Protective Effects of Collagen Tripeptides in Human Aortic Endothelial Cells by Restoring ROS-Induced Transcriptional Repression
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. DNA Microarray Analysis
2.4. Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-qPCR) Analysis
2.5. Production of Lentiviral Vector Encoding shRNA Sequences against Interleukin-3 Receptor Subunit Alpha (IL3RA)
2.6. Transduction of Lentiviral Vector
2.7. Statistical Analysis
3. Results
3.1. Microarray Analysis of Gene Expression Changes under Oxidative Stress and Effects of CTP in HAECs and SMCs
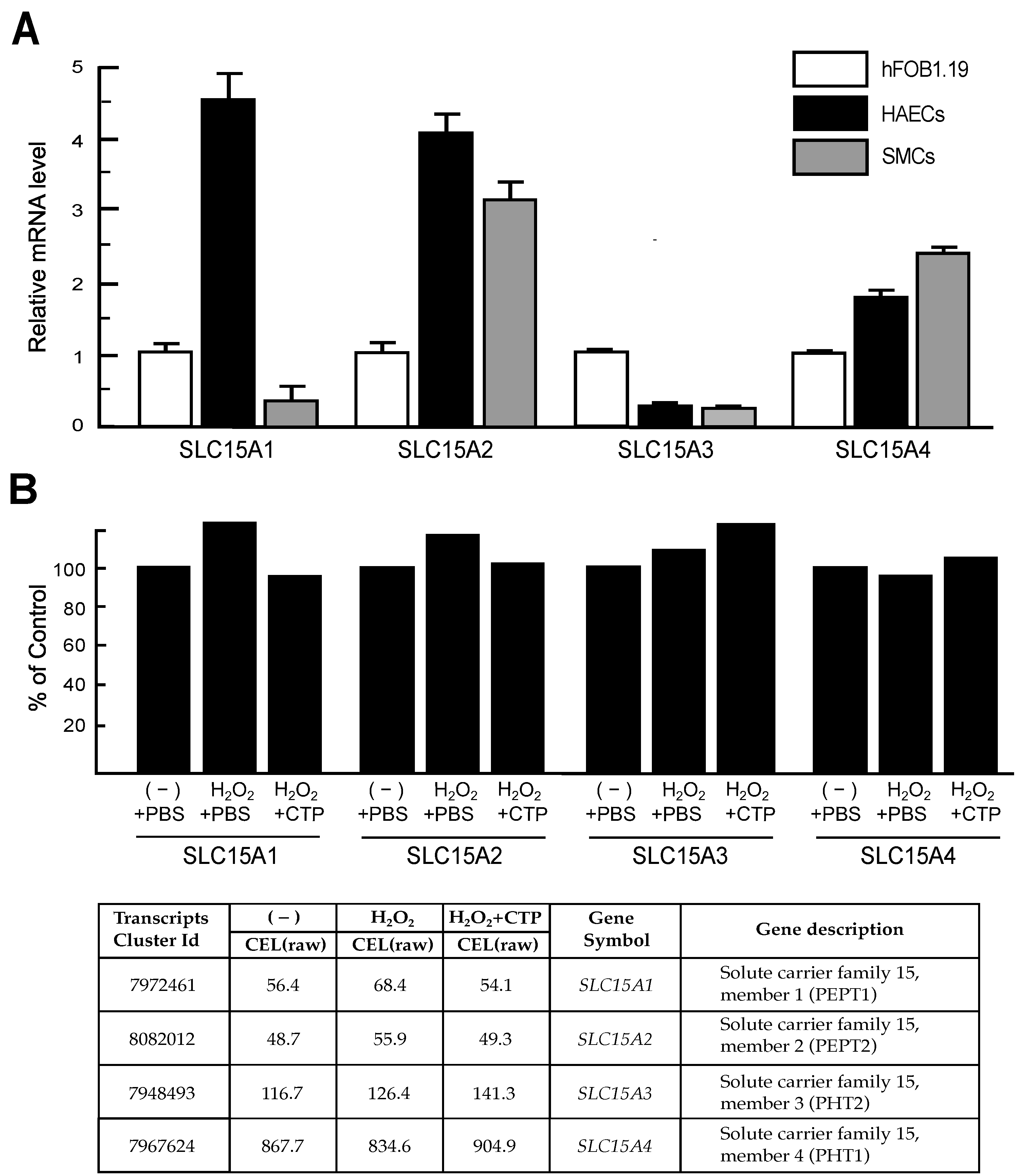
3.2. RT-qPCR Analysis of Genes Recovered in CTP-Treated HAECs
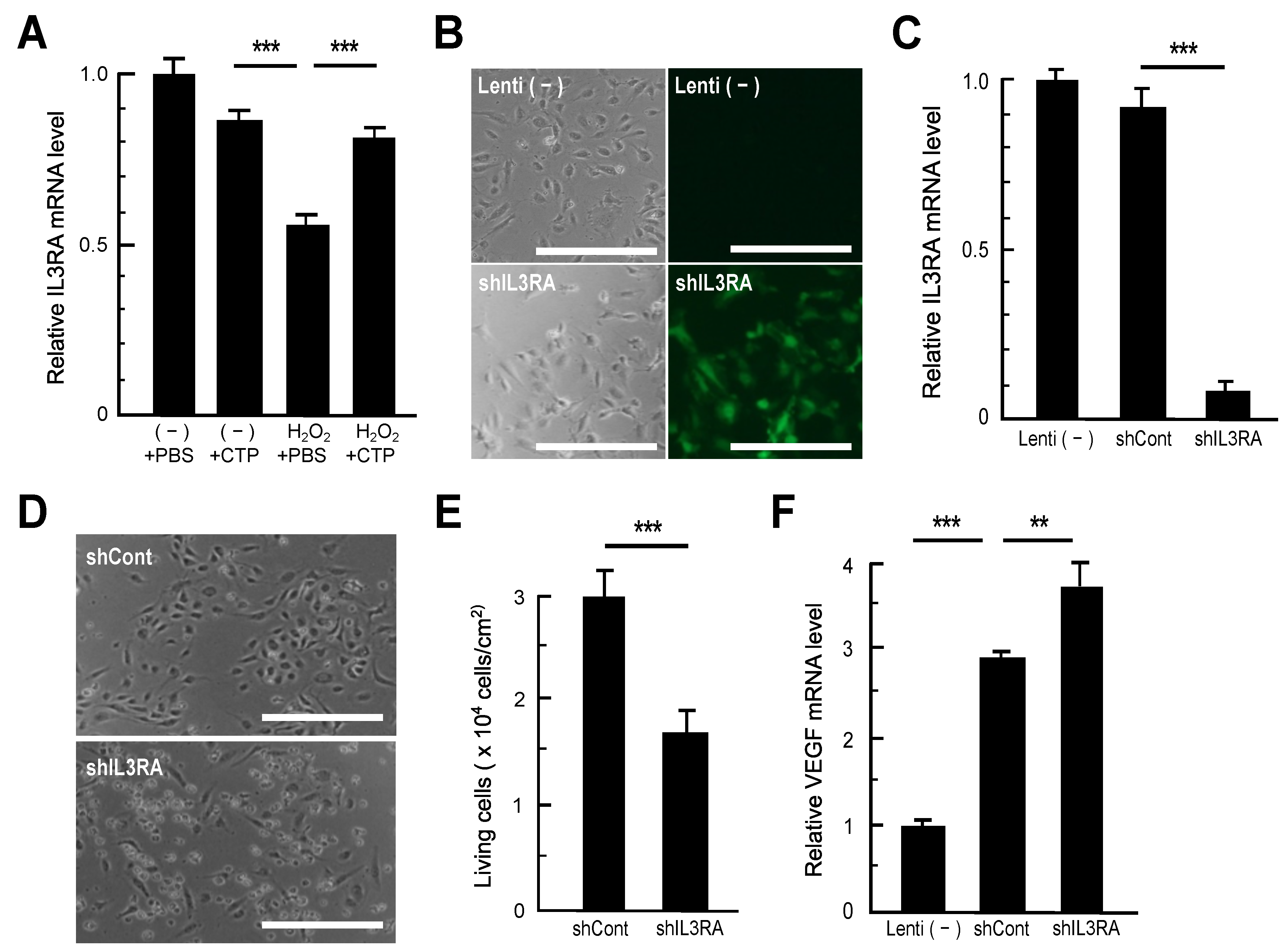
3.3. Impacts of IL3RA Suppression on HAECs
3.4. Peptide Transporter Expression in HAECs and SMCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakai, Y.; Yamato, R.; Onuma, M.; Kikuta, T.; Watanabe, M.; Nakayama, T. Non-antigenic and low allergic gelatin produced by specific digestion with an enzyme-coupled matrix. Biol. Pharm. Bull. 1998, 21, 330–334. [Google Scholar] [CrossRef]
- Tsuruoka, N.; Yamato, R.; Sakai, Y.; Yoshitake, Y.; Yonekura, H. Promotion by collagen tripeptide of type I collagen gene expression in human osteoblastic cells and fracture healing of rat femur. Biosci. Biotechnol. Biochem. 2007, 71, 2680–2687. [Google Scholar] [CrossRef]
- Yamamoto, S.; Deguchi, K.; Onuma, M.; Numata, N.; Sakai, Y. Absorption and urinary excretion of peptides after collagen tripeptide ingestion in humans. Biol. Pharm. Bull. 2016, 39, 428–434. [Google Scholar] [CrossRef]
- Yamamoto, S.; Hayasaka, F.; Deguchi, K.; Okudera, T.; Furusawa, T.; Sakai, Y. Absorption and plasma kinetics of collagen tripeptide after peroral or intraperitoneal administration in rats. Biosci. Biotechnol. Biochem. 2015, 79, 2026–2033. [Google Scholar] [CrossRef] [PubMed]
- Naraoka, T.; Ishibashi, Y.; Tsuda, E.; Yamamoto, Y.; Kusumi, T.; Toh, S. Periodic knee injections of collagen tripeptide delay cartilage degeneration in rabbit experimental osteoarthritis. Arthritis Res. Ther. 2013, 15, R32. [Google Scholar] [CrossRef] [PubMed]
