Integrating Dietary Data into Microbiome Studies: A Step Forward for Nutri-Metaomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. 24-h Dietary Recall: The Pilot Study
2.3. Design and Development of the sFFQ
2.4. sFFQ Administration
2.5. Analysis of the sFFQ Responses
2.6. Identification of Unreliable sFFQs and 24HRs
2.7. Statistical Analysis to Evaluate the Validity and Reproducibility of the sFFQ
2.8. Microbiome and Statistical Analyses
2.9. Deposition of Sequences Data
3. Results
3.1. Study Design
3.2. Participants’ Characteristics
3.3. Validation of the sFFQ
3.4. Reproducibility of the sFFQ
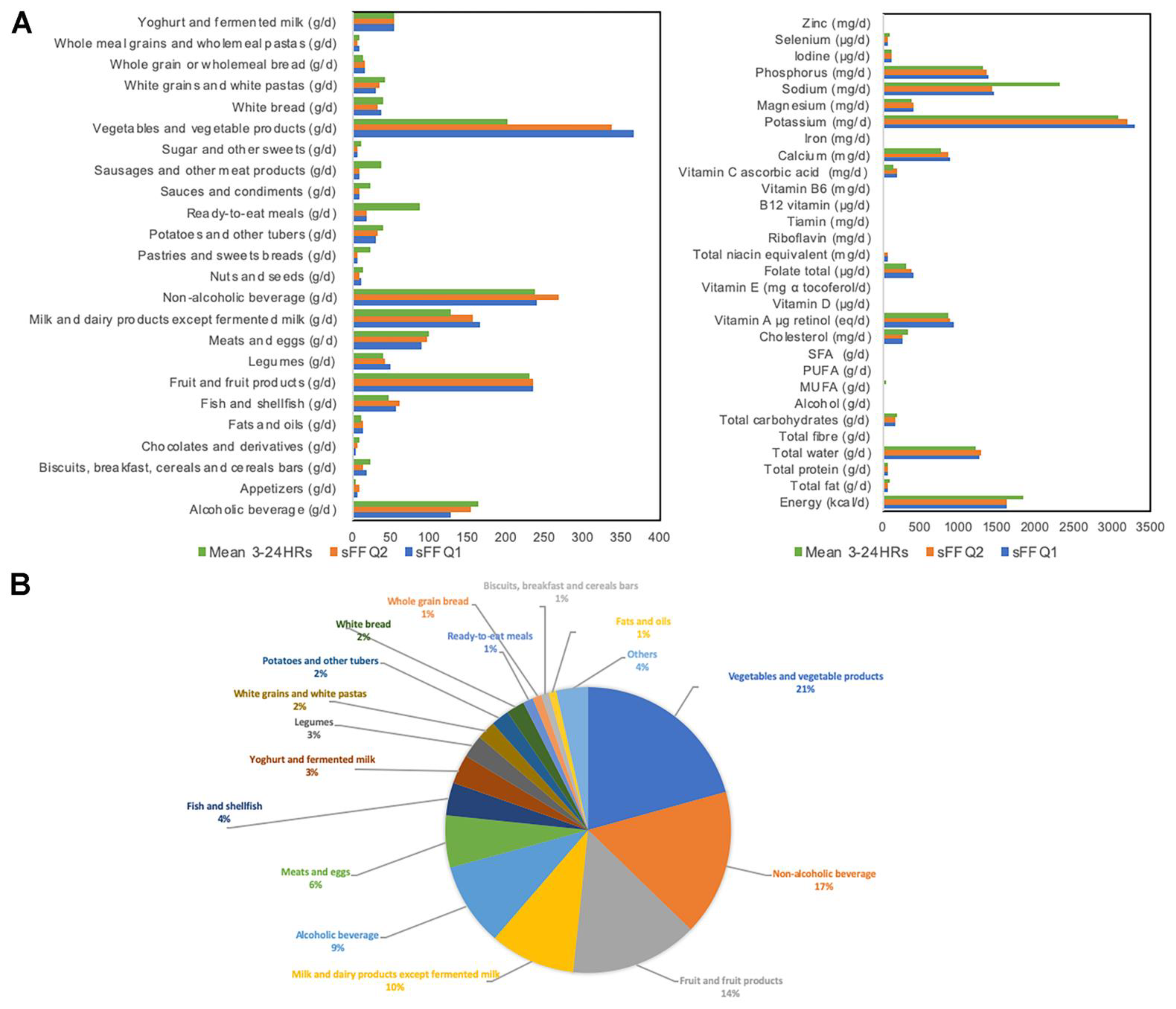
3.5. Participants’ Dietary Profile
3.6. Correlation between Participants’ Characteristics and Microbial Diversity and Taxa
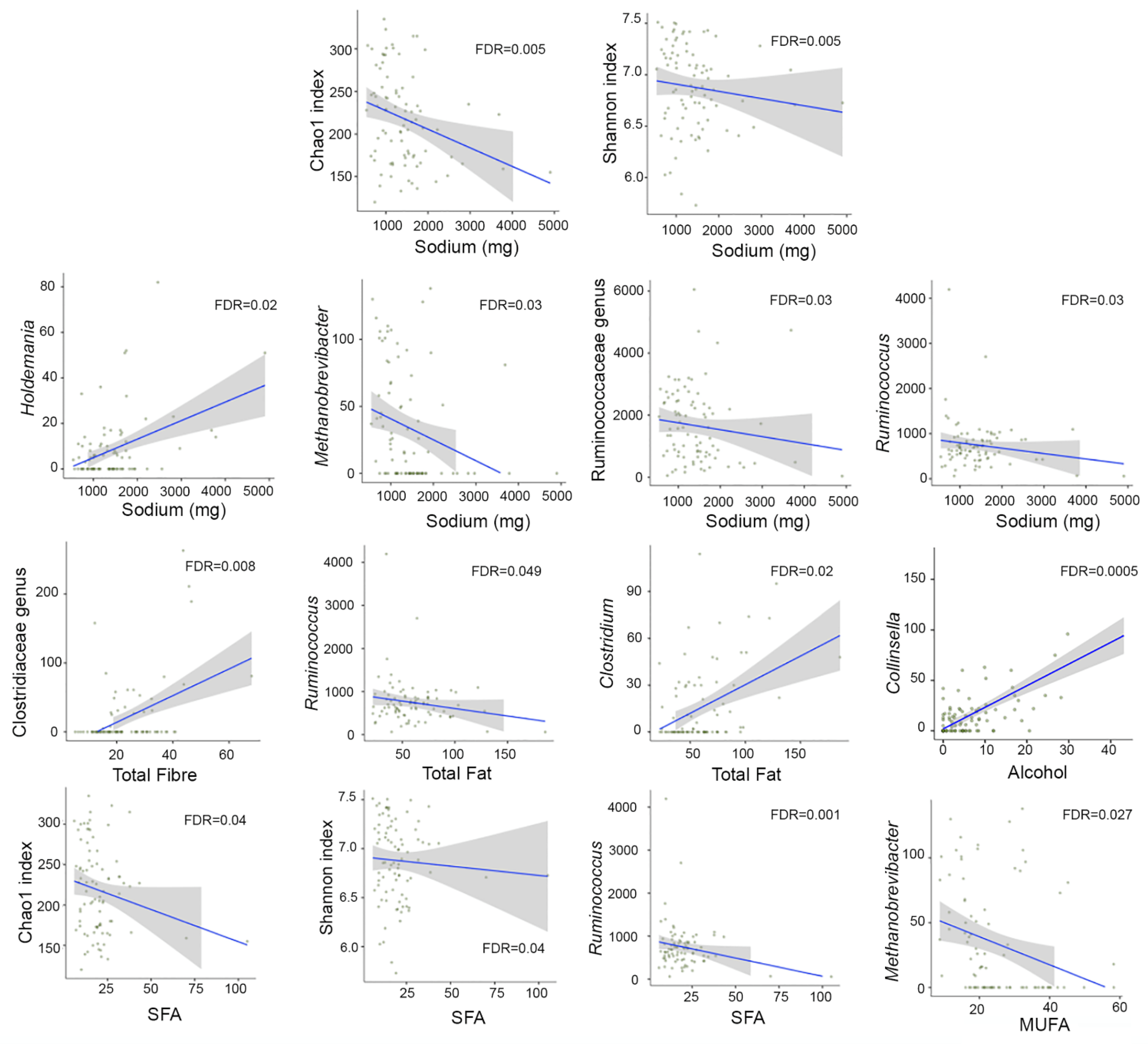
3.7. Correlation between Dietary Intake and Microbial Diversity and Taxa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gowlett, J.A. The discovery of fire by humans: A long and convoluted process. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150164. [Google Scholar] [CrossRef] [Green Version]
- Ragir, S. Diet and Food Preparation: Rethinking Early Hominid Behavior. Evol. Anthropol. 2000, 9, 153–155. [Google Scholar] [CrossRef]
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef]
