Epigenetics in Food Allergy and Immunomodulation
Abstract
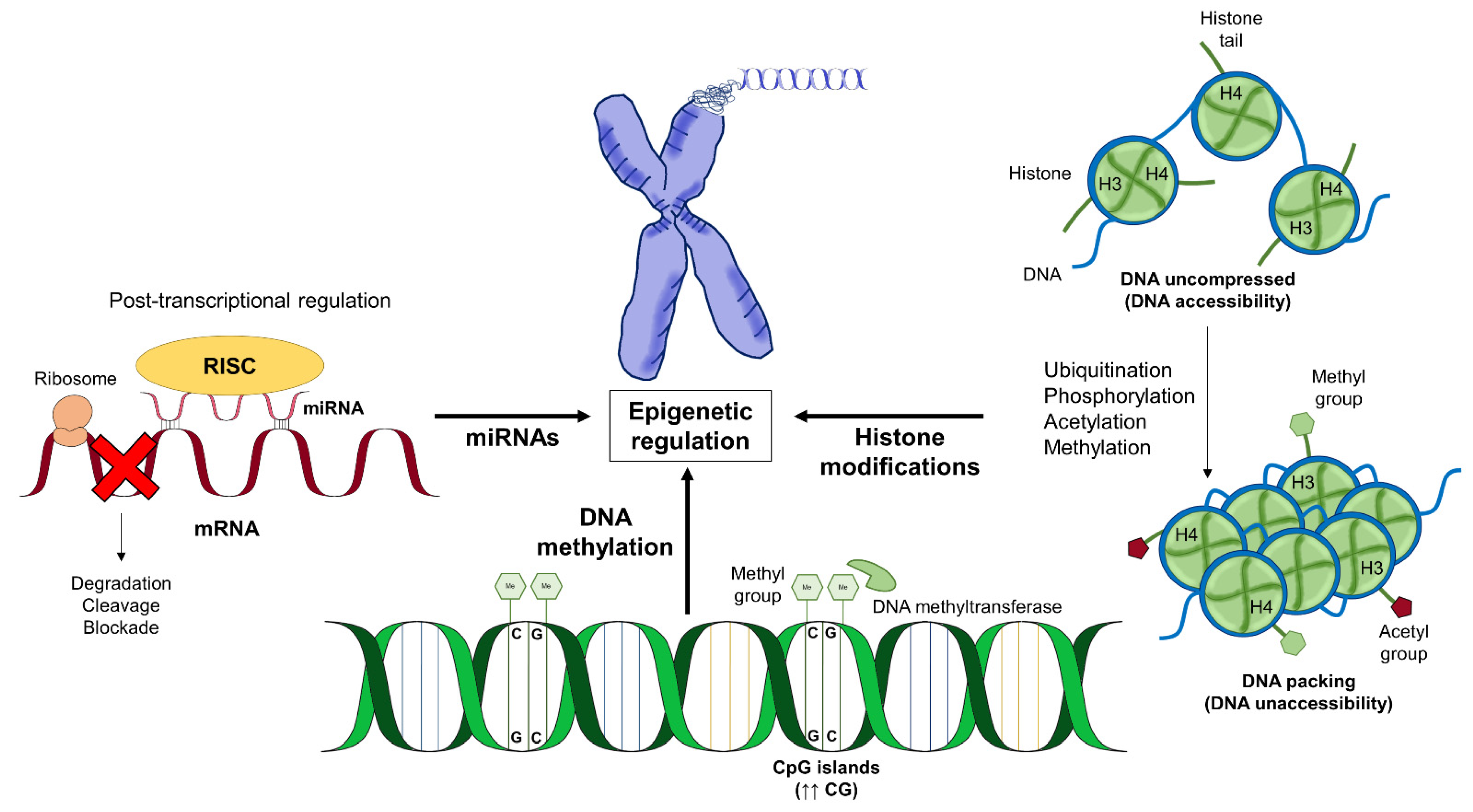
:1. Introduction
2. Epigenetics in Food Allergy
2.1. Cow‘s Milk Allergy
2.2. Peanut Allergy
3. Epigenetics in Food Allergy Immunomodulation
3.1. Epigenetics and Immunomodulation
3.2. The Role of Vitamin D Epigenetics in Food Allergy
3.3. Butyrate
3.4. Methyl Group Donors Folic Acid
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Prescott, S.; Allen, K.J. Food allergy: Riding the second wave of the allergy epidemic. Pediatr. Allergy Immunol. 2011, 22, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Mayorga, C.; Palomares, F.; Cañas, J.A.; Pérez-Sánchez, N.; Núñez, R.; Torres, M.J.; Gómez, F. New insights in therapy for food allergy. Foods 2021, 10, 1037. [Google Scholar] [CrossRef]
- Tsai, H.J.; Kumar, R.; Pongracic, J.; Liu, X.; Story, R.; Yu, Y.; Caruso, D.; Costello, J.; Schroeder, A.; Fang, Y.; et al. Familial aggregation of food allergy and sensitization to food allergens: A family-based study. Clin. Exp. Allergy 2009, 39, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Martino, D.J.; Prescott, S.L. Progress in understanding the epigenetic basis for immune development, immune function, and the rising incidence of allergic disease. Curr. Allergy Asthma Rep. 2013, 13, 85–92. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987, 3, 342–347. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schubeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of histone modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Guenther, M.G.; Levine, S.S.; Boyer, L.A.; Jaenisch, R.; Young, R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 2007, 130, 77–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [Green Version]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef]
- Kimura, H. Histone modifications for human epigenome analysis. J. Hum. Genet. 2013, 58, 439–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 2014, 1839, 627–643. [Google Scholar] [CrossRef] [Green Version]
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 24–29. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Pager, C.T.; Wehner, K.A.; Fuchs, G.; Sarnow, P. MicroRNA-mediated gene silencing. Prog. Mol. Biol. Transl. Sci. 2009, 90, 187–210. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Aure, M.R.; Fleischer, T.; Bjorklund, S.; Ankill, J.; Castro-Mondragon, J.A.; Osbreac; Borresen-Dale, A.L.; Tost, J.; Sahlberg, K.K.; Mathelier, A.; et al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.; Renzini, A.; Adamo, S.; Moresi, V. Coordinated actions of microRNAs with other epigenetic factors regulate skeletal muscle development and adaptation. Int. J. Mol. Sci. 2017, 18, 840. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, S.; Pisano, D.G.; Serrano, M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 2008, 7, 2601–2608. [Google Scholar] [CrossRef]
