Epigenetics in Food Allergy and Immunomodulation
Abstract
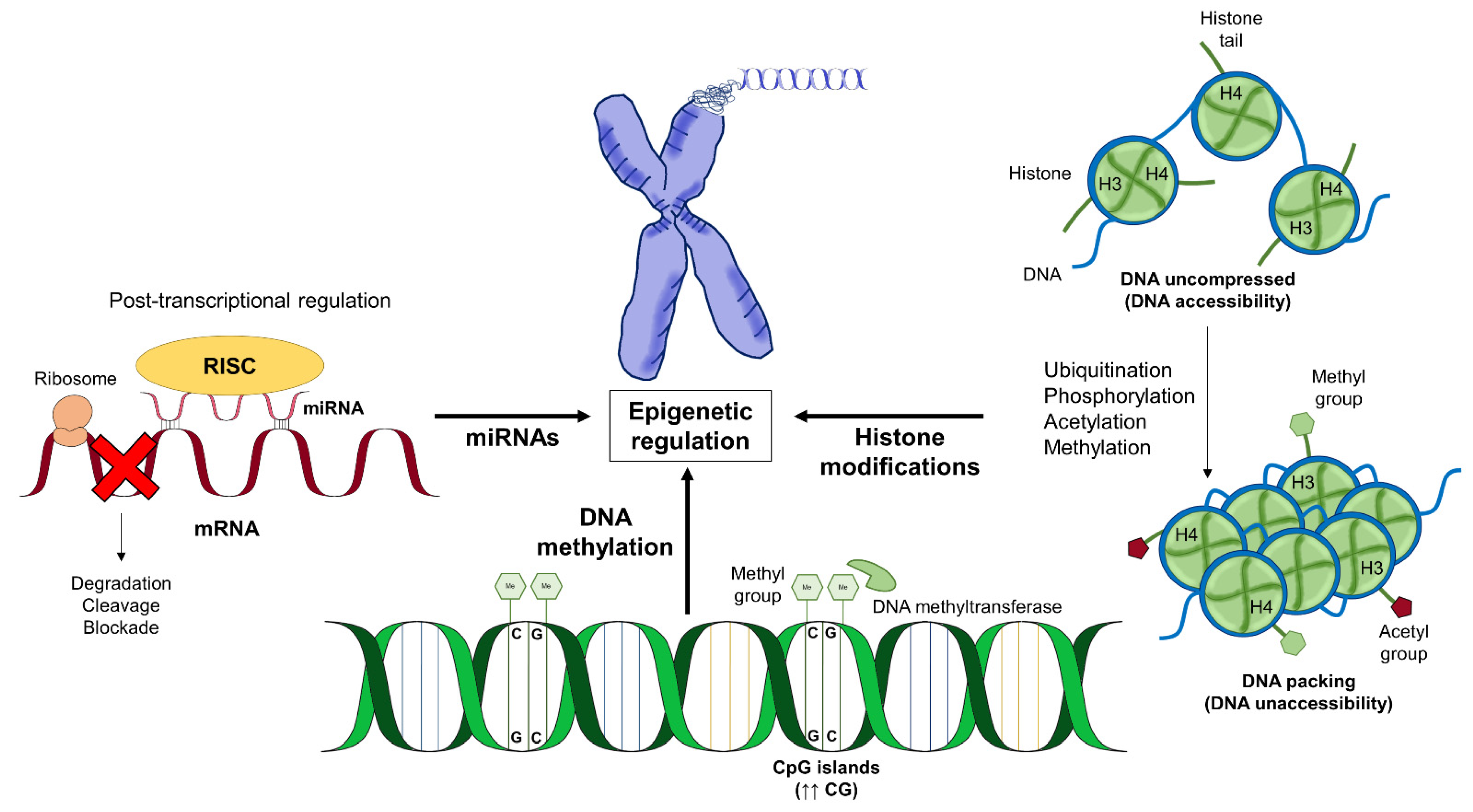
1. Introduction
2. Epigenetics in Food Allergy
2.1. Cow‘s Milk Allergy
2.2. Peanut Allergy
3. Epigenetics in Food Allergy Immunomodulation
3.1. Epigenetics and Immunomodulation
3.2. The Role of Vitamin D Epigenetics in Food Allergy
3.3. Butyrate
3.4. Methyl Group Donors Folic Acid
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Prescott, S.; Allen, K.J. Food allergy: Riding the second wave of the allergy epidemic. Pediatr. Allergy Immunol. 2011, 22, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Mayorga, C.; Palomares, F.; Cañas, J.A.; Pérez-Sánchez, N.; Núñez, R.; Torres, M.J.; Gómez, F. New insights in therapy for food allergy. Foods 2021, 10, 1037. [Google Scholar] [CrossRef]
- Tsai, H.J.; Kumar, R.; Pongracic, J.; Liu, X.; Story, R.; Yu, Y.; Caruso, D.; Costello, J.; Schroeder, A.; Fang, Y.; et al. Familial aggregation of food allergy and sensitization to food allergens: A family-based study. Clin. Exp. Allergy 2009, 39, 101–109. [Google Scholar] [CrossRef]
- Martino, D.J.; Prescott, S.L. Progress in understanding the epigenetic basis for immune development, immune function, and the rising incidence of allergic disease. Curr. Allergy Asthma Rep. 2013, 13, 85–92. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987, 3, 342–347. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schubeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of histone modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Guenther, M.G.; Levine, S.S.; Boyer, L.A.; Jaenisch, R.; Young, R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 2007, 130, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef]
- Kimura, H. Histone modifications for human epigenome analysis. J. Hum. Genet. 2013, 58, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 2014, 1839, 627–643. [Google Scholar] [CrossRef]
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 24–29. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Pager, C.T.; Wehner, K.A.; Fuchs, G.; Sarnow, P. MicroRNA-mediated gene silencing. Prog. Mol. Biol. Transl. Sci. 2009, 90, 187–210. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Aure, M.R.; Fleischer, T.; Bjorklund, S.; Ankill, J.; Castro-Mondragon, J.A.; Osbreac; Borresen-Dale, A.L.; Tost, J.; Sahlberg, K.K.; Mathelier, A.; et al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.; Renzini, A.; Adamo, S.; Moresi, V. Coordinated actions of microRNAs with other epigenetic factors regulate skeletal muscle development and adaptation. Int. J. Mol. Sci. 2017, 18, 840. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Gonzalez, S.; Pisano, D.G.; Serrano, M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 2008, 7, 2601–2608. [Google Scholar] [CrossRef]