- Pittelkow, C.M.; Liang, X.; Linquist, B.A.; van Groenigen, K.J.; Lee, J.; Lundy, M.E.; van Gestel, N.; Six, J.; Venterea, R.T.; van Kessel, C. Productivity limits and potentials of the principles of conservation agriculture. Nature 2015, 517, 365–368. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zuo, T.; Cheung, C.P.; Gu, W.; Wan, Y.; Zhang, F.; Chen, N.; Zhan, H.; Yeoh, Y.K.; Niu, J.; et al. Population-Level Configurations of Gut Mycobiome Across 6 Ethnicities in Urban and Rural China. Gastroenterology 2021, 160, 272–286.e11. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, A.J.; Zheng, J.J.; Kang, J.W.; Saboe, A.; Knights, D.; Zivkovic, A.M. A Guide to Diet-Microbiome Study Design. Front. Nutr. 2020, 7, 79. [Google Scholar] [CrossRef]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef] [Green Version]
- Gibson, R.S. Principles of Nutritional Assessment, 2nd ed.; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Thompson, F.E.; Subar, A.F. Dietary Assessment Methodology; Chapter 1; Academic Press: San Diego, CA, USA, 2017. [Google Scholar]
- Cade, J.E.; Burley, V.J.; Warm, D.L.; Thompson, R.L.; Margetts, B.M. Food-frequency questionnaires: A review of their design, validation and utilisation. Nutr. Res. Rev. 2004, 17, 5–22. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, M.D.; Martín-Lagos, R.A. Guía Para Estudios Dietéticos. Album Fotográfico de Alimentos, 1st ed.; Universidad de Granada: Granada, Spain, 2010. [Google Scholar]
- Hercberg, S.; Deheeger, M.; Preziosi, P. Portions Alimentaries: Manuel Photos Pour L’Estimation des Quantités, 1st ed.; Economica: Paris, France, 2002. [Google Scholar]
- Moreiras, O.; Carbajal, A.; Cabrera, L. Tabla de Composición de Alimentos, 19th ed.; Pirámide: Madrid, Spain, 2019. [Google Scholar]