- Acevedo, N.; Scala, G.; Merid, S.K.; Frumento, P.; Bruhn, S.; Andersson, A.; Ogris, C.; Bottai, M.; Pershagen, G.; Koppelman, G.H.; et al. DNA methylation levels in mononuclear leukocytes from the mother and her child are associated with IgE sensitization to allergens in early life. Int. J. Mol. Sci. 2021, 22, 801. [Google Scholar] [CrossRef] [PubMed]
- Martino, D.; Joo, J.E.; Sexton-Oates, A.; Dang, T.; Allen, K.; Saffery, R.; Prescott, S. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics 2014, 9, 998–1006. [Google Scholar] [CrossRef]
- Martino, D.; Neeland, M.; Dang, T.; Cobb, J.; Ellis, J.; Barnett, A.; Tang, M.; Vuillermin, P.; Allen, K.; Saffery, R. Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy. Nat. Commun. 2018, 9, 3308. [Google Scholar] [CrossRef]
- Izuhara, K. Genetic or epigenetic regulations in immune responses and allergic diseases. Allergol. Int. 2016, 65, 121–122. [Google Scholar] [CrossRef] [Green Version]
- Martino, D.; Dang, T.; Sexton-Oates, A.; Prescott, S.; Tang, M.L.; Dharmage, S.; Gurrin, L.; Koplin, J.; Ponsonby, A.L.; Allen, K.J.; et al. Blood DNA methylation biomarkers predict clinical reactivity in food-sensitized infants. J. Allergy Clin. Immunol. 2015, 135, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Krajewski, D.; Kaczenski, E.; Rovatti, J.; Polukort, S.; Thompson, C.; Dollard, C.; Ser-Dolansky, J.; Schneider, S.S.; Kinney, S.R.M.; Mathias, C.B. Epigenetic regulation via altered histone acetylation results in suppression of mast cell function and mast cell-mediated food allergic responses. Front. Immunol. 2018, 9, 2414. [Google Scholar] [CrossRef]
- Bunning, B.J.; DeKruyff, R.H.; Nadeau, K.C. Epigenetic changes during food-specific immunotherapy. Curr. Allergy Asthma Rep. 2016, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Quake, C.; Nadeau, K.C. The role of epigenetic mediation and the future of food allergy research. Semin. Cell Dev. Biol. 2015, 43, 125–130. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berni Canani, R.; Paparo, L.; Nocerino, R.; Cosenza, L.; Pezzella, V.; Di Costanzo, M.; Capasso, M.; Del Monaco, V.; D’Argenio, V.; Greco, L.; et al. Differences in DNA methylation profile of Th1 and Th2 cytokine genes are associated with tolerance acquisition in children with IgE-mediated cow’s milk allergy. Clin. Epigenetics 2015, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Del Monaco, V.; Paparo, L.; De Palma, F.D.E.; Nocerino, R.; D’Alessio, F.; Visconte, F.; Discepolo, V.; Del Vecchio, L.; Salvatore, F.; et al. Altered miR-193a-5p expression in children with cow’s milk allergy. Allergy 2018, 73, 379–386. [Google Scholar] [CrossRef]
