Genistein Regulates Lipid Metabolism via Estrogen Receptor β and Its Downstream Signal Akt/mTOR in HepG2 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture and Treatment
2.3. Cell Viability Assay
2.4. Lipid Content Assay
2.5. Western Blot Analysis
2.6. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.7. Statistical Analysis
3. Results
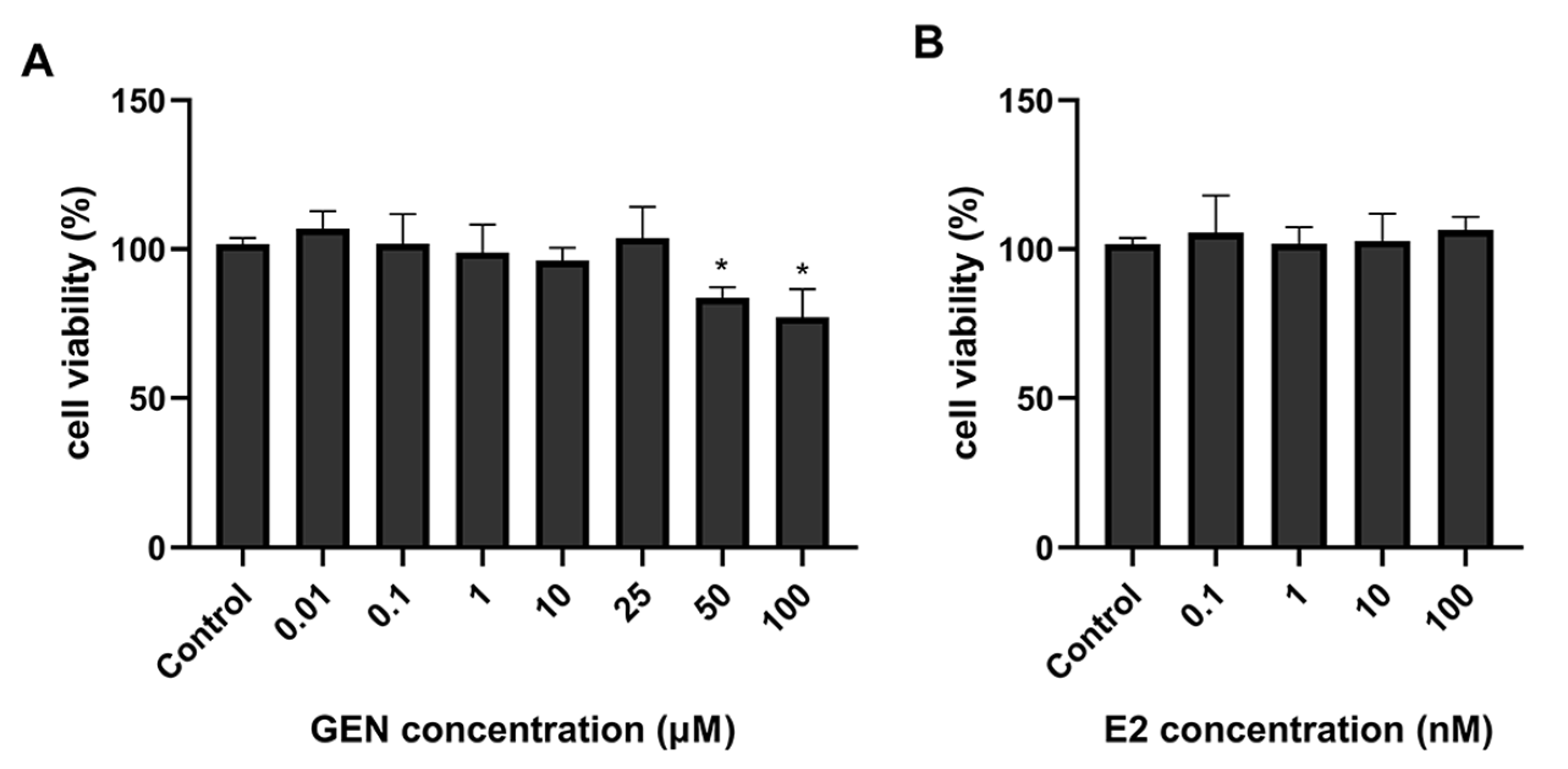
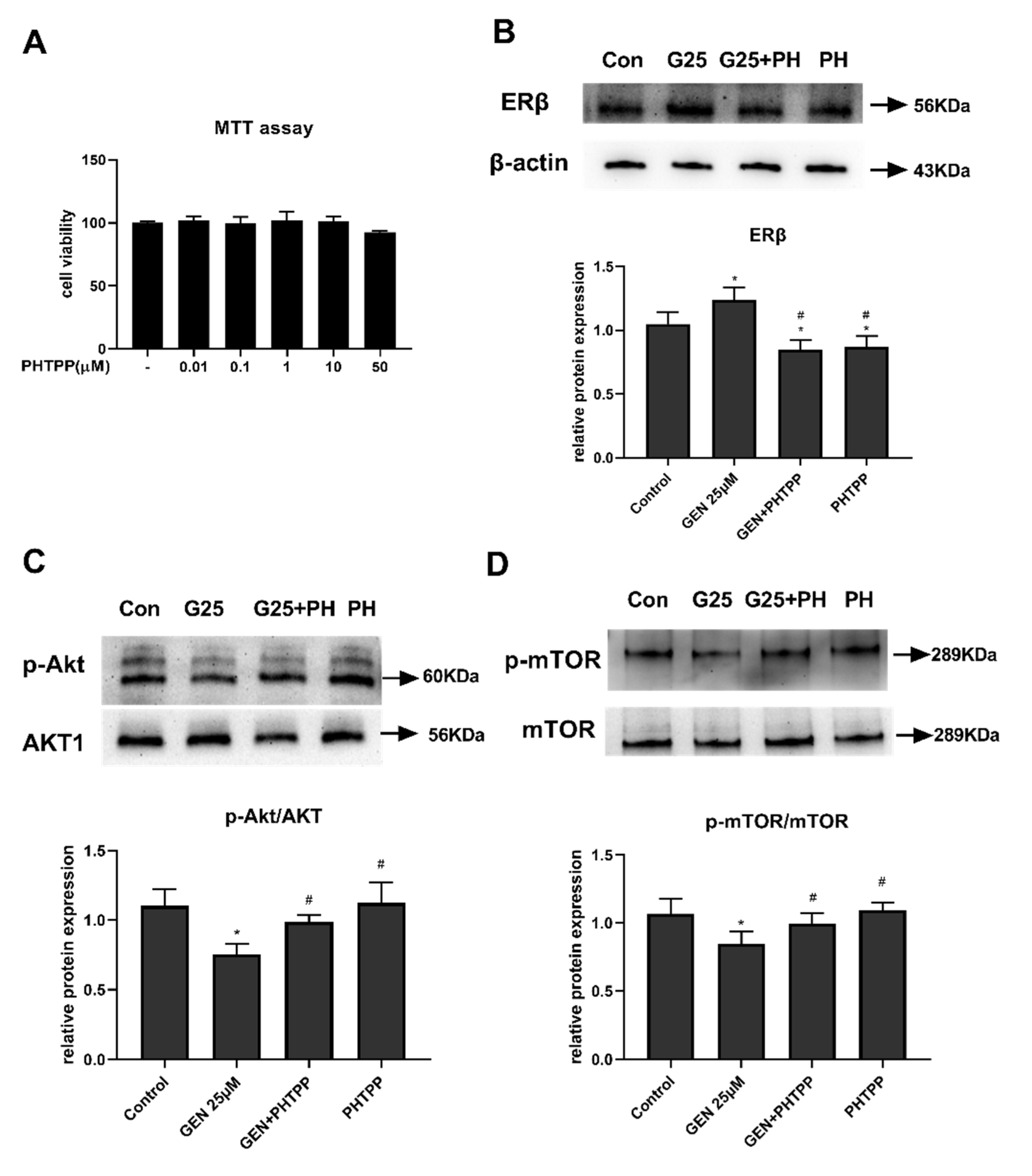
3.1. Effects of GEN on HepG2 Cell Viability
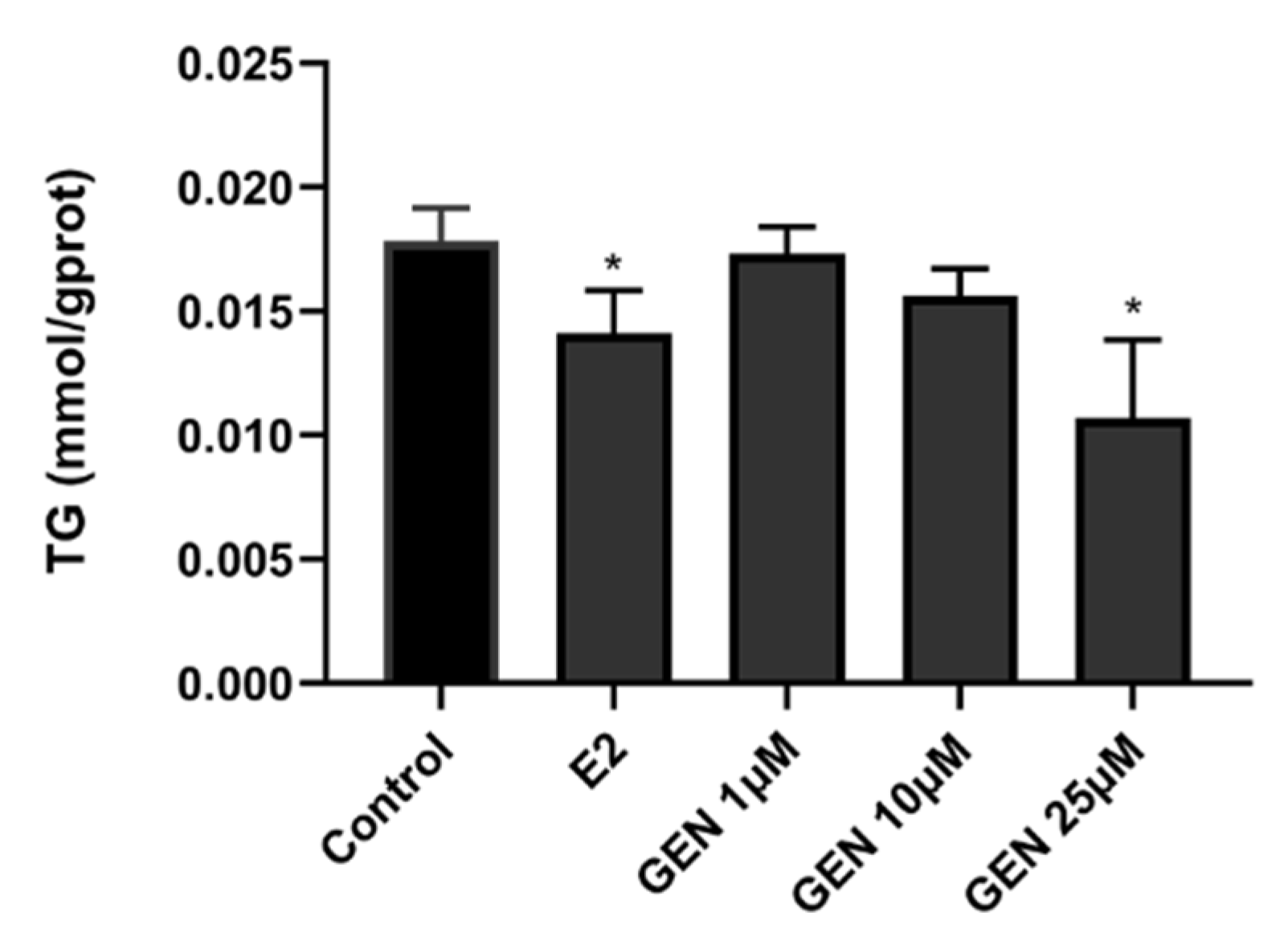
3.2. Effects of GEN on Lipid Accumulation
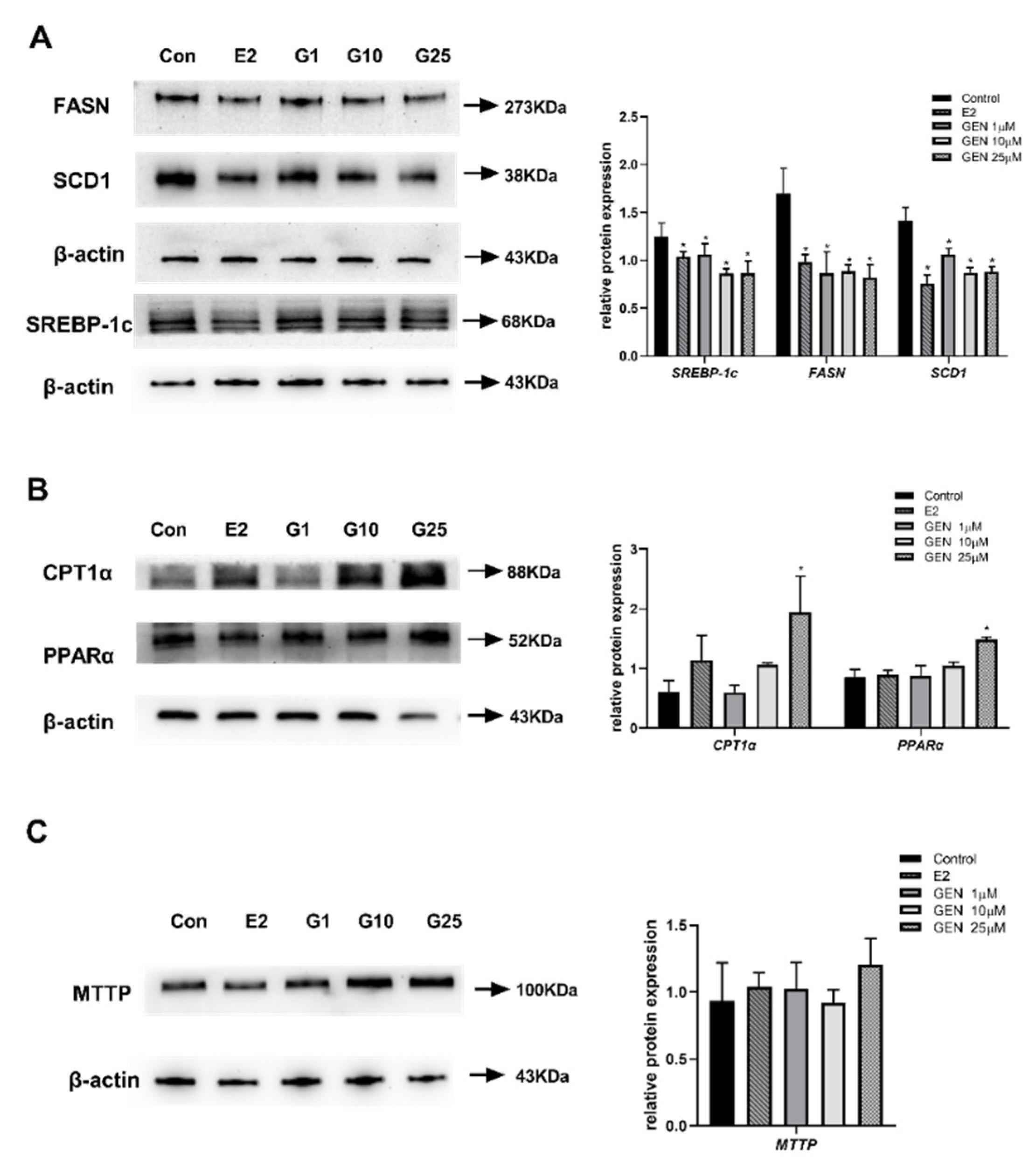