- Acevedo, N.; Scala, G.; Merid, S.K.; Frumento, P.; Bruhn, S.; Andersson, A.; Ogris, C.; Bottai, M.; Pershagen, G.; Koppelman, G.H.; et al. DNA methylation levels in mononuclear leukocytes from the mother and her child are associated with IgE sensitization to allergens in early life. Int. J. Mol. Sci. 2021, 22, 801. [Google Scholar] [CrossRef] [PubMed]
- Martino, D.; Joo, J.E.; Sexton-Oates, A.; Dang, T.; Allen, K.; Saffery, R.; Prescott, S. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics 2014, 9, 998–1006. [Google Scholar] [CrossRef]
- Martino, D.; Neeland, M.; Dang, T.; Cobb, J.; Ellis, J.; Barnett, A.; Tang, M.; Vuillermin, P.; Allen, K.; Saffery, R. Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy. Nat. Commun. 2018, 9, 3308. [Google Scholar] [CrossRef]
- Izuhara, K. Genetic or epigenetic regulations in immune responses and allergic diseases. Allergol. Int. 2016, 65, 121–122. [Google Scholar] [CrossRef][Green Version]
- Martino, D.; Dang, T.; Sexton-Oates, A.; Prescott, S.; Tang, M.L.; Dharmage, S.; Gurrin, L.; Koplin, J.; Ponsonby, A.L.; Allen, K.J.; et al. Blood DNA methylation biomarkers predict clinical reactivity in food-sensitized infants. J. Allergy Clin. Immunol. 2015, 135, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Krajewski, D.; Kaczenski, E.; Rovatti, J.; Polukort, S.; Thompson, C.; Dollard, C.; Ser-Dolansky, J.; Schneider, S.S.; Kinney, S.R.M.; Mathias, C.B. Epigenetic regulation via altered histone acetylation results in suppression of mast cell function and mast cell-mediated food allergic responses. Front. Immunol. 2018, 9, 2414. [Google Scholar] [CrossRef]
- Bunning, B.J.; DeKruyff, R.H.; Nadeau, K.C. Epigenetic changes during food-specific immunotherapy. Curr. Allergy Asthma Rep. 2016, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Quake, C.; Nadeau, K.C. The role of epigenetic mediation and the future of food allergy research. Semin. Cell Dev. Biol. 2015, 43, 125–130. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Paparo, L.; Nocerino, R.; Cosenza, L.; Pezzella, V.; Di Costanzo, M.; Capasso, M.; Del Monaco, V.; D’Argenio, V.; Greco, L.; et al. Differences in DNA methylation profile of Th1 and Th2 cytokine genes are associated with tolerance acquisition in children with IgE-mediated cow’s milk allergy. Clin. Epigenetics 2015, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Del Monaco, V.; Paparo, L.; De Palma, F.D.E.; Nocerino, R.; D’Alessio, F.; Visconte, F.; Discepolo, V.; Del Vecchio, L.; Salvatore, F.; et al. Altered miR-193a-5p expression in children with cow’s milk allergy. Allergy 2018, 73, 379–386. [Google Scholar] [CrossRef]
- Paparo, L.; Nocerino, R.; Cosenza, L.; Aitoro, R.; D’Argenio, V.; Del Monaco, V.; Di Scala, C.; Amoroso, A.; Di Costanzo, M.; Salvatore, F.; et al. Epigenetic features of FoxP3 in children with cow’s milk allergy. Clin. Epigenetics 2016, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Paparo, L.; di Costanzo, M.; di Scala, C.; Cosenza, L.; Leone, L.; Nocerino, R.; Canani, R.B. The influence of early life nutrition on epigenetic regulatory mechanisms of the immune system. Nutrients 2014, 6, 4706–4719. [Google Scholar] [CrossRef]