- Raper, N.; Perloff, B.; Ingwersen, L.; Steinfeldt, L.; Anand, J. An overview of USDA’s Dietary Intake Data System. J. Food Compos. Anal. 2004, 17, 545–555. [Google Scholar] [CrossRef]
- The USDA National Nutrient Database for Standard Reference. Release 27 ed.; 2015. Available online: http://www.ars.usda.gov/nea/bhnrc/mafcl (accessed on 25 March 2021).
- Cade, J.; Thompson, R.; Burley, V.; Warm, D. Development, validation and utilisation of food-frequency questionnaires—A review. Public Health Nutr. 2002, 5, 567–587. [Google Scholar] [CrossRef] [Green Version]
- Lombard, M.J.; Steyn, N.P.; Charlton, K.E.; Senekal, M. Application and interpretation of multiple statistical tests to evaluate validity of dietary intake assessment methods. Nutr. J. 2015, 14, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, W. Nutritional Epidemiology, 3rd ed.; Oxford University Press: New York, NY, USA, 2012. [Google Scholar]
- Willett, W.C.; Sampson, L.; Stampfer, M.J.; Rosner, B.; Bain, C.; Witschi, J.; Hennekens, C.H.; Speizer, F.E. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am. J. Epidemiol. 1985, 122, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.V.; Mañas, R.J.; Fernández, S.R.; de Dios, R. National Food Survey on Adults, the Elderly and Pregnant Women (ENALIA2). 2016. Available online: http://www.aesan.gob.es/AECOSAN/docs/documentos/seguridad_alimentaria/evaluacion_riesgos/Spanish_National_dietary_survey_adults_elderly_pregnant.pdf (accessed on 17 February 2021).