- Paparo, L.; Nocerino, R.; Cosenza, L.; Aitoro, R.; D’Argenio, V.; Del Monaco, V.; Di Scala, C.; Amoroso, A.; Di Costanzo, M.; Salvatore, F.; et al. Epigenetic features of FoxP3 in children with cow’s milk allergy. Clin. Epigenetics 2016, 8, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paparo, L.; di Costanzo, M.; di Scala, C.; Cosenza, L.; Leone, L.; Nocerino, R.; Canani, R.B. The influence of early life nutrition on epigenetic regulatory mechanisms of the immune system. Nutrients 2014, 6, 4706–4719. [Google Scholar] [CrossRef] [Green Version]
- Paparo, L.; Nocerino, R.; Bruno, C.; Di Scala, C.; Cosenza, L.; Bedogni, G.; Di Costanzo, M.; Mennini, M.; D’Argenio, V.; Salvatore, F.; et al. Randomized controlled trial on the influence of dietary intervention on epigenetic mechanisms in children with cow’s milk allergy: The EPICMA study. Sci. Rep. 2019, 9, 2828. [Google Scholar] [CrossRef] [PubMed]
- Paparo, L.; Picariello, G.; Bruno, C.; Pisapia, L.; Canale, V.; Sarracino, A.; Nocerino, R.; Carucci, L.; Cosenza, L.; Cozzolino, T.; et al. Tolerogenic effect elicited by protein fraction derived from different formulas for dietary treatment of cow’s milk allergy in human cells. Front. Immunol. 2020, 11, 604075. [Google Scholar] [CrossRef] [PubMed]
- Petrus, N.C.M.; Henneman, P.; Venema, A.; Mul, A.; van Sinderen, F.; Haagmans, M.; Mook, O.; Hennekam, R.C.; Sprikkelman, A.B.; Mannens, M. Cow’s milk allergy in Dutch children: An epigenetic pilot survey. Clin. Transl. Allergy 2016, 6, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbring, S.; Wolf, J.; Ayechu-Muruzabal, V.; Diks, M.A.P.; Alhamwe, B.A.; Alhamdan, F.; Harb, H.; Renz, H.; Garn, H.; Garssen, J.; et al. Raw cow’s milk reduces allergic symptoms in a murine model for food allergy—A potential role for epigenetic modifications. Nutrients 2019, 11, 1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alashkar Alhamwe, B.; Meulenbroek, L.; Veening-Griffioen, D.H.; Wehkamp, T.M.D.; Alhamdan, F.; Miethe, S.; Harb, H.; Hogenkamp, A.; Knippels, L.M.J.; Pogge von Strandmann, E.; et al. Decreased histone acetylation levels at Th1 and regulatory loci after induction of food allergy. Nutrients 2020, 12, 3193. [Google Scholar] [CrossRef]
- Hong, X.; Hao, K.; Ladd-Acosta, C.; Hansen, K.D.; Tsai, H.J.; Liu, X.; Xu, X.; Thornton, T.A.; Caruso, D.; Keet, C.A.; et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat. Commun. 2015, 6, 6304. [Google Scholar] [CrossRef]
- Zhou, X.; Han, X.; Lyu, S.C.; Bunning, B.; Kost, L.; Chang, I.; Cao, S.; Sampath, V.; Nadeau, K.C. Targeted DNA methylation profiling reveals epigenetic signatures in peanut allergy. JCI Insight 2021, 6, e143058. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Wang, X. Early life precursors, epigenetics, and the development of food allergy. Semin. Immunopathol. 2012, 34, 655–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, A.N.; Watson, C.T.; Cohain, A.T.; Griffin, R.S.; Grishin, A.; Wood, R.A.; Wesley Burks, A.; Jones, S.M.; Scurlock, A.; Leung, D.Y.M.; et al. Dual transcriptomic and epigenomic study of reaction severity in peanut-allergic children. J. Allergy Clin. Immunol. 2020, 145, 1219–1230. [Google Scholar] [CrossRef]
