Hepatic Nfe2l2 Is Not an Essential Mediator of the Metabolic Phenotype Produced by Dietary Methionine Restriction
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Experiment 1
2.3. Experiment 2
2.4. Analysis of Energy Expenditure
2.5. RNA Isolation and qPCR of Nfe2l2 and ER Stress Target Genes
2.6. RNAseq Analysis
2.7. Bioinformatics Analysis
2.8. Serum Metabolite Analyses
2.9. Data Analysis
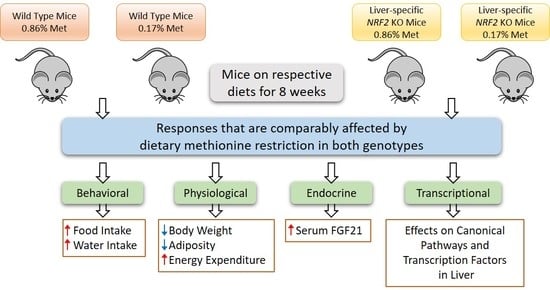
3. Results
3.1. Experiment 1
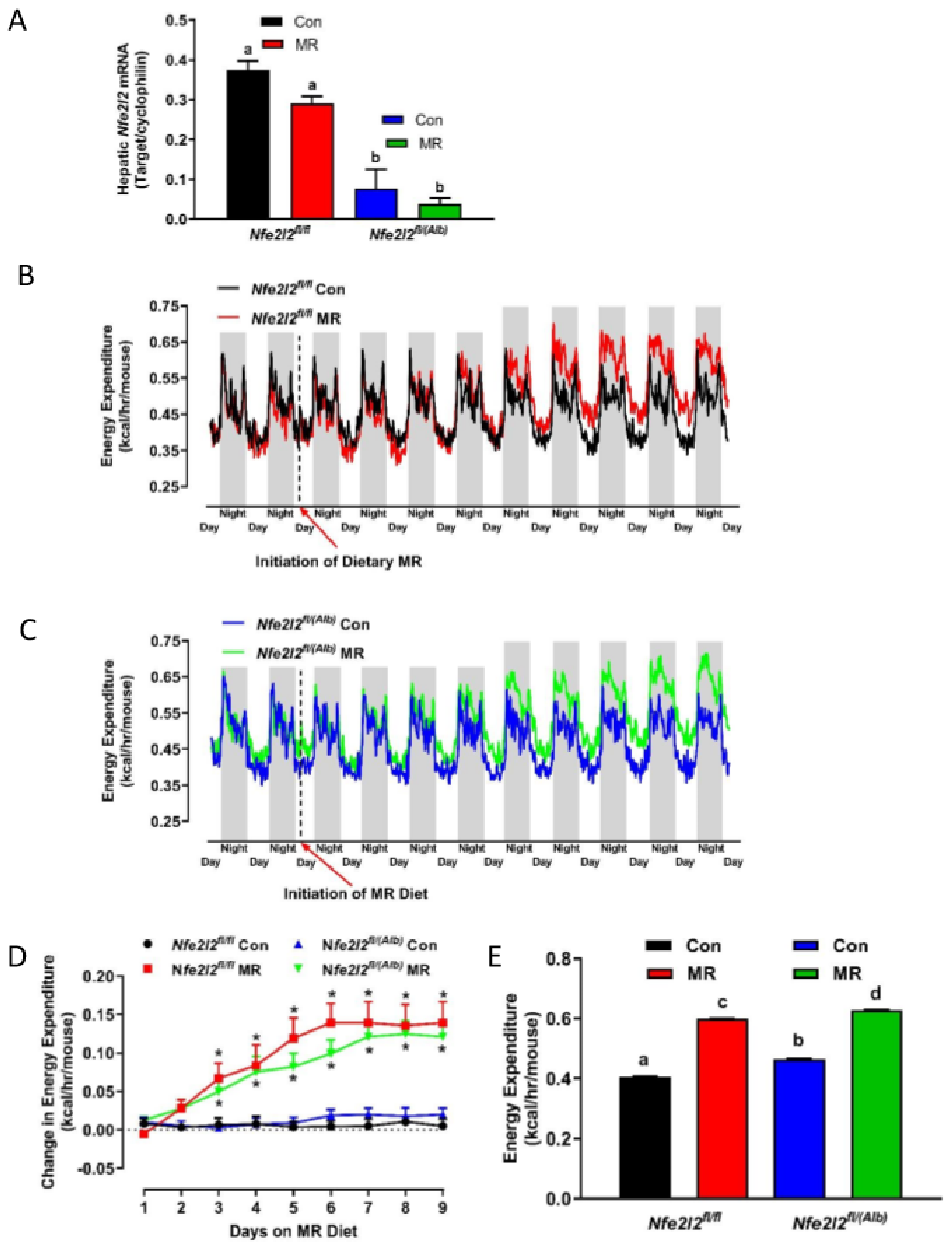
3.2. Experiment 2
3.3. Real Time qPCR
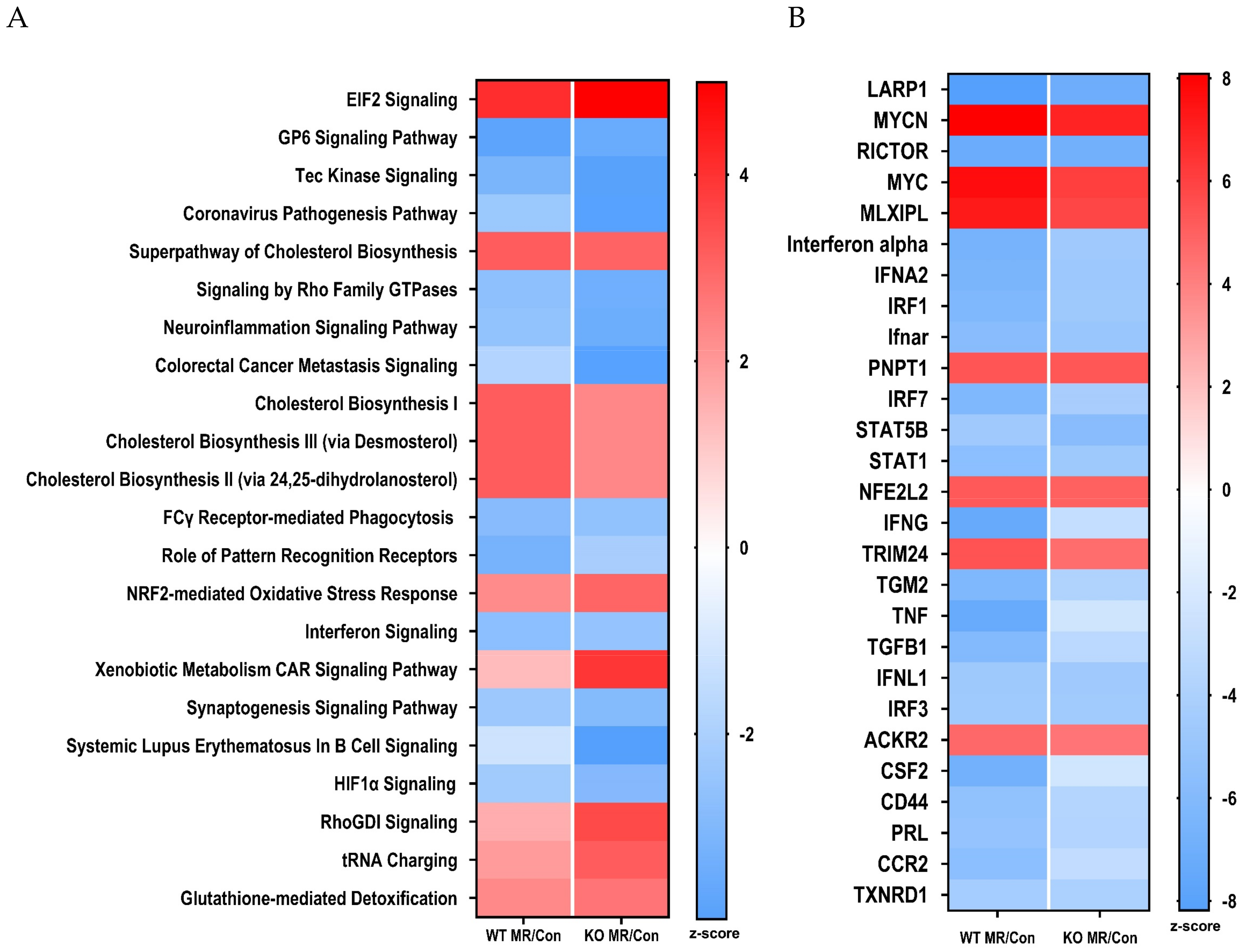
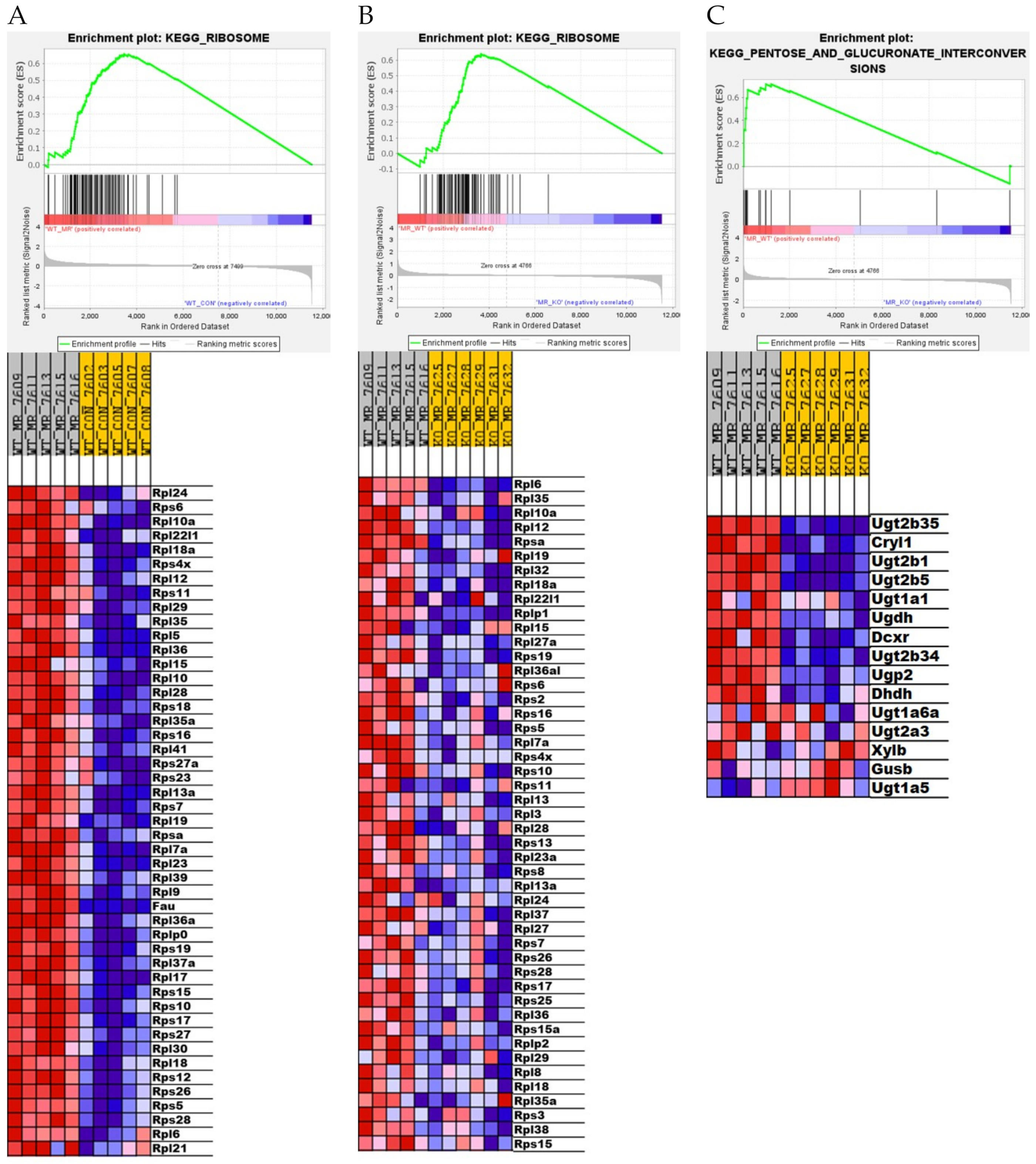
3.4. Differential Gene Expression in the Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orentreich, N.; Matias, J.R.; DeFelice, A.; Zimmerman, J.A. Low methionine ingestion by rats extends life span. J. Nutr. 1993, 123, 269–274. [Google Scholar] [PubMed]
- Richie, J.P., Jr.; Leutzinger, Y.; Parthasarathy, S.; Malloy, V.; Orentreich, N.; Zimmerman, J.A. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994, 8, 1302–1307. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Buehner, G.; Chang, Y.; Harper, J.M.; Sigler, R.; Smith-Wheelock, M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 2005, 4, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Malloy, V.L.; Krajcik, R.A.; Bailey, S.J.; Hristopoulos, G.; Plummer, J.D.; Orentreich, N. Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell 2006, 5, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Hasek, B.E.; Stewart, L.K.; Henagan, T.M.; Boudreau, A.; Lenard, N.R.; Black, C.; Shin, J.; Huypens, P.; Malloy, V.L.; Plaisance, E.P.; et al. Dietary methionine restriction enhances metabolic flexibility and increases uncoupled respiration in both fed and fasted states. Am. J. Physiol. Integr. Comp. Physiol. 2010, 299, R728–R739. [Google Scholar] [CrossRef]
- Perrone, C.E.; Malloy, V.L.; Orentreich, D.S.; Orentreich, N. Metabolic adaptations to methionine restriction that benefit health and lifespan in rodents. Exp. Gerontol. 2013, 48, 654–660. [Google Scholar] [CrossRef]