- Hakuta, A.; Yamaguchi, Y.; Okawa, T.; Yamamoto, S.; Sakai, Y.; Aihara, M. Anti-inflammatory effect of collagen tripeptide in atopic dermatitis. J. Dermatol. Sci. 2017, 88, 357–364. [Google Scholar] [CrossRef]
- Pyun, H.B.; Kim, M.; Park, J.; Sakai, Y.; Numata, N.; Shin, J.Y.; Shin, H.J.; Kim, D.U.; Hwang, J.K. Effects of collagen tripeptide supplement on photoaging and epidermal skin barrier in UVB-exposed hairless mice. Prev. Nutr. Food Sci. 2012, 17, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Okawa, T.; Yamaguchi, Y.; Takada, S.; Sakai, Y.; Numata, N.; Nakamura, F.; Nagashima, Y.; Ikezawa, Z.; Aihara, M. Oral administration of collagen tripeptide improves dryness and pruritus in the acetone-induced dry skin model. J. Dermatol. Sci. 2012, 66, 136–143. [Google Scholar] [CrossRef]
- Tak, Y.J.; Shin, D.K.; Kim, A.H.; Kim, J.I.; Lee, Y.L.; Ko, H.C.; Kim, Y.W.; Lee, S.Y. Effect of collagen tripeptide and adjusting for climate change on skin hydration in middle-aged women: A randomized, double-blind, placebo-controlled trial. Front. Med. 2021, 7, 1–11. [Google Scholar] [CrossRef]
- Tang, L.; Sakai, Y.; Ueda, Y.; Katsuda, S. Effects of oral administration of tripeptides derived from type I collagen (collagen tripeptide) on atherosclerosis development in hypercholesterolemic rabbits. J. Biosci. Bioeng. 2015, 119, 558–563. [Google Scholar] [CrossRef]
- Tomosugi, N.; Yamamoto, S.; Takeuchi, M.; Yonekura, H.; Ishigaki, Y.; Numata, N.; Katsuda, S.; Sakai, Y. Effect of collagen tripeptide on atherosclerosis in healthy humans. J. Atheroscler. Thromb. 2017, 24, 530–538. [Google Scholar] [CrossRef]
- Hansson, G.K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005, 352, 1685–1695. [Google Scholar] [CrossRef]
- Daiber, A.; Steven, S.; Weber, A.; Shuvaev, V.V.; Muzykantov, V.R.; Laher, I.; Li, H.; Lamas, S.; Münzel, T. Targeting vascular (endothelial) dysfunction. Br. J. Pharmacol. 2017, 174, 1591–1619. [Google Scholar] [CrossRef]