- Arnolds, K.L.; Martin, C.G.; Lozupone, C.A. Blood type and the microbiome- untangling a complex relationship with lessons from pathogens. Curr. Opin. Microbiol. 2020, 56, 59–66. [Google Scholar] [CrossRef]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.; Man, W.H.; Chu, M.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karl, J.P.; Armstrong, N.J.; McClung, H.L.; Player, R.A.; Rood, J.C.; Racicot, K.; Soares, J.W.; Montain, S.J. A diet of U.S. military food rations alters gut microbiota composition and does not increase intestinal permeability. J. Nutr. Biochem. 2019, 72, 108217. [Google Scholar] [CrossRef]
- Zimmer, J.; Lange, B.; Frick, J.S.; Sauer, H.; Zimmermann, K.; Schwiertz, A.; Rusch, K.; Klosterhalfen, S.; Enck, P. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur. J. Clin. Nutr. 2012, 66, 53–60. [Google Scholar] [CrossRef]
- Cani, P.D.; Everard, A. Talking microbes: When gut bacteria interact with diet and host organs. Mol. Nutr. Food Res. 2016, 60, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- McDonald, D.; Hyde, E.; Debelius, J.W.; Morton, J.T.; Gonzalez, A.; Ackermann, G.; Aksenov, A.A.; Behsaz, B.; Brennan, C.; Chen, Y.; et al. American Gut: An Open Platform for Citizen Science Microbiome Research. MSystems 2018, 3, e00031-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Battaglioli, E.J.; Kashyap, P.C. Diet Effects on Gut Microbiome Composition, Function, and Host Physiology; Elsevier Inc.: London, UK, 2018; pp. 755–766. [Google Scholar]
- SENC (Sociedad Española de Nutrición Comunitaria). Dietary guidelines for the Spanish population. Nutr. Hosp. 2016, 33, 1–48. [Google Scholar]
- Russolillo, G.; Baladia, E.; Moñino, M.; Marques-Lopes, I.; Farran, A.; Bonany, J.; Gilabert, V.; Astiasarán, I.; Raigón, M.D.; Martínez, A.; et al. Establecimiento del tamaño de raciones de consumo de frutas y hortalizas para su uso en guías alimentarias en el entorno español: Propuesta del Comité Científico de la Asociación 5 al día. Rev. Esp. Nutr. Hum. Diet. 2019, 23, 205–221. [Google Scholar] [CrossRef]
- Nelson, M.; Atkinson, M.; Darbyshire, S. Food photography. I: The perception of food portion size from photographs. Br. J. Nutr. 1994, 72, 649–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, M.; Atkinson, M.; Darbyshire, S. Food photography II: Use of food photographs for estimating portion size and the nutrient content of meals. Br. J. Nutr. 1996, 76, 31–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subar, A.F.; Thompson, F.E.; Smith, A.F.; Jobe, J.B.; Ziegler, R.G.; Potischman, N.; Schatzkin, A.; Hartman, A.; Swanson, C.; Kruse, L.; et al. Improving food frequency questionnaires: A qualitative approach using cognitive interviewing. J. Am. Diet. Assoc. 1995, 95, 781–788, quiz 789–790. [Google Scholar] [CrossRef]
- Subar, A.F.; Thompson, F.E.; Kipnis, V.; Midthune, D.; Hurwitz, P.; McNutt, S.; McIntosh, A.; Rosenfeld, S. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires: The Eating at America’s Table Study. Am. J. Epidemiol. 2001, 154, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Verger, E.O.; Armstrong, P.; Nielsen, T.; Chakaroun, R.; Aron-Wisnewsky, J.; Gobel, R.J.; Schutz, T.; Delaere, F.; Gausseres, N.; Clement, K.; et al. Dietary Assessment in the MetaCardis Study: Development and Relative Validity of an Online Food Frequency Questionnaire. J. Acad. Nutr. Diet. 2017, 117, 878–888. [Google Scholar] [CrossRef]
