mysteries. Proc. Natl. Acad. Sci. USA 2014, 111, 7165. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Couzin, J. Cell biology: The ins and outs of exosomes. Science 2005, 308, 1862–1863. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Hata, T.; Murakami, K.; Nakatani, H.; Yamamoto, Y.; Matsuda, T.; Aoki, N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem. Biophys. Res. Commun. 2010, 396, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Kunz, C.; Rudloff, S.; Baier, W.; Klein, N.; Strobel, S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000, 20, 699–722. [Google Scholar] [CrossRef]
- Lucas, A.; Cole, T.J. Breast milk and neonatal necrotising enterocolitis. Lancet 1990, 336, 1519–1523. [Google Scholar] [CrossRef]
- Klement, E.; Reif, S. Breastfeeding and risk of inflammatory bowel disease. Am. J. Clin. Nutr. 2005, 82, 486. [Google Scholar] [CrossRef] [PubMed]
- Probert, F.; Mitchell, D.A.; Dixon, A.M. NMR evidence for oligosaccharide release from the dendritic-cell specific intercellular adhesion molecule 3-grabbing non-integrin-related (CLEC4M) carbohydrate recognition domain at low pH. FEBS J. 2014, 281, 3739–3750. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, S.; Kling, D.E.; Leone, S.; Lawlor, N.T.; Huang, Y.; Feinberg, S.B.; Hill, D.R.; Newburg, D.S. The human milk oligosaccharide 2′-fucosyllactose modulates CD14 expression in human enterocytes, thereby attenuating LPS-induced inflammation. Gut 2016, 65, 33–46. [Google Scholar] [CrossRef]
- Bain, C.C.; Mowat, A.M. Macrophages in intestinal homeostasis and inflammation. Immunol. Rev. 2014, 260, 102–117. [Google Scholar] [CrossRef]
- Sheikh, S.Z.; Plevy, S.E. The role of the macrophage in sentinel responses in intestinal immunity. Curr. Opin. Gastroenterol. 2010, 26, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Mowat, A.M.; Bain, C.C. Mucosal macrophages in intestinal homeostasis and inflammation. J. Innate Immun. 2011, 3, 550–564. [Google Scholar] [CrossRef]