- Vanhoutte, P.M.; Shimokawa, H.; Feletou, M.; Tang, E.H.C. Endothelial dysfunction and vascular disease—A 30th anniversary update. Acta Physiol. 2017, 219, 22–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, Y.; Li, Y.; Ren, X.; Zhang, X.; Hu, D.; Gao, Y.; Xing, Y.; Shang, H. Oxidative stress-mediated atherosclerosis: Mechanisms and therapies. Front. Physiol. 2017, 8, 600. [Google Scholar] [CrossRef]
- Ikeda, T.; Yoshitomi, Y.; Saito, H.; Shimasaki, T.; Yamaya, H.; Kobata, T.; Ishigaki, Y.; Tomosugi, N.; Yoshitake, Y.; Yonekura, H. Regulation of soluble Flt-1 (VEGFR-1) production by hnRNP D and protein arginine methylation. Mol. Cell. Biochem. 2016, 413, 155–164. [Google Scholar] [CrossRef]
- Yoshitomi, Y.; Ikeda, T.; Saito, H.; Yoshitake, Y.; Ishigaki, Y.; Hatta, T.; Kato, N.; Yonekura, H. JunB regulates angiogenesis and neurovascular parallel alignment in mouse embryonic skin. J. Cell Sci. 2017, 130, 916–926. [Google Scholar] [CrossRef]
- Coyle, C.H.; Kader, K.N. Mechanisms of H2O2-induced oxidative stress in endothelial cells exposed to physiologic shear stress. ASAIO J. 2007, 53, 17–22. [Google Scholar] [CrossRef]
- Kremer, E.; Baker, E.; D’Andrea, R.J.; Slim, R.; Phillips, H.; Moretti, P.A.B.; Lopez, A.F.; Petit, C.; Vadas, M.A.; Sutherland, G.R.; et al. A cytokine receptor gene cluster in the X-Y pseudoautosomal region? Blood 1993, 82, 22–28. [Google Scholar] [CrossRef]
- Stratman, A.N.; Davis, M.J.; Davis, G.E. VEGF and FGF prime vascular tube morphogenesis and sprouting directed by hematopoietic stem cell cytokines. Blood 2011, 117, 3709–3719. [Google Scholar] [CrossRef]
- Smith, A.O.; Bowers, S.L.K.; Stratman, A.N.; Davis, G.E. Hematopoietic stem cell cytokines and fibroblast growth factor-2 stimulate human endothelial cell-pericyte tube co-assembly in 3D fibrin matrices under serum-free defined conditions. PLoS ONE 2013, 8, e85147. [Google Scholar] [CrossRef]