- Paparo, L.; Nocerino, R.; Bruno, C.; Di Scala, C.; Cosenza, L.; Bedogni, G.; Di Costanzo, M.; Mennini, M.; D’Argenio, V.; Salvatore, F.; et al. Randomized controlled trial on the influence of dietary intervention on epigenetic mechanisms in children with cow’s milk allergy: The EPICMA study. Sci. Rep. 2019, 9, 2828. [Google Scholar] [CrossRef] [PubMed]
- Paparo, L.; Picariello, G.; Bruno, C.; Pisapia, L.; Canale, V.; Sarracino, A.; Nocerino, R.; Carucci, L.; Cosenza, L.; Cozzolino, T.; et al. Tolerogenic effect elicited by protein fraction derived from different formulas for dietary treatment of cow’s milk allergy in human cells. Front. Immunol. 2020, 11, 604075. [Google Scholar] [CrossRef] [PubMed]
- Petrus, N.C.M.; Henneman, P.; Venema, A.; Mul, A.; van Sinderen, F.; Haagmans, M.; Mook, O.; Hennekam, R.C.; Sprikkelman, A.B.; Mannens, M. Cow’s milk allergy in Dutch children: An epigenetic pilot survey. Clin. Transl. Allergy 2016, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Abbring, S.; Wolf, J.; Ayechu-Muruzabal, V.; Diks, M.A.P.; Alhamwe, B.A.; Alhamdan, F.; Harb, H.; Renz, H.; Garn, H.; Garssen, J.; et al. Raw cow’s milk reduces allergic symptoms in a murine model for food allergy—A potential role for epigenetic modifications. Nutrients 2019, 11, 1721. [Google Scholar] [CrossRef] [PubMed]
- Alashkar Alhamwe, B.; Meulenbroek, L.; Veening-Griffioen, D.H.; Wehkamp, T.M.D.; Alhamdan, F.; Miethe, S.; Harb, H.; Hogenkamp, A.; Knippels, L.M.J.; Pogge von Strandmann, E.; et al. Decreased histone acetylation levels at Th1 and regulatory loci after induction of food allergy. Nutrients 2020, 12, 3193. [Google Scholar] [CrossRef]
- Hong, X.; Hao, K.; Ladd-Acosta, C.; Hansen, K.D.; Tsai, H.J.; Liu, X.; Xu, X.; Thornton, T.A.; Caruso, D.; Keet, C.A.; et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat. Commun. 2015, 6, 6304. [Google Scholar] [CrossRef]
- Zhou, X.; Han, X.; Lyu, S.C.; Bunning, B.; Kost, L.; Chang, I.; Cao, S.; Sampath, V.; Nadeau, K.C. Targeted DNA methylation profiling reveals epigenetic signatures in peanut allergy. JCI Insight 2021, 6, e143058. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Wang, X. Early life precursors, epigenetics, and the development of food allergy. Semin. Immunopathol. 2012, 34, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Do, A.N.; Watson, C.T.; Cohain, A.T.; Griffin, R.S.; Grishin, A.; Wood, R.A.; Wesley Burks, A.; Jones, S.M.; Scurlock, A.; Leung, D.Y.M.; et al. Dual transcriptomic and epigenomic study of reaction severity in peanut-allergic children. J. Allergy Clin. Immunol. 2020, 145, 1219–1230. [Google Scholar] [CrossRef]