- Gillette, D.D.; Tridandapani, S.; Butchar, J.P. Monocyte/macrophage inflammatory response pathways to combat Francisella infection: Possible therapeutic targets? Front. Cell. Infect. Microbiol. 2014, 4, 18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stein, M.; Keshav, S.; Harris, N.; Gordon, S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J. Exp. Med. 1992, 176, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Brancato, S.K.; Albina, J.E. Wound macrophages as key regulators of repair: Origin, phenotype, and function. Am. J. Pathol. 2011, 178, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Danielson, K.M.; Estanislau, J.; Tigges, J.; Toxavidis, V.; Camacho, V.; Felton, E.J.; Khoory, J.; Kreimer, S.; Ivanov, A.R.; Mantel, P.Y.; et al. Diurnal variations of circulating extracellular vesicles measured by nano flow cytometry. PLoS ONE 2016, 11, e0144678. [Google Scholar] [CrossRef]
- Takashiba, S.; Van Dyke, T.E.; Amar, S.; Murayama, Y.; Soskolne, A.W.; Shapira, L. Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor kappaB. Infect. Immun. 1999, 67, 5573–5578. [Google Scholar] [CrossRef]
- Bigge, J.C.; Patel, T.P.; Bruce, J.A.; Goulding, P.N.; Charles, S.M.; Parekh, R.B. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 1995, 230, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Marino, K.; Lane, J.A.; Abrahams, J.L.; Struwe, W.B.; Harvey, D.J.; Marotta, M.; Hickey, R.M.; Rudd, P.M. Method for milk oligosaccharide profiling by 2-aminobenzamide labeling and hydrophilic interaction chromatography. Glycobiology 2011, 21, 1317–1330. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Denizot, J.; Dreux, N.; Robin, F.; Billard, E.; Bonnet, R.; Darfeuille-Michaud, A.; Barnich, N. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 2014, 63, 116–124. [Google Scholar] [CrossRef]
- Small, C.L.; Reid-Yu, S.A.; McPhee, J.B.; Coombes, B.K. Persistent infection with Crohn’s disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat. Commun. 2013, 4, 1957. [Google Scholar] [CrossRef]