- Imran, S.; Neeland, M.R.; Koplin, J.; Dharmage, S.; Tang, M.L.; Sawyer, S.; Dang, T.; McWilliam, V.; Peters, R.; Perrett, K.P.; et al. Epigenetic programming underpins B-cell dysfunction in peanut and multi-food allergy. Clin. Transl. Immunol. 2021, 10, e1324. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.; Song, Y.; Brown, H.; Hart, P.H.; Zhang, G.B. Cellular and molecular mechanisms of vitamin D in food allergy. J. Cell. Mol. Med. 2018, 22, 3270–3277. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Liu, C.; Hui, Y.; Srivastava, K.; Zhou, Z.; Chen, J.; Miller, R.L.; Finkelman, F.D.; Li, X.M. Maternal allergy increases susceptibility to offspring allergy in association with TH2-biased epigenetic alterations in a mouse model of peanut allergy. J. Allergy Clin. Immunol. 2014, 134, 1339–1345. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yang, I.V.; Davidson, E.J.; Joetham, A.; Takeda, K.; O’Connor, B.P.; Gelfand, E.W. Forkhead box protein 3 demethylation is associated with tolerance induction in peanut-induced intestinal allergy. J. Allergy Clin. Immunol. 2018, 141, 659–670. [Google Scholar] [CrossRef] [Green Version]
- Syed, A.; Garcia, M.A.; Lyu, S.C.; Bucayu, R.; Kohli, A.; Ishida, S.; Berglund, J.P.; Tsai, M.; Maecker, H.; O’Riordan, G.; et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3). J. Allergy Clin. Immunol. 2014, 133, 500–510. [Google Scholar] [CrossRef] [Green Version]
- Mondoulet, L.; Dioszeghy, V.; Busato, F.; Plaquet, C.; Dhelft, V.; Bethune, K.; Leclere, L.; Daviaud, C.; Ligouis, M.; Sampson, H.; et al. Gata3 hypermethylation and Foxp3 hypomethylation are associated with sustained protection and bystander effect following epicutaneous immunotherapy in peanut-sensitized mice. Allergy 2019, 74, 152–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondoulet, L.; Dioszeghy, V.; Puteaux, E.; Ligouis, M.; Dhelft, V.; Plaquet, C.; Dupont, C.; Benhamou, P.H. Specific epicutaneous immunotherapy prevents sensitization to new allergens in a murine model. J. Allergy Clin. Immunol. 2015, 135, 1546–1557. [Google Scholar] [CrossRef]
- Swamy, R.S.; Reshamwala, N.; Hunter, T.; Vissamsetti, S.; Santos, C.B.; Baroody, F.M.; Hwang, P.H.; Hoyte, E.G.; Garcia, M.A.; Nadeau, K.C. Epigenetic modifications and improved regulatory T-cell function in subjects undergoing dual sublingual immunotherapy. J. Allergy Clin. Immunol. 2012, 130, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holick, M.F. Vitamin D deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Suaini, N.H.; Zhang, Y.; Vuillermin, P.J.; Allen, K.J.; Harrison, L.C. Immune modulation by vitamin D and its relevance to food allergy. Nutrients 2015, 7, 6088–6108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benetti, C.; Comberiati, P.; Capristo, C.; Boner, A.L.; Peroni, D.G. Therapeutic effects of vitamin D in asthma and allergy. Mini Rev. Med. Chem. 2015, 15, 935–943. [Google Scholar] [CrossRef]