- Stone, K.P.; Wanders, D.; Orgeron, M.; Cortez, C.C.; Gettys, T.W. Mechanisms of Increased In Vivo Insulin Sensitivity by Dietary Methionine Restriction in Mice. Diabetes 2014, 63, 3721–3733. [Google Scholar] [CrossRef]
- Plaisance, E.P.; Henagan, T.M.; Echlin, H.; Boudreau, A.; Hill, K.L.; Lenard, N.R.; Hasek, B.E.; Orentreich, N.; Gettys, T.W. Role of ß-adrenergic receptors in the hyperphagic and hypermetabolic responses to dietary methionine restriction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R740–R750. [Google Scholar] [CrossRef]
- Orgeron, M.; Stone, K.P.; Wanders, D.; Cortez, C.C.; Van, N.T.; Gettys, T.W. The Impact of Dietary Methionine Restriction on Biomarkers of Metabolic Health. Prog. Mol. Biol. Transl. Sci. 2014, 121, 351–376. [Google Scholar]
- Wanders, D.; Ghosh, S.; Stone, K.P.; Van, N.T.; Gettys, T.W. Transcriptional impact of dietary methionine restriction on systemic inflammation: Relevance to biomarkers of metabolic disease during aging. BioFactors 2014, 40, 13–26. [Google Scholar] [CrossRef]
- Perrone, C.E.; Mattocks, D.A.; Plummer, J.D.; Chittur, S.V.; Mohney, R.; Vignola, K.; Orentreich, D.S.; Orentreich, N. Genomic and metabolic responses to methionine-restricted and methionine-restricted, cysteine-supplemented diets in Fischer 344 rat inguinal adipose tissue, liver and quadriceps muscle. J. Nutr. Nutr. 2012, 5, 132–157. [Google Scholar] [CrossRef]
- Wanders, D.; Forney, L.A.; Stone, K.P.; Burk, D.H.; Pierse, A.; Gettys, T.W. FGF21 Mediates the Thermogenic and Insulin-Sensitizing Effects of Dietary Methionine Restriction but Not Its Effects on Hepatic Lipid Metabolism. Diabetes 2017, 66, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Forney, L.A.; Fang, H.; Sims, L.C.; Stone, K.P.; Vincik, L.Y.; Vick, A.M.; Gibson, A.N.; Burk, D.H.; Gettys, T.W. Dietary Methionine Restriction Signals to the Brain Through Fibroblast Growth Factor 21 to Regulate Energy Balance and Remodeling of Adipose Tissue. Obesity 2020, 28, 1912–1921. [Google Scholar] [CrossRef] [PubMed]