- Koelink, P.J.; Wildenberg, M.E.; Stitt, L.W.; Feagan, B.G.; Koldijk, M.; van’t Wout, A.B.; Atreya, R.; Vieth, M.; Brandse, J.F.; Duijst, S.; et al. Development of reliable, valid and responsive scoring systems for endoscopy and histology in animal models for inflammatory bowel disease. J. Crohn Colitis 2018, 12, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Cvjetkovic, A.; Lotvall, J.; Lasser, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef]
- Verheijden, K.A.; Braber, S.; Leusink-Muis, T.; Thijssen, S.; Boon, L.; Kraneveld, A.D.; Garssen, J.; Folkerts, G.; Willemsen, L.E. Regulatory T cell depletion abolishes the protective effect of dietary galacto-oligosaccharides on eosinophilic airway inflammation in house dust mite-induced asthma in mice. J. Nutr. 2015, 146, 831–837. [Google Scholar] [CrossRef]
- Perdijk, O.; van Baarlen, P.; Fernandez-Gutierrez, M.M.; van den Brink, E.; Schuren, F.H.J.; Brugman, S.; Savelkoul, H.F.J.; Kleerebezem, M.; van Neerven, R.J.J. Sialyllactose and galactooligosaccharides promote epithelial barrier functioning and distinctly modulate microbiota composition and short chain fatty acid production in vitro. Front. Immunol. 2019, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Bain, C.C.; Scott, C.L.; Uronen-Hansson, H.; Gudjonsson, S.; Jansson, O.; Grip, O.; Guilliams, M.; Malissen, B.; Agace, W.W.; Mowat, A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013, 6, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Bujko, A.; Atlasy, N.; Landsverk, O.J.B.; Richter, L.; Yaqub, S.; Horneland, R.; Oyen, O.; Aandahl, E.M.; Aabakken, L.; Stunnenberg, H.G.; et al. Transcriptional and functional profiling defines human small intestinal macrophage subsets. J. Exp. Med. 2018, 215, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.D.; Smythies, L.E.; Shen, R.; Greenwell-Wild, T.; Gliozzi, M.; Wahl, S.M. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011, 4, 31–42. [Google Scholar] [CrossRef]
- Bretin, A.; Lucas, C.; Larabi, A.; Dalmasso, G.; Billard, E.; Barnich, N.; Bonnet, R.; Nguyen, H.T.T. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci. Rep. 2018, 8, 12301. [Google Scholar] [CrossRef] [PubMed]