- Comberiati, P.; Tsabouri, S.; Piacentini, G.L.; Moser, S.; Minniti, F.; Peroni, D.G. Is vitamin D deficiency correlated with childhood wheezing and asthma? Front. Biosci. 2014, 6, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Matsui, T.; Tanaka, K.; Yamashita, H.; Saneyasu, K.I.; Tanaka, H.; Takasato, Y.; Sugiura, S.; Inagaki, N.; Ito, K. Food allergy is linked to season of birth, sun exposure, and vitamin D deficiency. Allergol. Int. 2019, 68, 172–177. [Google Scholar] [CrossRef]
- Umar, M.; Sastry, K.S.; Al Ali, F.; Al-Khulaifi, M.; Wang, E.; Chouchane, A.I. Vitamin D and the pathophysiology of inflammatory skin diseases. Skin Pharmacol. Physiol. 2018, 31, 74–86. [Google Scholar] [CrossRef]
- Allen, K.J.; Koplin, J.J.; Ponsonby, A.L.; Gurrin, L.C.; Wake, M.; Vuillermin, P.; Martin, P.; Matheson, M.; Lowe, A.; Robinson, M.; et al. Vitamin D insufficiency is associated with challenge-proven food allergy in infants. J. Allergy Clin. Immunol. 2013, 131, 1109–1116. [Google Scholar] [CrossRef]
- Rosendahl, J.; Pelkonen, A.S.; Helve, O.; Hauta-Alus, H.; Holmlund-Suila, E.; Valkama, S.; Enlund-Cerullo, M.; Viljakainen, H.; Hytinantti, T.; Mäkitie, O.; et al. High-dose vitamin D supplementation does not prevent allergic sensitization of infants. J. Pediatr. 2019, 209, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Pae, M.; Wu, D. Nutritional modulation of age-related changes in the immune system and risk of infection. Nutr. Res. 2017, 41, 14–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, M.T.; Lee, Y.K.; Maynard, C.L.; Oliver, J.R.; Bikle, D.D.; Jetten, A.M.; Weaver, C.T. Lineage-specific effects of 1,25-dihydroxyvitamin D(3) on the development of effector CD4 T cells. J. Biol. Chem. 2011, 286, 997–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heine, G.; Niesner, U.; Chang, H.D.; Steinmeyer, A.; Zügel, U.; Zuberbier, T.; Radbruch, A.; Worm, M. 1,25-dihydroxyvitamin D(3) promotes IL-10 production in human B cells. Eur. J. Immunol. 2008, 38, 2210–2218. [Google Scholar] [CrossRef]
- Hyppönen, E.; Berry, D.J.; Wjst, M.; Power, C. Serum 25-hydroxyvitamin D and IgE—A significant but nonlinear relationship. Allergy 2009, 64, 613–620. [Google Scholar] [CrossRef]
- Baeke, F.; van Etten, E.; Gysemans, C.; Overbergh, L.; Mathieu, C. Vitamin D signaling in immune-mediated disorders: Evolving insights and therapeutic opportunities. Mol. Aspects Med. 2008, 29, 376–387. [Google Scholar] [CrossRef]
- Dimeloe, S.; Nanzer, A.; Ryanna, K.; Hawrylowicz, C. Regulatory T cells, inflammation and the allergic response—The role of glucocorticoids and vitamin D. J. Steroid Biochem. Mol. 2010, 120, 86–95. [Google Scholar] [CrossRef]