3.3. Effects of GEN on Protein Expression Involved in Lipid Metabolism in HepG2 Cells
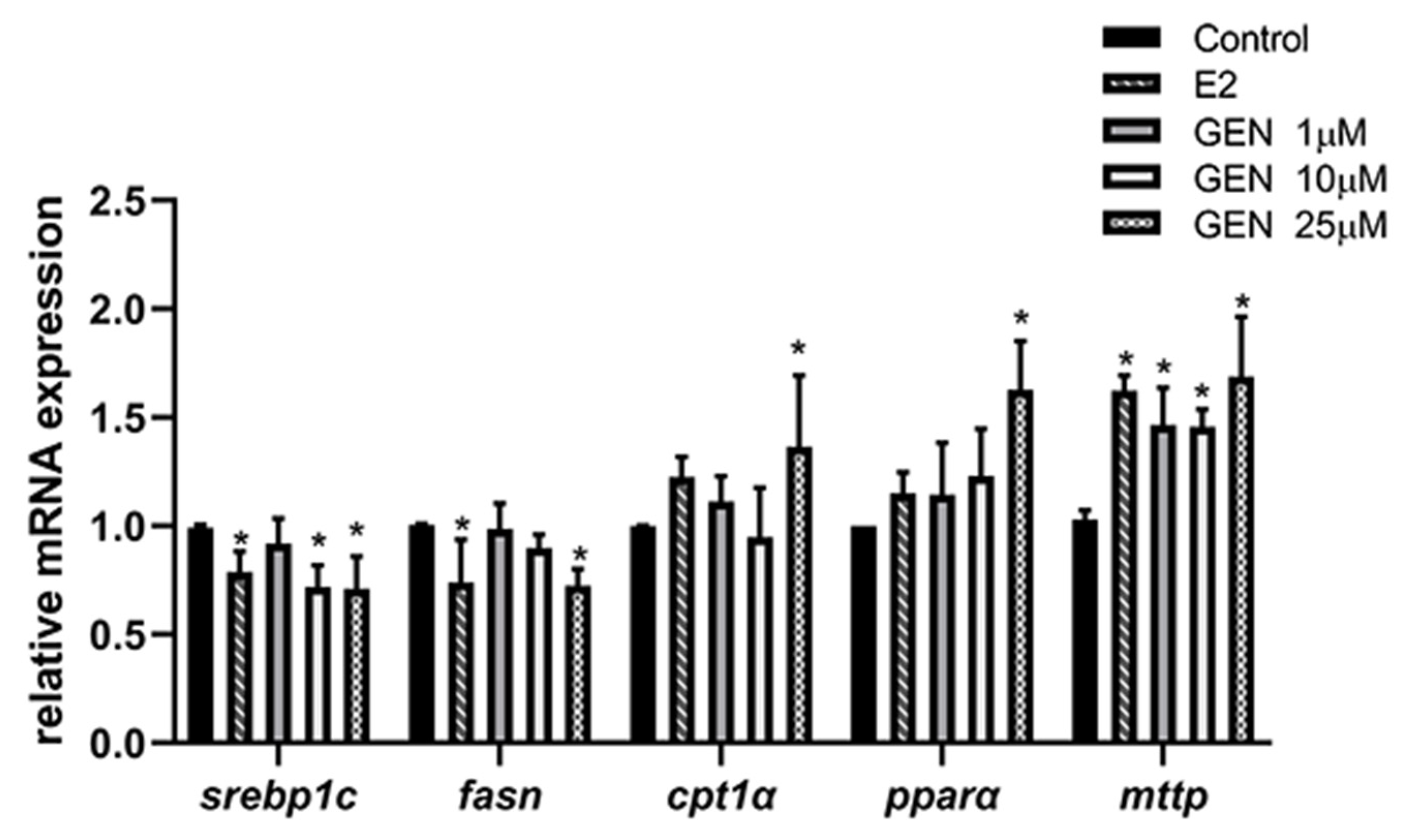
3.4. Effects of GEN on mRNA Expression Involved in Lipid Metabolism in HepG2 Cells
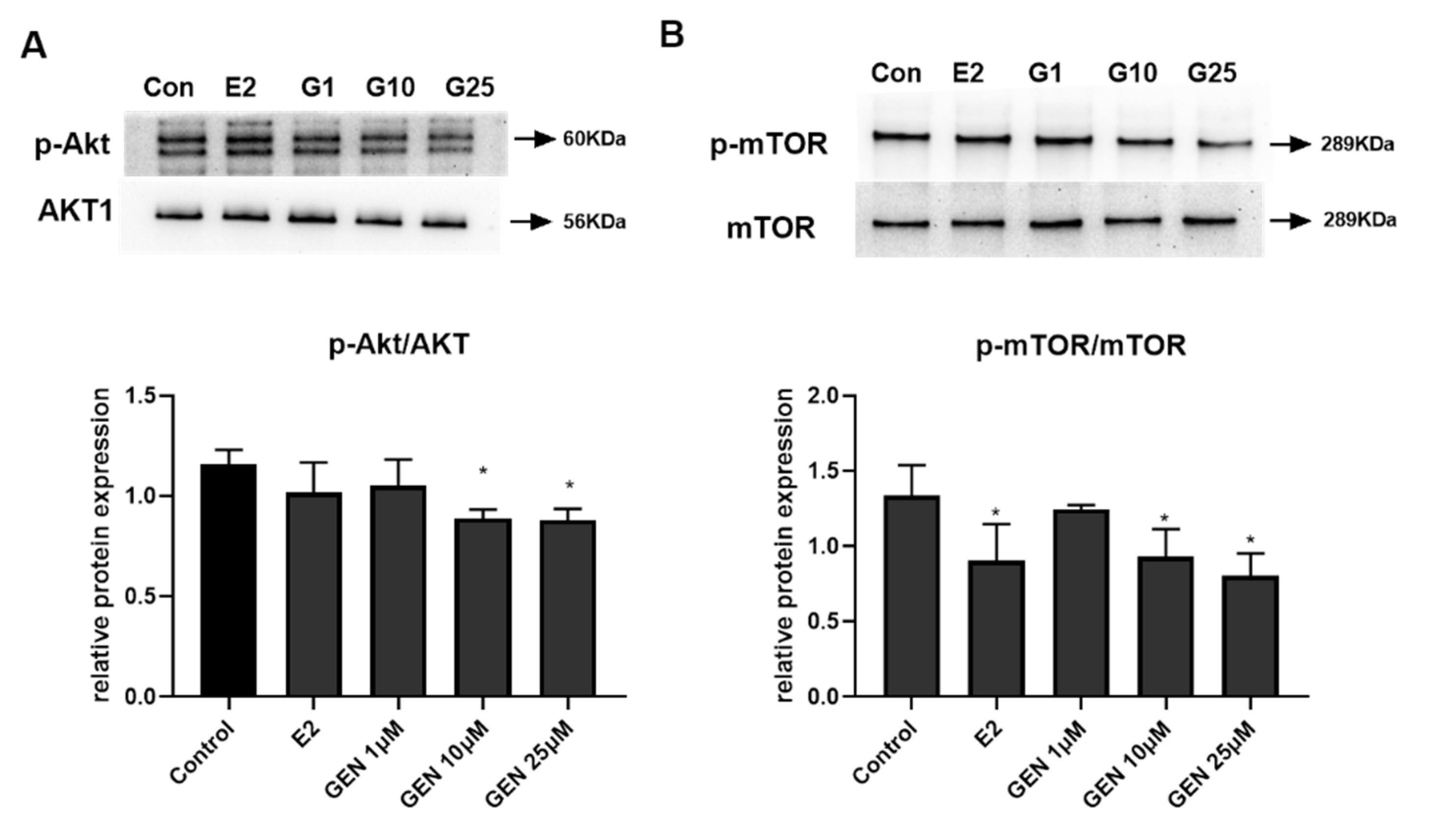
3.5. Effects of GEN on Expression of Key Metabolic Regulators in HepG2 Cells
3.6. The Role of ERβ on Akt/mTOR Signal in GEN-Treated HepG2 Cells
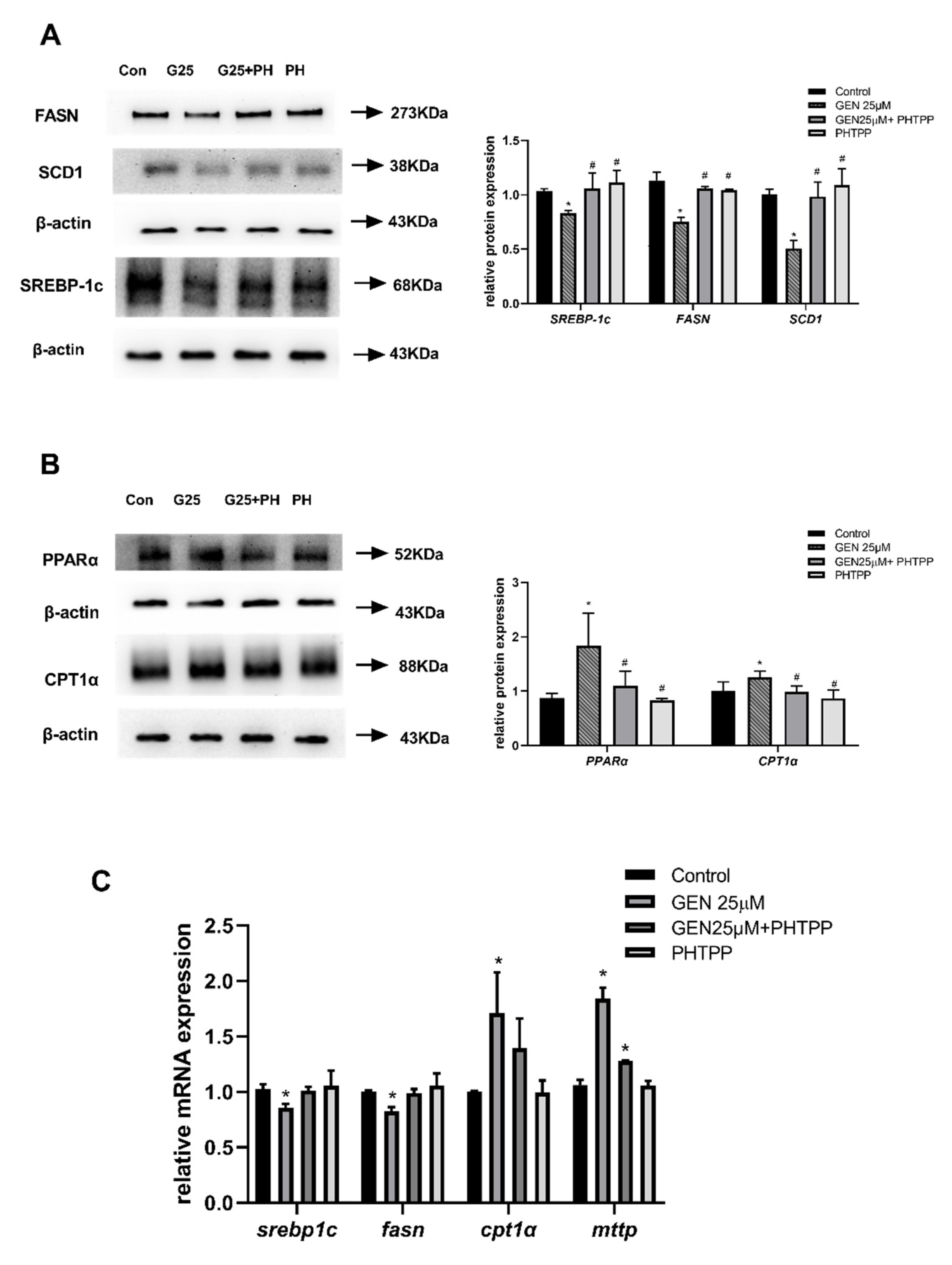
3.7. The Role of ERβ on Hepatic Lipid Metabolism in GEN-Treated HepG2 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sanghavi, M.; Turer, A.; Neeland, I.; Ayers, C.; de Lemos, J.; Khera, A. Postmenopausal status is