- Wanders, D.; Burk, D.H.; Cortez, C.C.; Van, N.T.; Stone, K.P.; Baker, M.; Mendoza, T.; Mynatt, R.L.; Gettys, T.W. UCP1 is an essential mediator of the effects of methionine restriction on energy balance but not insulin sensitivity. FASEB J. 2015, 29, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Laeger, T.; Henagan, T.M.; Albarado, D.C.; Redman, L.M.; Bray, G.A.; Noland, R.C.; Münzberg, H.; Hutson, S.M.; Gettys, T.W.; Schwartz, M.W.; et al. FGF21 is an endocrine signal of protein restriction. J. Clin. Investig. 2014, 124, 3913–3922. [Google Scholar] [CrossRef] [PubMed]
- Deval, C.; Chaveroux, C.; Maurin, A.-C.; Cherasse, Y.; Parry, L.; Carraro, V.; Milenkovic, D.; Ferrara, M.; Bruhat, A.; Jousse, C.; et al. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 2009, 276, 707–718. [Google Scholar] [CrossRef]
- Shan, J.X.; Ord, D.; Ord, T.; Kilberg, M.S. Elevated ATF4 Expression, in the Absence of Other Signals, Is Sufficient for Transcriptional Induction via CCAAT Enhancer-binding Protein-activating Transcription Factor Response Elements. J. Biol. Chem. 2009, 284, 21241–21248. [Google Scholar] [CrossRef] [PubMed]
- Palii, S.S.; Kays, C.E.; Deval, C.; Bruhat, A.; Fafournoux, P.; Kilberg, M.S. Specificity of amino acid regulated gene expression: Analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids 2009, 37, 79–88. [Google Scholar] [CrossRef][Green Version]
- Palam, L.R.; Baird, T.D.; Wek, R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011, 286, 10939–10949. [Google Scholar] [CrossRef]
- Maruyama, R.; Shimizu, M.; Li, J.; Inoue, J.; Sato, R. Fibroblast growth factor 21 induction by activating transcription factor 4 is regulated through three amino acid response elements in its promoter region. Biosci. Biotechnol. Biochem. 2016, 80, 929–934. [Google Scholar] [CrossRef]