- Imran, S.; Neeland, M.R.; Koplin, J.; Dharmage, S.; Tang, M.L.; Sawyer, S.; Dang, T.; McWilliam, V.; Peters, R.; Perrett, K.P.; et al. Epigenetic programming underpins B-cell dysfunction in peanut and multi-food allergy. Clin. Transl. Immunol. 2021, 10, e1324. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.; Song, Y.; Brown, H.; Hart, P.H.; Zhang, G.B. Cellular and molecular mechanisms of vitamin D in food allergy. J. Cell. Mol. Med. 2018, 22, 3270–3277. [Google Scholar] [CrossRef]
- Song, Y.; Liu, C.; Hui, Y.; Srivastava, K.; Zhou, Z.; Chen, J.; Miller, R.L.; Finkelman, F.D.; Li, X.M. Maternal allergy increases susceptibility to offspring allergy in association with TH2-biased epigenetic alterations in a mouse model of peanut allergy. J. Allergy Clin. Immunol. 2014, 134, 1339–1345. [Google Scholar] [CrossRef]
- Wang, M.; Yang, I.V.; Davidson, E.J.; Joetham, A.; Takeda, K.; O’Connor, B.P.; Gelfand, E.W. Forkhead box protein 3 demethylation is associated with tolerance induction in peanut-induced intestinal allergy. J. Allergy Clin. Immunol. 2018, 141, 659–670. [Google Scholar] [CrossRef]
- Syed, A.; Garcia, M.A.; Lyu, S.C.; Bucayu, R.; Kohli, A.; Ishida, S.; Berglund, J.P.; Tsai, M.; Maecker, H.; O’Riordan, G.; et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3). J. Allergy Clin. Immunol. 2014, 133, 500–510. [Google Scholar] [CrossRef]
- Mondoulet, L.; Dioszeghy, V.; Busato, F.; Plaquet, C.; Dhelft, V.; Bethune, K.; Leclere, L.; Daviaud, C.; Ligouis, M.; Sampson, H.; et al. Gata3 hypermethylation and Foxp3 hypomethylation are associated with sustained protection and bystander effect following epicutaneous immunotherapy in peanut-sensitized mice. Allergy 2019, 74, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Mondoulet, L.; Dioszeghy, V.; Puteaux, E.; Ligouis, M.; Dhelft, V.; Plaquet, C.; Dupont, C.; Benhamou, P.H. Specific epicutaneous immunotherapy prevents sensitization to new allergens in a murine model. J. Allergy Clin. Immunol. 2015, 135, 1546–1557. [Google Scholar] [CrossRef]
- Swamy, R.S.; Reshamwala, N.; Hunter, T.; Vissamsetti, S.; Santos, C.B.; Baroody, F.M.; Hwang, P.H.; Hoyte, E.G.; Garcia, M.A.; Nadeau, K.C. Epigenetic modifications and improved regulatory T-cell function in subjects undergoing dual sublingual immunotherapy. J. Allergy Clin. Immunol. 2012, 130, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. Vitamin D deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Suaini, N.H.; Zhang, Y.; Vuillermin, P.J.; Allen, K.J.; Harrison, L.C. Immune modulation by vitamin D and its relevance to food allergy. Nutrients 2015, 7, 6088–6108. [Google Scholar] [CrossRef] [PubMed]
- Benetti, C.; Comberiati, P.; Capristo, C.; Boner, A.L.; Peroni, D.G. Therapeutic effects of vitamin D in asthma and allergy. Mini Rev. Med. Chem. 2015, 15, 935–943. [Google Scholar] [CrossRef]