- Yoo, J.; Lee, H.N.; Choi, I.; Choi, D.; Chung, H.K.; Kim, K.E.; Lee, S.; Aguilar, B.; Kang, J.; Park, E.; et al. Opposing regulation of PROX1 by interleukin-3 receptor and NOTCH directs differential host cell fate reprogramming by kaposi sarcoma herpes virus. PLoS Pathog. 2012, 8, e1002770. [Google Scholar] [CrossRef]
- Ikeda, T.; Sun, L.; Tsuruoka, N.; Ishigaki, Y.; Yoshitomi, Y.; Yoshitake, Y.; Yonekura, H. Hypoxia down-regulates sFlt-1 (sVEGFR-1) expression in human microvascular endothelial cells by a mechanism involving mRNA alternative processing. Biochem. J. 2011, 436, 399–407. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Kaur, H.; Li, J.J.; Bay, B.H.; Yung, L.Y.L. Investigating the antiproliferative activity of high affinity DNA aptamer on cancer cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Brown, J.M. Tumor hypoxia in cancer therapy. Methods Enzymol. 2007, 435, 295–321. [Google Scholar] [CrossRef]
- Adibi, S.A. Regulation of expression of the intestinal oligopeptide transporter (Pept-1) in health and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G779–G788. [Google Scholar] [CrossRef]
- Smith, D.E.; Clémençon, B.; Hediger, M.A. Proton-coupled oligopeptide transporter family SLC15: Physiological, pharmacological and pathological implications. Mol. Asp. Med. 2013, 34, 323–336. [Google Scholar] [CrossRef]
- Morikiri, Y.; Matsuta, E.; Inoue, H. The collagen-derived compound collagen tripeptide induces collagen expression and extends lifespan via a conserved p38 mitogen-activated protein kinase cascade. Biochem. Biophys. Res. Commun. 2018, 505, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, T.; Kawakami, K.; Uraji, M. Inhibitory effect of collagen-derived tripeptides on dipeptidylpeptidase-IV activity. J. Enzyme Inhib. Med. Chem. 2014, 29, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Dentelli, P.; Del Sorbo, L.; Rosso, A.; Molinar, A.; Garbarino, G.; Camussi, G.; Pegoraro, L.; Brizzi, M.F. Human IL-3 stimulates endothelial cell motility and promotes in vivo new vessel formation. J. Immunol. 1999, 163, 2151–2159. [Google Scholar]