- De Sousa-Coelho, A.L.; Marrero, P.F.; Haro, D. Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. Biochem. J. 2012, 443, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Bunpo, P.; Cundiff, J.K.; Reinert, R.B.; Wek, R.C.; Aldrich, C.J.; Anthony, T.G. The eIF2 Kinase GCN2 Is Essential for the Murine Immune System to Adapt to Amino Acid Deprivation by Asparaginase. J. Nutr. 2010, 140, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.J.; Lennox, B.A.; She, P.X.; Mirek, E.T.; Al Baghdadi, R.J.T.; Fusakio, M.E.; Dixon, J.L.; Henderson, G.C.; Wek, R.C.; Anthony, T.G. GCN2 is required to increase fibroblast growth factor 21 and maintain hepatic triglyceride homeostasis during asparaginase treatment. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E283–E293. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.S.; Lu, X.H.; Xiao, Y.C.; Lin, Y.; Zhu, H.; Ding, T.; Yang, Y.; Huang, Y.; Zhang, Y.; Liu, Y.L.; et al. ATF4-and CHOP-Dependent Induction of FGF21 through Endoplasmic Reticulum Stress. BioMed Res. Int. 2014, 2014, 807874. [Google Scholar] [CrossRef] [PubMed]
- Wanders, D.; Stone, K.P.; Forney, L.A.; Cortez, C.C.; Dille, K.N.; Simon, J.; Xu, M.; Hotard, E.C.; Nikonorova, I.A.; Pettit, A.P.; et al. Role of GCN2-independent signaling through a non-canonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes 2016, 65, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Pettit, A.P.; Jonsson, W.O.; Bargoud, A.R.; Mirek, E.T.; Peelor, F.F., III; Wang, Y.; Gettys, T.W.; Kimball, S.R.; Miller, B.F.; Hamilton, K.L.; et al. Dietary Methionine Restriction Regulates Liver Protein Synthesis and Gene Expression Independently of Eukaryotic Initiation Factor 2 Phosphorylation in Mice. J. Nutr. 2017, 147, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, W.O.; Margolies, N.S.; Mirek, E.T.; Zhang, Q.; Linden, M.; Hill, C.; Link, C.; Bithi, N.; Zalma, B.; Levy, J.; et al. Physiological responses to dietary sulfur amino acid restriction are influenced by Atf4 status and biological sex. J. Nutr. 2021, 151, 785–799. [Google Scholar] [CrossRef]
- He, C.H.; Gong, P.F.; Hu, B.; Stewart, D.; Choi, M.E.; Choi, A.M.K.; Alam, J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein—Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001, 276, 20858–20865. [Google Scholar] [CrossRef] [PubMed]