- Brown, C.C.; Kipnis, V.; Freedman, L.S.; Hartman, A.M.; Schatzkin, A.; Wacholder, S. Energy adjustment methods for nutritional epidemiology: The effect of categorization. Am. J. Epidemiol. 1994, 139, 323–338. [Google Scholar] [CrossRef] [Green Version]
- Shrout, P.E.; Fleiss, J.L. Intraclass correlations: Uses in assessing rater reliability. Psychol. Bull. 1979, 86, 420–428. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masson, L.F.; McNeill, G.; Tomany, J.O.; Simpson, J.A.; Peace, H.S.; Wei, L.; Grubb, D.A.; Bolton-Smith, C. Statistical approaches for assessing the relative validity of a food-frequency questionnaire: Use of correlation coefficients and the kappa statistic. Public Health Nutr. 2003, 6, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [Google Scholar] [CrossRef]
- Giavarina, D. Understanding Bland Altman analysis. Biochem. Med. 2015, 25, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Sarrabayrouse, G.; Landolfi, S.; Pozuelo, M.; Willamil, J.; Varela, E.; Clark, A.; Campos, D.; Herrera, C.; Santiago, A.; Machiels, K.; et al. Mucosal microbial load in Crohn’s disease: A potential predictor of response to faecal microbiota transplantation. EBioMedicine 2020, 51, 102611. [Google Scholar] [CrossRef] [Green Version]
- Santiago, A.; Sanchez, E.; Clark, A.; Pozuelo, M.; Calvo, M.; Yáñez, F.; Sarrabayrouse, G.; Perea, L.; Vidal, S.; Gallardo, A.; et al. Sequential Changes in the Mesenteric Lymph Node Microbiome and Immune Response during Cirrhosis Induction in Rats. MSystems 2019, 4, e00278-18. [Google Scholar] [CrossRef] [Green Version]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-scale Meta-omics Studies. BioRxiv 2021. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurer, J.; Taren, D.L.; Teixeira, P.J.; Thomson, C.A.; Lohman, T.G.; Going, S.B.; Houtkooper, L.B. The psychosocial and behavioral characteristics related to energy misreporting. Nutr. Rev. 2006, 64, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Tooze, J.A.; Subar, A.F.; Thompson, F.E.; Troiano, R.; Schatzkin, A.; Kipnis, V. Psychosocial predictors of energy underreporting in a large doubly labeled water study. Am. J. Clin. Nutr. 2004, 79, 795–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, A.; Pryer, J.; Shetty, P. The problem of accuracy in dietary surveys. Analysis of the over 65 UK National Diet and Nutrition Survey. J. Epidemiol. Community Health 2000, 54, 611–616. [Google Scholar] [CrossRef]
- Hebert, J.R.; Hurley, T.G.; Peterson, K.E.; Resnicow, K.; Thompson, F.E.; Yaroch, A.L.; Ehlers, M.; Midthune, D.; Williams, G.C.; Greene, G.W.; et al. Social desirability trait influences on self-reported dietary measures among diverse participants in a multicenter multiple risk factor trial. J. Nutr. 2008, 138, 226S–234S. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.M.; Abdel-Maksoud, M.F.; Crane, L.A.; Marcus, A.C.; Byers, T.E. Effects of social approval bias on self-reported fruit and vegetable consumption: A randomized controlled trial. Nutr. J. 2008, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Bohlscheid-Thomas, S.; Hoting, I.; Boeing, H.; Wahrendorf, J. Reproducibility and relative validity of food group intake in a food frequency questionnaire developed for the German part of the EPIC project. European Prospective Investigation into Cancer and Nutrition. Int. J. Epidemiol. 1997, 26 (Suppl. S1), S59–S70. [Google Scholar] [CrossRef]