- Zeoli, A.; Dentelli, P.; Rosso, A.; Togliatto, G.; Trombetta, A.; Damiano, L.; Di Celle, P.F.; Pegoraro, L.; Altruda, F.; Brizzi, M.F. Interleukin-3 promotes expansion of hemopoietic-derived CD45+ angiogenic cells and their arterial commitment via STAT5 activation. Blood 2008, 112, 350–361. [Google Scholar] [CrossRef][Green Version]
- Bowers, S.L.K.; Kemp, S.S.; Aguera, K.N.; Koller, G.M.; Forgy, J.C.; Davis, G.E. Defining an upstream VEGF (vascular endothelial growth factor) priming signature for downstream factor-induced endothelial cell-pericyte tube network coassembly. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2891–2909. [Google Scholar] [CrossRef]
- Takahashi, M. NLRP3 inflammasome as a key driver of vascular disease. Cardiovasc. Res. 2021, 1–14. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Chou, M.M.; Blenis, J. The 70 kDa S6 kinase: Regulation of a kinase with multiple roles in mitogenic signalling. Curr. Opin. Cell Biol. 1995, 7, 806–814. [Google Scholar] [CrossRef]

| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| IL3RA | 5′-CAGGCGTCAACAGTACGAGT-3′ | 5′-TTGCAGTCATGTTGGGTGGA-3′ |
| VEGF-A | 5′-GCCTCCGAAACATGAACTTTCTGCTG-3′ | 5′-TGGTGATGTTGGACTCCTCA-3′ |
| SLC15A1 (PEPT1) | 5′-ATTCCGCCACAATGTCAACC-3′ | 5′-CCGTGACAGAGAAGACCACT-3′ |
| SLC15A2 (PEPT2) | 5′-CCCTGAAGGAAACATAGTGGCTC-3′ | 5′-TAGCCAGTGCTGTCGCTTTGGA-3′ |
| SLC15A3 (PHT2) | 5′-GCAAGAGGACATCGCCAACTTC-3′ | 5′-TTGGGATGTGGAGGTGAAGACC-3′ |
| SLC15A4 (PHT1) | 5′-CCTCTGAAGGACAAACTGGTCG-3′ | 5′-ACAAGGTTCAGCCTTTTACTCTCC-3′ |
| Transcript Cluster ID | CEL (Raw) | Fold Change by CTP | Gene Symbol | Gene Description | ||
|---|---|---|---|---|---|---|
| (−) | H2O2 | H2O2 + CTP | ||||
| 7896750 1 | 144.8 | 44.2 | 94.9 | 2.15 | ||
| 8165663 1 | 204.4 | 56.2 | 125 | 2.22 | ||
| 8086538 | 160.9 | 72.7 | 155.7 | 2.14 | LIMD1-AS1 | LIMD1 antisense RNA 1 |
| 8113356 1 | 100.6 | 46.3 | 93.2 | 2.02 | ||
| 8059852 | 318.1 | 154.3 | 279.3 | 1.81 | MSL3P1 | Male-specific lethal 3 homolog (Drosophila) pseudogene 1 |
| 8176323 | 246 | 119.2 | 213.1 | 1.79 | IL3RA | Interleukin 3 receptor, alpha (low affinity) |