- Széles, L.; Keresztes, G.; Töröcsik, D.; Balajthy, Z.; Krenács, L.; Póliska, S.; Steinmeyer, A.; Zuegel, U.; Pruenster, M.; Rot, A.; et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J. Immunol. 2009, 182, 2074–2083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koplin, J.J.; Suaini, N.H.; Vuillermin, P.; Ellis, J.A.; Panjari, M.; Ponsonby, A.L.; Peters, R.L.; Matheson, M.C.; Martino, D.; Dang, T.; et al. Polymorphisms affecting vitamin D-binding protein modify the relationship between serum vitamin D (25[OH]D3) and food allergy. J. Allergy Clin. Immunol. 2016, 137, 500–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junge, K.M.; Bauer, T.; Geissler, S.; Hirche, F.; Thürmann, L.; Bauer, M.; Trump, S.; Bieg, M.; Weichenhan, D.; Gu, L.; et al. Increased vitamin D levels at birth and in early infancy increase offspring allergy risk-evidence for involvement of epigenetic mechanisms. J. Allergy Clin. Immunol. 2016, 137, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Fetahu, I.S.; Höbaus, J.; Kállay, E. Vitamin D and the epigenome. Front. Physiol. 2014, 5, 164. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Wang, X.; Shi, H.; Su, S.; Harshfield, G.A.; Gutin, B.; Snieder, H.; Dong, Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J. Pediatr. 2013, 162, 1004–1009. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.M.; Gillespie, S.L.; Thiele, D.K.; Ralph, J.L.; Ohm, J.E. Effects of maternal vitamin D supplementation on the maternal and infant epigenome. Breastfeed. Med. 2018, 13, 371–380. [Google Scholar] [CrossRef]
- Jiao, X.; Wang, L.; Wei, Z.; Liu, B.; Liu, X.; Yu, X. Vitamin D deficiency during pregnancy affects the function of Th1/Th2 cells and methylation of IFN-γ gene in offspring rats. Immunol. Lett. 2019, 212, 98–105. [Google Scholar] [CrossRef]
- Lee, K.H.; Song, Y.; O’Sullivan, M.; Pereira, G.; Loh, R.; Zhang, G.B. The implications of DNA methylation on food allergy. Int. Arch. Allergy Immunol. 2017, 173, 183–192. [Google Scholar] [CrossRef]
- Giannetti, A.; Bernardini, L.; Cangemi, J.; Gallucci, M.; Masetti, R.; Ricci, G. Role of vitamin D in prevention of food allergy in infants. Front. Pediatr. 2020, 8, 447. [Google Scholar] [CrossRef]
- Peters, R.L.; Neeland, M.R.; Allen, K.J. Primary prevention of food allergy. Curr. Allergy Asthma Rep. 2017, 17, 52. [Google Scholar] [CrossRef]
- Aitoro, R.; Paparo, L.; Amoroso, A.; Di Costanzo, M.; Cosenza, L.; Granata, V.; Di Scala, C.; Nocerino, R.; Trinchese, G.; Montella, M.; et al. Gut microbiota as a target for preventive and therapeutic intervention against food allergy. Nutrients 2017, 9, 672. [Google Scholar] [CrossRef] [Green Version]
- Astbury, S.M.; Corfe, B.M. Uptake and metabolism of the short-chain fatty acid butyrate, a critical review of the literature. Curr. Drug Metab. 2012, 13, 815–821. [Google Scholar] [CrossRef]