- Mimura, J.; Inose-Maruyama, A.; Taniuchi, S.; Kosaka, K.; Yoshida, H.; Yamazaki, H.; Kasai, S.; Harada, N.; Kaufman, R.J.; Oyadomari, S.; et al. Concomitant Nrf2- and ATF4-activation by Carnosic Acid Cooperatively Induces Expression of Cytoprotective Genes. Int. J. Mol. Sci. 2019, 20, 1706. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Diehl, J.A. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J. Biol. Chem. 2004, 279, 20108–20117. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Zhang, D.; Hannink, M.; Arvisais, E.; Kaufman, R.J.; Diehl, J.A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003, 23, 7198–7209. [Google Scholar] [CrossRef]
- Chanas, S.A.; Jiang, Q.; McMahon, M.; McWalter, G.K.; McLellan, L.I.; Elcombe, C.R.; Henderson, C.J.; Wolf, C.R.; Moffat, G.J.; Itoh, K.; et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 2002, 365, 405–416. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef] [PubMed]
- Elshorbagy, A.K.; Valdivia-Garcia, M.; Refsum, H.; Smith, A.D.; Mattocks, D.A.; Perrone, C.E. Sulfur amino acids in methionine-restricted rats: Hyperhomocysteinemia. Nutrition 2010, 26, 1201–1204. [Google Scholar] [CrossRef]
- Elshorbagy, A.K.; Valdivia-Garcia, M.; Mattocks, D.A.L.; Plummer, J.D.; Smith, A.D.; Drevon, C.A.; Refsum, H.; Perrone, C.E. Cysteine supplementation reverses methionine restriction effects on rat adiposity: Significance of stearoyl-coenzyme A desaturase. J. Lipid Res. 2011, 52, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Forney, L.A.; Wanders, D.; Stone, K.P.; Gettys, T.W. An integrative analysis of tissue-specific transcriptomic and metabolomic responses to short-term dietary methionine restriction in mice. PLoS ONE 2017, 12, e0177513. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.R.; Cho, J.M.; Shin, D.H.; Yong, C.S.; Choi, H.G.; Wakabayashi, N.; Kwak, M.K. Adaptive response to GSH depletion and resistance to L-buthionine-(S,R)-sulfoximine: Involvement of Nrf2 activation. Mol. Cell. Biochem. 