- Nadalian, B.; Yadegar, A.; Houri, H.; Olfatifar, M.; Shahrokh, S.; Asadzadeh Aghdaei, H.; Suzuki, H.; Zali, M.R. Prevalence of the pathobiont adherent-invasive Escherichia coli and inflammatory bowel disease: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2021, 36, 852–863. [Google Scholar] [CrossRef] [PubMed]
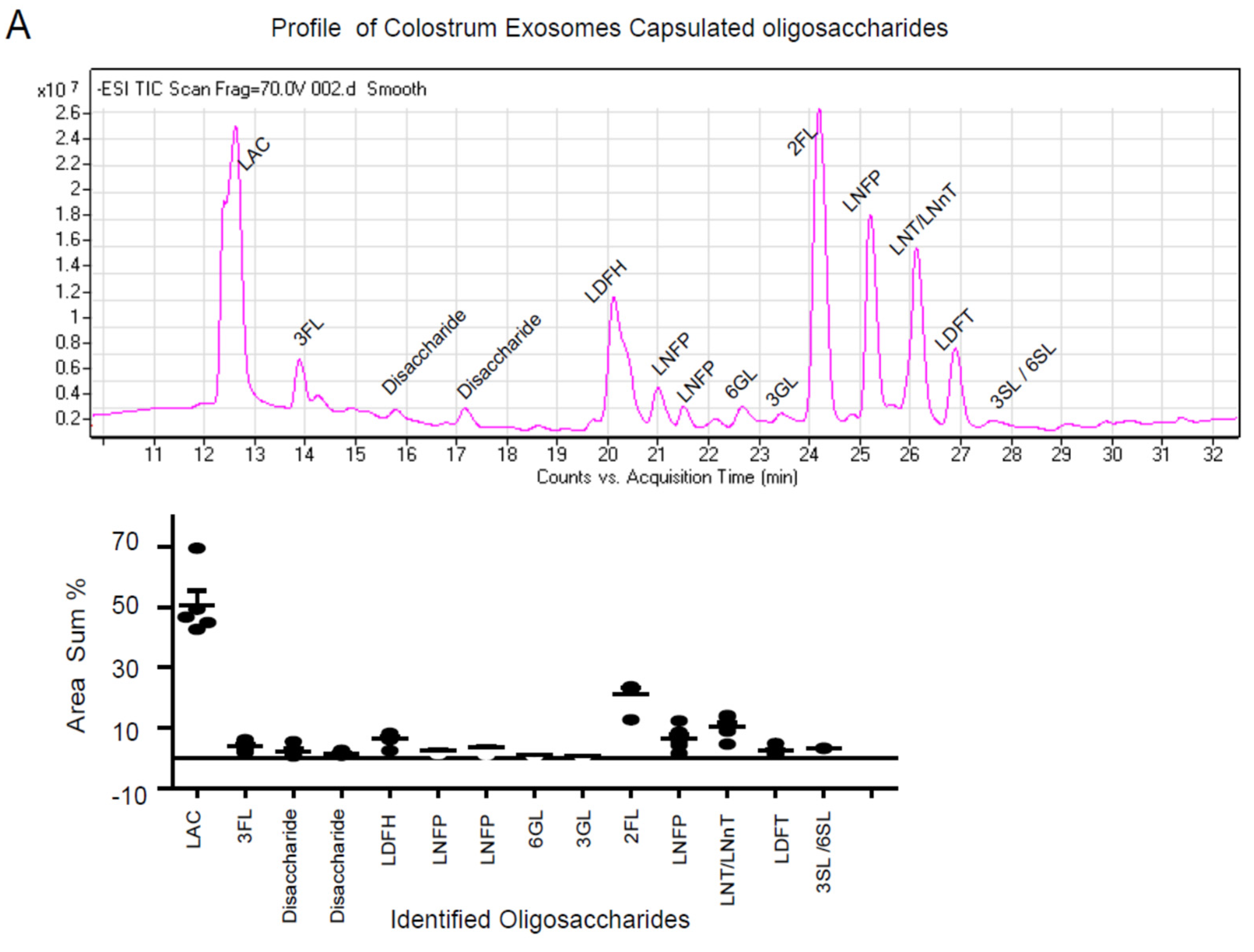


| Sample | Peak | Abbreviation | Full Name | Area Sum (%) |
|---|---|---|---|---|
| Colostrum exosomes | 1 | LAC | Lactose | 46.85 ± 5.50 |
| Colostrum exosomes | 2 | 3-FL | 3-Fucosyllactose | 2.29 ± 0.71 |
| Colostrum exosomes | 3 | Disaccharide | Disaccharide | 1.45 ± 0.28 |
| Colostrum exosomes | 4 | Disaccharide | Disaccharide | 3.93 ± 0.66 |
| Colostrum exosomes | 5 | LDFH | Lacto-N-difucohexaose | 6.29 ± 0.82 |
| Colostrum exosomes | 6 | LNFP II | Lacto-N-Fucopentaose II | 2.55 ± 0.34 |
| Colostrum exosomes | 7 | LNFP | Lacto-N-Fucopentaose | 3.38 ± 0.63 |
| Colostrum exosomes | 8 | 6-GL | 6′-galactosyllactose | 0.91 ± 0.09 |
| Colostrum exosomes | 9 | 3-GL | 3′-galactosyllactose | 0.72 ± 0.07 |
| Colostrum exosomes | 10 | 2’-FL | 2-Fucosyllactose | 21.08 ± 2.09 |
| Colostrum exosomes | 11 | LNFP I | Lacto-N-Fucopentaose I | 7.43 ± 1.52 |
| Colostrum exosomes | 12 | LNT/LNnT | Lacto-N-Tetraose | 6.44 ± 1.43 |
| Colostrum exosomes | 13 | LDFT | Lacto-Difucotetraose | 2.32 ± 0.56 |
| Colostrum exosomes | 14 | 3SL/6SL | 3′/6-Sialyllactose | 3.18 ± 0.04 |
| Mature milk exosomes | 1 | LAC | Lactose | 50.30 ± 4.15 |
| Mature milk exosomes | 2 | Disaccharide | Disaccharide | 2.61 ± 0.65 |
| Mature milk exosomes | 3 | Disaccharide | Disaccharide | 1.69 ± 0.67 |
| Mature milk exosomes | 4 | 2-FL | 2-Fucosyllactose | 12.24 ± 0.47 |
| Mature milk exosomes | 5 | LNFP I | Lacto-N-Fucopentaose I | 8.43 ± 1.03 |
| Mature milk exosomes | 6 | LNT/LNnT | Lacto-N-tetraose/Neotetraose | 5.70 ± 0.45 |
| Mature milk exosomes | 7 | M FLNH | Monofucosyl-lacto-N-hexaose | 4.06 ± 0.90 |
| Mature milk exosomes | 8 | LNH | Lacto-N-hexaose | 1.05 ± 0.48 |
| Mature milk exosomes | 9 | LNnH | Lacto-N-neohexaose | 1.08 ± 0.64 |
| Description | Size | Leading Edge Number | ES | NES | p-Value | FDR |