- Haftenberger, M.; Heuer, T.; Heidemann, C.; Kube, F.; Krems, C.; Mensink, G.B. Relative validation of a food frequency questionnaire for national health and nutrition monitoring. Nutr. J. 2010, 9, 36. [Google Scholar] [CrossRef] [Green Version]
- Harmouche-Karaki, M.; Mahfouz, M.; Obeyd, J.; Salameh, P.; Mahfouz, Y.; Helou, K. Development and validation of a quantitative food frequency questionnaire to assess dietary intake among Lebanese adults. Nutr. J. 2020, 19, 65. [Google Scholar] [CrossRef]
- Sierra-Ruelas, E.; Bernal-Orozco, M.F.; Macedo-Ojeda, G.; Marquez-Sandoval, Y.F.; Altamirano-Martinez, M.B.; Vizmanos, B. Validation of semiquantitative FFQ administered to adults: A systematic review. Public Health Nutr. 2020, 24, 3399–3418. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra382. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Bjorksten, B.; Engstrand, L.; Andersson, A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busi, S.B.; De Nies, L.; Habier, J.; Wampach, L.; Fritz, J.V.; Heintz-Buschart, A.; May, P.; Halder, R.; de Beaufort, C.; Wilmes, P. Persistence of birth mode-dependent effects on gut microbiome composition, immune system stimulation and antimicrobial resistance during the first year of life. ISME Commun. 2021, 1, 8. [Google Scholar] [CrossRef]
- De la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. MSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Serrano, J.; Smith, K.R.; Crouch, A.L.; Sharma, V.; Yi, F.; Vargova, V.; LaMoia, T.E.; Dupont, L.M.; Serna, V.; Tang, F.; et al. High-dose saccharin supplementation does not induce gut microbiota changes or glucose intolerance in healthy humans and mice. Microbiome 2021, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-Schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A.; et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Deehan, E.C.; Walter, J.; Backhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.; Peters, B.A.; Friedlander, C.; Freiman, H.J.; Goedert, J.J.; Sinha, R.; Miller, G.; Bernstein, M.A.; Hayes, R.B.; Ahn, J. Association of dietary fibre intake and gut microbiota in adults. Br. J. Nutr. 2018, 120, 1014–1022. [Google Scholar] [CrossRef] [Green Version]
- Carmody, R.N.; Bisanz, J.E.; Bowen, B.P.; Maurice, C.F.; Lyalina, S.; Louie, K.B.; Treen, D.; Chadaideh, K.S.; Maini Rekdal, V.; Bess, E.N.; et al. Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol. 2019, 4, 2052–2063. [Google Scholar] [CrossRef]



| Total | Pilot Study | Validation Study | |
|---|---|---|---|
| n | 84 | 40 | 44 |
| Age (years) | 34.2 ± 12.7 | 32.6 ± 11.1 | 35.7 ± 14.0 |
| 18–29 years, n (%) | 44 (52.4) | 21 (52.5) | 23 (52.3) |
| 30–39 years, n (%) | 19 (22.6) | 13 (32.5) | 6 (13.6) |