2008, 318, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Cvetko, F.; Caldwell, S.T.; Higgins, M.; Suzuki, T.; Yamamoto, M.; Prag, H.A.; Hartley, R.C.; Dinkova-Kostova, A.T.; Murphy, M.P. Nrf2 is activated by disruption of mitochondrial thiol homeostasis but not by enhanced mitochondrial superoxide production. J. Biol. Chem. 2020, 296, 100169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Reiner, A.; Yekutieli, D.; Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003, 19, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Wanders, D.; Forney, L.A.; Stone, K.P.; Hasek, B.E.; Johnson, W.D.; Gettys, T.W. The Components of Age-Dependent Effects of Dietary Methionine Restriction on Energy Balance in Rats. Obesity 2018, 26, 740–746. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Stone, K.P.; Ghosh, S.; Kovalik, J.P.; Orgeron, M.; Wanders, D.; Sims, L.C.; Gettys, T.W. The acute transcriptional responses to dietary methionine restriction are triggered by inhibition of ternary complex formation and linked to Erk1/2, mTOR, and ATF4. Sci. Rep. 2021, 11, 3765. [Google Scholar] [CrossRef]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Nguyen, T.; Sherratt, P.J.; Huang, H.C.; Yang, C.S.; Pickett, C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element—Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003, 278, 4536–4541. [Google Scholar] [CrossRef]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef]
- Levonen, A.L.; Hill, B.G.; Kansanen, E.; Zhang, J.; Darley-Usmar, V.M. Redox regulation of antioxidants, autophagy, and the response to stress: Implications for electrophile therapeutics. Free Radic. Biol. Med. 2014, 71, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Bryan, H.K.; Olayanju, A.; Goldring, C.E.; Park, B.K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 2013, 85, 705–717. [Google Scholar] [CrossRef]