|---|---|---|---|---|---|---|
| Immune System | 22 | 13 | 0.4598 | 2.2262 | 0.0038241 | 0.11357 |
| Human Thyroid Stimulating Hormone (TSH) signaling pathway | 5 | 5 | 0.72976 | 1.9786 | 0.0039761 | 0.30352 |
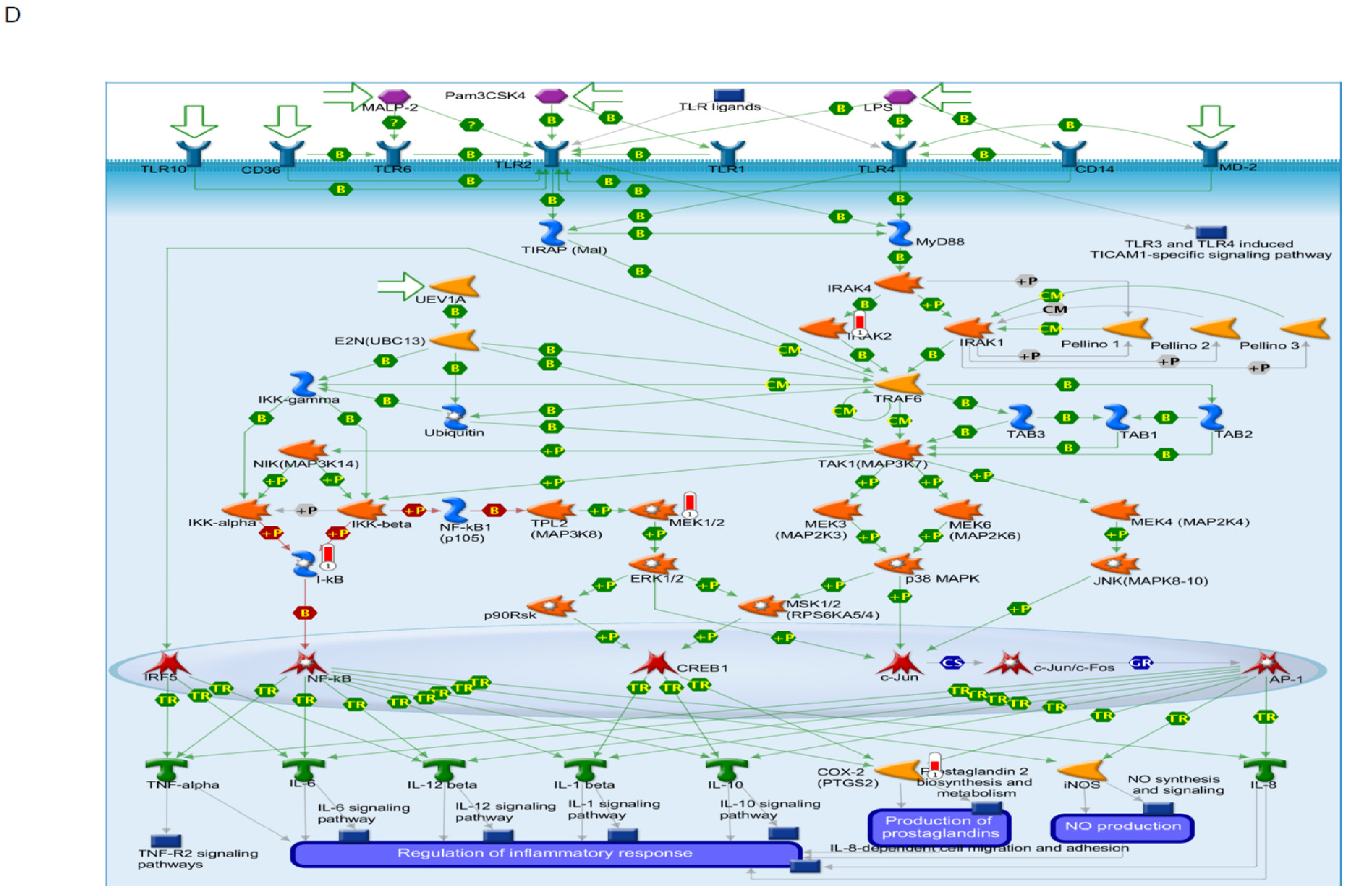
| Toll-like Receptor Signaling related to MyD88 | 5 | 5 | −0.65919 | −1.8344 | 0.012848 | 0.64332 |
| Neural Crest Cell Migration during Development | 7 | 6 | 0.55526 | 1.7762 | 0.015009 | 0.4129 |
| Apoptosis Modulation and Signaling | 7 | 7 | −0.53998 | −1.8065 | 0.018908 | 0.38316 |
| Neural Crest Differentiation | 11 | 7 | 0.45688 | 1.8374 | 0.018939 | 0.49888 |
| Alzheimer’s Disease | 7 | 7 | 0.5241 | 1.6724 | 0.025688 | 0.51279 |
| Chemokine signaling pathway | 14 | 14 | 0.36888 | 1.6563 | 0.033582 | 0.43128 |
| Angiopoietin Like Protein 8 Regulatory Pathway | 11 | 10 | −0.39309 | −1.5854 | 0.042735 | 0.78929 |
| AMP-activated Protein Kinase (AMPK) Signaling | 8 | 8 | 0.45753 | 1.5804 | 0.04717 | 0.50515 |
| RAC1/PAK1/p38/MMP2 Pathway | 6 | 4 | 0.53976 | 1.6085 | 0.048319 | 0.48671 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; He, Z.; Leone, S.; Liu, S. Milk Exosomes Transfer Oligosaccharides into Macrophages to Modulate Immunity and Attenuate Adherent-Invasive E. coli (AIEC) Infection. Nutrients 2021, 13, 3198. https://doi.org/10.3390/nu13093198
He Y, He Z, Leone S, Liu S. Milk Exosomes Transfer Oligosaccharides into Macrophages to Modulate Immunity and Attenuate Adherent-Invasive E. coli (AIEC) Infection. Nutrients. 2021; 13(9):3198. https://doi.org/10.3390/nu13093198
Chicago/Turabian StyleHe, Yingying, Zhicheng He, Serena Leone, and Shubai Liu. 2021. "Milk Exosomes Transfer Oligosaccharides into Macrophages to Modulate Immunity and Attenuate Adherent-Invasive E. coli (AIEC) Infection" Nutrients 13, no. 9: 3198. https://doi.org/10.3390/nu13093198
APA StyleHe, Y., He, Z., Leone, S., & Liu, S. (2021). Milk Exosomes Transfer Oligosaccharides into Macrophages to Modulate Immunity and Attenuate Adherent-Invasive E. coli (AIEC) Infection. Nutrients, 13(9), 3198. https://doi.org/10.3390/nu13093198