| 8097058 | 322.2 | 158.8 | 284 | 1.79 | CEP170L | Centrosomal protein 170 kDa-like |
| 8165752 | 246.2 | 119.6 | 213.4 | 1.78 | IL3RA | Interleukin 3 receptor, alpha (low affinity) |
| 7916898 | 407.7 | 81.1 | 132.1 | 1.63 | DEPDC1 | DEP domain containing 1 |
| 8165672 1 | 466.3 | 158.9 | 255.7 | 1.61 | ||
| 7914878 | 276.8 | 74.2 | 117.5 | 1.58 | CLSPN | Claspin |
| 7971653 | 225.1 | 80.1 | 126.3 | 1.58 | DLEU2 | Deleted in lymphocytic leukemia 2 (non-protein coding) |
| 7979307 | 936.5 | 242.2 | 373.1 | 1.54 | DLGAP5 | Discs, large (Drosophila) homolog-associated protein 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito-Takatsuji, H.; Yoshitomi, Y.; Ishigaki, Y.; Yamamoto, S.; Numata, N.; Sakai, Y.; Takeuchi, M.; Tomosugi, N.; Katsuda, S.; Yonekura, H.; et al. Protective Effects of Collagen Tripeptides in Human Aortic Endothelial Cells by Restoring ROS-Induced Transcriptional Repression. Nutrients 2021, 13, 2226. https://doi.org/10.3390/nu13072226
Saito-Takatsuji H, Yoshitomi Y, Ishigaki Y, Yamamoto S, Numata N, Sakai Y, Takeuchi M, Tomosugi N, Katsuda S, Yonekura H, et al. Protective Effects of Collagen Tripeptides in Human Aortic Endothelial Cells by Restoring ROS-Induced Transcriptional Repression. Nutrients. 2021; 13(7):2226. https://doi.org/10.3390/nu13072226
Chicago/Turabian StyleSaito-Takatsuji, Hidehito, Yasuo Yoshitomi, Yasuhito Ishigaki, Shoko Yamamoto, Noriaki Numata, Yasuo Sakai, Masayoshi Takeuchi, Naohisa Tomosugi, Shogo Katsuda, Hideto Yonekura, and et al. 2021. "Protective Effects of Collagen Tripeptides in Human Aortic Endothelial Cells by Restoring ROS-Induced Transcriptional Repression" Nutrients 13, no. 7: 2226. https://doi.org/10.3390/nu13072226
APA StyleSaito-Takatsuji, H., Yoshitomi, Y., Ishigaki, Y., Yamamoto, S., Numata, N., Sakai, Y., Takeuchi, M., Tomosugi, N., Katsuda, S., Yonekura, H., & Ikeda, T. (2021). Protective Effects of Collagen Tripeptides in Human Aortic Endothelial Cells by Restoring ROS-Induced Transcriptional Repression. Nutrients, 13(7), 2226. https://doi.org/10.3390/nu13072226