- Donnelly, N.; Gorman, A.M.; Gupta, S.; Samali, A. The eIF2alpha kinases: Their structures and functions. Cell. Mol. Life Sci. 2013, 70, 3493–3511. [Google Scholar] [CrossRef] [PubMed]
- Pomatto, L.C.D.; Dill, T.; Carboneau, B.; Levan, S.; Kato, J.; Mercken, E.M.; Pearson, K.J.; Bernier, M.; de Cabo, R. Deletion of Nrf2 shortens lifespan in C57BL6/J male mice but does not alter the health and survival benefits of caloric restriction. Free Radic. Biol. Med. 2020, 152, 650–658. [Google Scholar] [CrossRef]
- Yoh, K.; Itoh, K.; Enomoto, A.; Hirayama, A.; Yamaguchi, N.; Kobayashi, M.; Morito, N.; Koyama, A.; Yamamoto, M.; Takahashi, S. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001, 60, 1343–1353. [Google Scholar] [CrossRef]
- Tullet, J.M.A.; Green, J.W.; Au, C.; Benedetto, A.; Thompson, M.A.; Clark, E.; Gilliat, A.F.; Young, A.; Schmeisser, K.; Gems, D. The SKN-1/Nrf2 transcription factor can protect against oxidative stress and increase lifespan in C. elegans by distinct mechanisms. Aging Cell 2017, 16, 1191–1194. [Google Scholar] [CrossRef] [PubMed]
- Bhide, S.; Trujillo, A.S.; O’Connor, M.T.; Young, G.H.; Cryderman, D.E.; Chandran, S.; Nikravesh, M.; Wallrath, L.L.; Melkani, G.C. Increasing autophagy and blocking Nrf2 suppress laminopathy-induced age-dependent cardiac dysfunction and shortened lifespan. Aging Cell 2018, 17, e12747. [Google Scholar] [CrossRef]


| Gene | Signaling | Nfe2l2fl/fl | Nfe2l2fl/(Alb) | ||
|---|---|---|---|---|---|
| Symbol 1 | Pathway | Control | MR | Control | MR |
| Aox1 | Nfe2l2 | 1 ± 0.25 a | 1.28 ± 0.20 a | 0.35 ± 0.16 b | 0.40 ± 0.09 b |
| Gclc | Nfe2l2 | 1 ± 0.13 a | 0.90 ± 0.07 a | 0.45 ± 0.04 b | 0.38 ± 0.03 b |
| Gsr | Nfe2l2 | 1 ± 0.08 a,b | 1.20 ± 0.11 a | 0.73 ± 0.05 b | 0.86 ± 0.12 b |
| Txnrd1 | Nfe2l2 | 1 ± 0.04 a | 1.20 ± 0.18 a | 0.68 ± 0.05 b | 0.80 ± 0.05 a,b |
| Trb3 | ER stress | 1 ± 0.22 a | 1.24 ± 0.13 a | 0.37 ± 0.05 b | 0.71 ± 0.08 a,b |
| Atf4 | ER Stress | 1 ± 0.05 a,b | 1.11 ± 0.08 b | 0.68 ± 0.04 c | 0.86 ± 0.04 a,c |
| Xbp1s | ER Stress | 1 ± 0.19 a | 0.66 ± 0.13 a,b | 0.45 ± 0.06 b,c | 0.44 ± 0.07 b,c |