- Comberiati, P.; Tsabouri, S.; Piacentini, G.L.; Moser, S.; Minniti, F.; Peroni, D.G. Is vitamin D deficiency correlated with childhood wheezing and asthma? Front. Biosci. 2014, 6, 31–39. [Google Scholar] [CrossRef][Green Version]
- Matsui, T.; Tanaka, K.; Yamashita, H.; Saneyasu, K.I.; Tanaka, H.; Takasato, Y.; Sugiura, S.; Inagaki, N.; Ito, K. Food allergy is linked to season of birth, sun exposure, and vitamin D deficiency. Allergol. Int. 2019, 68, 172–177. [Google Scholar] [CrossRef]
- Umar, M.; Sastry, K.S.; Al Ali, F.; Al-Khulaifi, M.; Wang, E.; Chouchane, A.I. Vitamin D and the pathophysiology of inflammatory skin diseases. Skin Pharmacol. Physiol. 2018, 31, 74–86. [Google Scholar] [CrossRef]
- Allen, K.J.; Koplin, J.J.; Ponsonby, A.L.; Gurrin, L.C.; Wake, M.; Vuillermin, P.; Martin, P.; Matheson, M.; Lowe, A.; Robinson, M.; et al. Vitamin D insufficiency is associated with challenge-proven food allergy in infants. J. Allergy Clin. Immunol. 2013, 131, 1109–1116. [Google Scholar] [CrossRef]
- Rosendahl, J.; Pelkonen, A.S.; Helve, O.; Hauta-Alus, H.; Holmlund-Suila, E.; Valkama, S.; Enlund-Cerullo, M.; Viljakainen, H.; Hytinantti, T.; Mäkitie, O.; et al. High-dose vitamin D supplementation does not prevent allergic sensitization of infants. J. Pediatr. 2019, 209, 139–145. [Google Scholar] [CrossRef]
- Pae, M.; Wu, D. Nutritional modulation of age-related changes in the immune system and risk of infection. Nutr. Res. 2017, 41, 14–35. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.T.; Lee, Y.K.; Maynard, C.L.; Oliver, J.R.; Bikle, D.D.; Jetten, A.M.; Weaver, C.T. Lineage-specific effects of 1,25-dihydroxyvitamin D(3) on the development of effector CD4 T cells. J. Biol. Chem. 2011, 286, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Heine, G.; Niesner, U.; Chang, H.D.; Steinmeyer, A.; Zügel, U.; Zuberbier, T.; Radbruch, A.; Worm, M. 1,25-dihydroxyvitamin D(3) promotes IL-10 production in human B cells. Eur. J. Immunol. 2008, 38, 2210–2218. [Google Scholar] [CrossRef]
- Hyppönen, E.; Berry, D.J.; Wjst, M.; Power, C. Serum 25-hydroxyvitamin D and IgE—A significant but nonlinear relationship. Allergy 2009, 64, 613–620. [Google Scholar] [CrossRef]
- Baeke, F.; van Etten, E.; Gysemans, C.; Overbergh, L.; Mathieu, C. Vitamin D signaling in immune-mediated disorders: Evolving insights and therapeutic opportunities. Mol. Aspects Med. 2008, 29, 376–387. [Google Scholar] [CrossRef]
- Dimeloe, S.; Nanzer, A.; Ryanna, K.; Hawrylowicz, C. Regulatory T cells, inflammation and the allergic response—The role of glucocorticoids and vitamin D. J. Steroid Biochem. Mol. 2010, 120, 86–95. [Google Scholar] [CrossRef]
- Széles, L.; Keresztes, G.; Töröcsik, D.; Balajthy, Z.; Krenács, L.; Póliska, S.; Steinmeyer, A.; Zuegel, U.; Pruenster, M.; Rot, A.; et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J. Immunol. 2009, 182, 2074–2083. [Google Scholar] [CrossRef] [PubMed]