independently associated with advanced hepatic steatosis: The Dallas heart study. J. Am. Coll. Cardiol. 2013, 61 (Suppl. S10), E1429. [Google Scholar] [CrossRef] [Green Version]
- Klair, J.S.; Yang, J.D.; Abdelmalek, M.F.; Guy, C.D.; Gill, R.M.; Yates, K.; Unalp-Arida, A.; Lavine, J.E.; Clark, J.M.; Diehl, A.M.; et al. A longer duration of estrogen deficiency increases fibrosis risk among postmenopausal women with nonalcoholic fatty liver disease. Hepatology 2016, 64, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Ropero, A.B.; Alonso-Magdalena, P.; Quesada, I.; Nadal, A. The role of estrogen receptors in the control of energy and glucose homeostasis. Steroids 2008, 73, 874–879. [Google Scholar] [CrossRef]
- Morán-Costoya, A.; Proenza, A.M.; Gianotti, M.; Lladó, I.; Valle, A. Sex Differences in Nonalcoholic Fatty Liver Disease: Estrogen Influence on the Liver-Adipose Tissue Crosstalk. Antioxid. Redox Signal. 2021, 35, 753–774. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, M.W.S. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet 2003, 362, 419–427. [Google Scholar]
- Tempfer, C.B.; Hilal, Z.; Kern, P.; Juhasz-Boess, I.; Rezniczek, G.A. Menopausal Hormone Therapy and Risk of Endometrial Cancer: A Systematic Review. Cancers 2020, 12, 2195. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.H.; Kim, H.S. Menopause-Associated Lipid Metabolic Disorders and Foods Beneficial for Postmenopausal Women. Nutrients 2020, 12, 202. [Google Scholar] [CrossRef] [Green Version]
- Nie, Q.; Xing, M.; Hu, J.; Hu, X.; Nie, S.; Xie, M. Metabolism and health effects of phyto-estrogens. Crit. Rev. Food Sci. Nutr. 2017, 57, 2432–2454. [Google Scholar] [CrossRef]
- Seidemann, L.; Krüger, A.; Kegel-Hübner, V.; Seehofer, D.; Damm, G. Influence of Genistein on Hepatic Lipid Metabolism in an In Vitro Model of Hepatic Steatosis. Molecules 2021, 26, 1156. [Google Scholar] [CrossRef]
- Weigt, C.; Hertrampf, T.; Zoth, N.; Fritzemeier, K.H.; Diel, P. Impact of estradiol, ER subtype specific agonists and genistein on energy homeostasis in a rat model of nutrition induced obesity. Mol. Cell. Endocrinol. 2012, 351, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, G.G.; Lemmen, J.G.; Carlsson, B.; Corton, J.C.; Safe, S.H.; van der Saag, P.T.; van der Burg, B.; Gustafsson, J.A. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology 1998, 139, 4252–4263. [Google Scholar] [CrossRef]
- Weigt, C.; Hertrampf, T.; Kluxen, F.M.; Flenker, U.; Hulsemann, F.; Fritzemeier, K.H.; Diel, P. Molecular