| Ephx1 | Nfe2l2 | 1 ± 0.17 a | 1.42 ± 0.07 b | 0.35 ± 0.11 c | 0.43 ± 0.05 c |
| Sod2 | Nfe2l2 | 1 ± 0.18 a | 0.60 ± 0.08 b | 0.54 ± 0.04 b | 0.62 ± 0.07 b |
| Gsta2 | Nfe2l2 | 1 ± 0.32 a | 4.86 ± 0.63 b | 0.15 ± 0.09 c | 0.66 ± 0.20 a,c |
| Cbr1 | Nfe2l2 | 1 ± 0.20 a | 3.45 ± 0.52 b | 0.93 ± 0.14 a | 1.10 ± 0.17 a |
| Mgst3 | Nfe2l2 | 1 ± 0.13 a | 1.97 ± 0.16 b | 0.92 ± 0.18 a | 1.34 ± 0.18 a |
| Nqo1 | Nfe2l2 | 1 ± 0.15 a | 2.28 ± 0.16 b | 0.39 ± 0.09 a | 0.71 ± 0.13 a |
| Asns | ER stress | 1 ± 0.32 a | 12.23 ± 1.49 b | 0.74 ± 0.16 a | 10.59 ± 1.59 b |
| Cyp4a14 | ER stress | 1 ± 0.21 a | 1.74 ± 0.23 b | 0.97 ± 0.25 a | 2.13 ± 0.19 b |
| Fgf21 | ER stress | 1 ± 0.36 a | 2.77 ± 0.60 b | 1.04 ± 0.20 a | 2.90 ± 0.76 b |
| Psat1 | ER stress | 1 ± 0.33 a | 7.54 ± 0.96 b | 0.48 ± 0.12 a | 3.27 ± 0.54 c |
| Vldlr | ER stress | 1 ± 0.21 a | 3.25 ± 0.76 b | 1.02 ± 0.10 a | 3.79 ± 0.59 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Stone, K.P.; Ghosh, S.; Forney, L.A.; Sims, L.C.; Vincik, L.; Gettys, T.W. Hepatic Nfe2l2 Is Not an Essential Mediator of the Metabolic Phenotype Produced by Dietary Methionine Restriction. Nutrients 2021, 13, 1788. https://doi.org/10.3390/nu13061788
Fang H, Stone KP, Ghosh S, Forney LA, Sims LC, Vincik L, Gettys TW. Hepatic Nfe2l2 Is Not an Essential Mediator of the Metabolic Phenotype Produced by Dietary Methionine Restriction. Nutrients. 2021; 13(6):1788. https://doi.org/10.3390/nu13061788
Chicago/Turabian StyleFang, Han, Kirsten P. Stone, Sujoy Ghosh, Laura A. Forney, Landon C. Sims, LeighAnn Vincik, and Thomas W. Gettys. 2021. "Hepatic Nfe2l2 Is Not an Essential Mediator of the Metabolic Phenotype Produced by Dietary Methionine Restriction" Nutrients 13, no. 6: 1788. https://doi.org/10.3390/nu13061788
APA StyleFang, H., Stone, K. P., Ghosh, S., Forney, L. A., Sims, L. C., Vincik, L., & Gettys, T. W. (2021). Hepatic Nfe2l2 Is Not an Essential Mediator of the Metabolic Phenotype Produced by Dietary Methionine Restriction. Nutrients, 13(6), 1788. https://doi.org/10.3390/nu13061788