- Koplin, J.J.; Suaini, N.H.; Vuillermin, P.; Ellis, J.A.; Panjari, M.; Ponsonby, A.L.; Peters, R.L.; Matheson, M.C.; Martino, D.; Dang, T.; et al. Polymorphisms affecting vitamin D-binding protein modify the relationship between serum vitamin D (25[OH]D3) and food allergy. J. Allergy Clin. Immunol. 2016, 137, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Junge, K.M.; Bauer, T.; Geissler, S.; Hirche, F.; Thürmann, L.; Bauer, M.; Trump, S.; Bieg, M.; Weichenhan, D.; Gu, L.; et al. Increased vitamin D levels at birth and in early infancy increase offspring allergy risk-evidence for involvement of epigenetic mechanisms. J. Allergy Clin. Immunol. 2016, 137, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Fetahu, I.S.; Höbaus, J.; Kállay, E. Vitamin D and the epigenome. Front. Physiol. 2014, 5, 164. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, X.; Shi, H.; Su, S.; Harshfield, G.A.; Gutin, B.; Snieder, H.; Dong, Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J. Pediatr. 2013, 162, 1004–1009. [Google Scholar] [CrossRef]
- Anderson, C.M.; Gillespie, S.L.; Thiele, D.K.; Ralph, J.L.; Ohm, J.E. Effects of maternal vitamin D supplementation on the maternal and infant epigenome. Breastfeed. Med. 2018, 13, 371–380. [Google Scholar] [CrossRef]
- Jiao, X.; Wang, L.; Wei, Z.; Liu, B.; Liu, X.; Yu, X. Vitamin D deficiency during pregnancy affects the function of Th1/Th2 cells and methylation of IFN-γ gene in offspring rats. Immunol. Lett. 2019, 212, 98–105. [Google Scholar] [CrossRef]
- Lee, K.H.; Song, Y.; O’Sullivan, M.; Pereira, G.; Loh, R.; Zhang, G.B. The implications of DNA methylation on food allergy. Int. Arch. Allergy Immunol. 2017, 173, 183–192. [Google Scholar] [CrossRef]
- Giannetti, A.; Bernardini, L.; Cangemi, J.; Gallucci, M.; Masetti, R.; Ricci, G. Role of vitamin D in prevention of food allergy in infants. Front. Pediatr. 2020, 8, 447. [Google Scholar] [CrossRef]
- Peters, R.L.; Neeland, M.R.; Allen, K.J. Primary prevention of food allergy. Curr. Allergy Asthma Rep. 2017, 17, 52. [Google Scholar] [CrossRef]
- Aitoro, R.; Paparo, L.; Amoroso, A.; Di Costanzo, M.; Cosenza, L.; Granata, V.; Di Scala, C.; Nocerino, R.; Trinchese, G.; Montella, M.; et al. Gut microbiota as a target for preventive and therapeutic intervention against food allergy. Nutrients 2017, 9, 672. [Google Scholar] [CrossRef]
- Astbury, S.M.; Corfe, B.M. Uptake and metabolism of the short-chain fatty acid butyrate, a critical review of the literature. Curr. Drug Metab. 2012, 13, 815–821. [Google Scholar] [CrossRef]
- Velázquez, O.C.; Lederer, H.M.; Rombeau, J.L. Butyrate and the colonocyte. Production, absorption, metabolism, and therapeutic implications. Adv. Exp. Med. Biol. 1997, 427, 123–134. [Google Scholar] [PubMed]