| 40–49 years, n (%) | 7 (8.3) | 2 (5.0) | 5 (11.4) |
| 50–59 years, n (%) | 9 (10.7) | 3 (7.5) | 6 (13.6) |
| >60 years, n (%) | 5 (4.2) | 1 (2.5) | 4 (9.1) |
| Female gender, n (%) | 47 (55.9) | 26 (65.0) | 21 (47.7) |
| BMI (kg/m2) | 22.5 ± 3.0 | 22.0 ± 2.6 | 23.1 ± 3.3 |
| Weight status, n (%) | |||
| Underweight (<18.5 kg/m2) | 5 (4.2) | 3 (7.5) | 2 (4.5) |
| Normal (18.5–24.9 kg/m2) | 67 (79.8) | 35 (87.5) | 32 (72.2) |
| Overweight (25–29.9 kg/m2) | 10 (11.9) | 2 (5.0) | 8 (18.1) |
| Obese (>30 kg/m2) | 2 (2.4) | 0 | 2 (4.5) |
| Nationality, n (%) | |||
| Spain | 68 (81.0) | 29 (72.5) | 39 (88.6) |
| European—non-Spanish | 8 (9.5) | 6 (15) | 2 (4.5) |
| Others | 8 (9.5) | 5 (12.5) | 3 (6.8) |
| Birth type, n (%) | |||
| Vaginal birth | 72 (85.7) | 35 (87.5) | 37 (84.1) |
| C-section | 12 (14.3) | 5 (12.5) | 7 (16.0) |
| Blood type, n (%) | |||
| A | 26 (30.9) | 14 (16.7) | 12 (14.3) |
| B | 4 (4.7) | 2 (2.4) | 2 (2.4) |
| AB | 1 (1.2) | 0 | 1 (1.2) |
| O | 33 (39.3) | 15 (17.9) | 18 (21.4) |
| Unknown | 20 (23.8) | 9 (10.7) | 11 (13.1) |
| Smoking status, n (%) | |||
| Non-smoker | 61 (72.6) | 32 (80) | 29 (65.9) |
| Smoker | 9 (10.7) | 2 (5) | 7 (15.9) |
| Former smoker | 8 (9.5) | 0 | 8 (18.2) |
| Unknown | 6 (7.1) | 6 (15) | 0 |
| Diet type, n (%) | |||
| Conventional | 67 (79.8) | 31 (77.5) | 37 (84.1) |
| Vegetarian diet | 6 (7.1) | 4 (10.0) | 2 (4.5) |
| Vegan diet | 2 (2.4) | 1 (2.5) | 1 (2.3) |
| Organic diet | 2 (2.4) | 2 (5.0) | 0 |
| Others diet | 7 (8.3) | 2 (5.0) | 4 (9.1) |
| Intake of ready-to-eat meals, n (%) | |||
| Yes | 51 (60.7) | 23 (57.5) | 28 (63.6) |
| No | 33 (39.3) | 17 (42.5) | 16 (36.4) |
| Intake of sweeteners, n (%) | |||
| Yes | - | ¯ | 11 (25.0) |
| No | - | ¯ | 33 (75.0) |
| Intake of supplements or drugs, n (%) | |||
| Dietary supplements | 20 (23.8) | 8 (20.0) | 12 (27.3) |
| Probiotics | 1 (1.2) | 1 (2.5) | 0 |
| Oral contraceptive | 6 (7.1) | 4 (10.0) | 2 (4.5) |
| ACE inhibidors | 3 (3.6) | 2 (5.0) | 1 (2.3) |
| Fibrate | 1 (1.2) | 1 (2.5) | 0 |
| Statin | 1 (1.2) | 1 (2.5) | 0 |
| Levothyroxine | 2 (2.4) | 1 (2.5) | 1 (2.3) |
| Other drugs | 8 (9.5) | 4 (10.0) | 4 (9.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yáñez, F.; Soler, Z.; Oliero, M.; Xie, Z.; Oyarzun, I.; Serrano-Gómez, G.; Manichanh, C. Integrating Dietary Data into Microbiome Studies: A Step Forward for Nutri-Metaomics. Nutrients 2021, 13, 2978. https://doi.org/10.3390/nu13092978
Yáñez F, Soler Z, Oliero M, Xie Z, Oyarzun I, Serrano-Gómez G, Manichanh C. Integrating Dietary Data into Microbiome Studies: A Step Forward for Nutri-Metaomics. Nutrients. 2021; 13(9):2978. https://doi.org/10.3390/nu13092978
Chicago/Turabian StyleYáñez, Francisca, Zaida Soler, Manon Oliero, Zixuan Xie, Iñigo Oyarzun, Gerard Serrano-Gómez, and Chaysavanh Manichanh. 2021. "Integrating Dietary Data into Microbiome Studies: A Step Forward for Nutri-Metaomics" Nutrients 13, no. 9: 2978. https://doi.org/10.3390/nu13092978
APA StyleYáñez, F., Soler, Z., Oliero, M., Xie, Z., Oyarzun, I., Serrano-Gómez, G., & Manichanh, C. (2021). Integrating Dietary Data into Microbiome Studies: A Step Forward for Nutri-Metaomics. Nutrients, 13(9), 2978. https://doi.org/10.3390/nu13092978