- Velázquez, O.C.; Lederer, H.M.; Rombeau, J.L. Butyrate and the colonocyte. Production, absorption, metabolism, and therapeutic implications. Adv. Exp. Med. Biol. 1997, 427, 123–134. [Google Scholar] [PubMed]
- Patil, P.; Bhandary, S.K.; Haridas, V.; Sarathkumar, E.; Shetty, P. Is butyrate a natural alternative to dexamethasone in the management of CoVID-19? F1000Researsh 2021, 10, 273. [Google Scholar] [CrossRef]
- Du Toit, G.; Tsakok, T.; Lack, S.; Lack, G. Prevention of food allergy. J. Allergy Clin. Immunol. 2016, 137, 998–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedelkopoulou, N.; Dhawan, A.; Xinias, I.; Gidaris, D.; Farmaki, E. Interleukin 10: The critical role of a pleiotropic cytokine in food allergy. Allergol. Immunopathol. 2020, 48, 401–408. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Luu, M.; Monning, H.; Visekruna, A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front. Immunol. 2020, 11, 1225. [Google Scholar] [CrossRef]
- Paparo, L.; Nocerino, R.; Ciaglia, E.; Di Scala, C.; De Caro, C.; Russo, R.; Trinchese, G.; Aitoro, R.; Amoroso, A.; Bruno, C.; et al. Butyrate as a bioactive human milk protective component against food allergy. Allergy 2021, 76, 1398–1415. [Google Scholar] [CrossRef]
- Fellows, R.; Varga-Weisz, P. Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk. Mol. Metab. 2020, 38, 100925. [Google Scholar] [CrossRef] [PubMed]
- McStay, C.L.; Prescott, S.L.; Bower, C.; Palmer, D.J. Maternal folic acid supplementation during pregnancy and childhood allergic disease outcomes: A question of timing? Nutrients 2017, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.M.; Ali, M.M. Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Xing, Y.; Yu, X.; Dou, Y.; Ma, D. Effect of folic acid intake on infant and child allergic diseases: Systematic review and meta-analysis. Front. Pediatr. 2020, 8, 615406. [Google Scholar] [CrossRef]
- Hong, X.; Ladd-Acosta, C.; Hao, K.; Sherwood, B.; Ji, H.; Keet, C.A.; Kumar, R.; Caruso, D.; Liu, X.; Wang, G.; et al. Epigenome-wide association study links site-specific DNA methylation changes with cow’s milk allergy. J. Allergy Clin. Immunol. 2016, 138, 908–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cañas, J.A.; Núñez, R.; Cruz-Amaya, A.; Gómez, F.; Torres, M.J.; Palomares, F.; Mayorga, C. Epigenetics in Food Allergy and Immunomodulation. Nutrients 2021, 13, 4345. https://doi.org/10.3390/nu13124345
Cañas JA, Núñez R, Cruz-Amaya A, Gómez F, Torres MJ, Palomares F, Mayorga C. Epigenetics in Food Allergy and Immunomodulation. Nutrients. 2021; 13(12):4345. https://doi.org/10.3390/nu13124345
Chicago/Turabian StyleCañas, José A., Rafael Núñez, Anyith Cruz-Amaya, Francisca Gómez, María J. Torres, Francisca Palomares, and Cristobalina Mayorga. 2021. "Epigenetics in Food Allergy and Immunomodulation" Nutrients 13, no. 12: 4345. https://doi.org/10.3390/nu13124345