- Patil, P.; Bhandary, S.K.; Haridas, V.; Sarathkumar, E.; Shetty, P. Is butyrate a natural alternative to dexamethasone in the management of CoVID-19? F1000Researsh 2021, 10, 273. [Google Scholar] [CrossRef]
- Du Toit, G.; Tsakok, T.; Lack, S.; Lack, G. Prevention of food allergy. J. Allergy Clin. Immunol. 2016, 137, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Nedelkopoulou, N.; Dhawan, A.; Xinias, I.; Gidaris, D.; Farmaki, E. Interleukin 10: The critical role of a pleiotropic cytokine in food allergy. Allergol. Immunopathol. 2020, 48, 401–408. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Luu, M.; Monning, H.; Visekruna, A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front. Immunol. 2020, 11, 1225. [Google Scholar] [CrossRef]
- Paparo, L.; Nocerino, R.; Ciaglia, E.; Di Scala, C.; De Caro, C.; Russo, R.; Trinchese, G.; Aitoro, R.; Amoroso, A.; Bruno, C.; et al. Butyrate as a bioactive human milk protective component against food allergy. Allergy 2021, 76, 1398–1415. [Google Scholar] [CrossRef]
- Fellows, R.; Varga-Weisz, P. Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk. Mol. Metab. 2020, 38, 100925. [Google Scholar] [CrossRef] [PubMed]
- McStay, C.L.; Prescott, S.L.; Bower, C.; Palmer, D.J. Maternal folic acid supplementation during pregnancy and childhood allergic disease outcomes: A question of timing? Nutrients 2017, 9, 123. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Ali, M.M. Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xing, Y.; Yu, X.; Dou, Y.; Ma, D. Effect of folic acid intake on infant and child allergic diseases: Systematic review and meta-analysis. Front. Pediatr. 2020, 8, 615406. [Google Scholar] [CrossRef]
- Hong, X.; Ladd-Acosta, C.; Hao, K.; Sherwood, B.; Ji, H.; Keet, C.A.; Kumar, R.; Caruso, D.; Liu, X.; Wang, G.; et al. Epigenome-wide association study links site-specific DNA methylation changes with cow’s milk allergy. J. Allergy Clin. Immunol. 2016, 138, 908–911. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cañas, J.A.; Núñez, R.; Cruz-Amaya, A.; Gómez, F.; Torres, M.J.; Palomares, F.; Mayorga, C. Epigenetics in Food Allergy and Immunomodulation. Nutrients 2021, 13, 4345. https://doi.org/10.3390/nu13124345
Cañas JA, Núñez R, Cruz-Amaya A, Gómez F, Torres MJ, Palomares F, Mayorga C. Epigenetics in Food Allergy and Immunomodulation. Nutrients. 2021; 13(12):4345. https://doi.org/10.3390/nu13124345
Chicago/Turabian StyleCañas, José A., Rafael Núñez, Anyith Cruz-Amaya, Francisca Gómez, María J. Torres, Francisca Palomares, and Cristobalina Mayorga. 2021. "Epigenetics in Food Allergy and Immunomodulation" Nutrients 13, no. 12: 4345. https://doi.org/10.3390/nu13124345
APA StyleCañas, J. A., Núñez, R., Cruz-Amaya, A., Gómez, F., Torres, M. J., Palomares, F., & Mayorga, C. (2021). Epigenetics in Food Allergy and Immunomodulation. Nutrients, 13(12), 4345. https://doi.org/10.3390/nu13124345