effects of ER alpha- and beta-selective agonists on regulation of energy homeostasis in obese female Wistar rats. Mol. Cell. Endocrinol. 2013, 377, 147–158. [Google Scholar] [CrossRef]
- Han, J.; Wang, Y. mTORC1 signaling in hepatic lipid metabolism. Protein Cell 2018, 9, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Cornu, M.; Albert, V.; Hall, M.N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013, 23, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Sabatini, D.M. A Central role for mTOR in lipid homeostasis. Cell Metab. 2013, 18, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.L.; He, B.; Erickson, A.; Moreau, R. Improvement of mTORC1-driven overproduction of apoB-containing triacylglyceride-rich lipoproteins by short-chain fatty acids, 4-phenylbutyric acid and (R)-α-lipoic acid, in human hepatocellular carcinoma cells. Biochim. Biophys. Acta 2016, 1861, 166–176. [Google Scholar] [CrossRef]
- Lindberg, K.; Helguero, L.A.; Omoto, Y.; Gustafsson, J.; Haldosén, L.A. Estrogen receptor β represses Akt signaling in breast cancer cells via downregulation of HER2/HER3 and upregulation of PTEN: Implications for tamoxifen sensitivity. Breast Cancer Res. BCR 2011, 13, R43. [Google Scholar] [CrossRef] [Green Version]
- Chaurasiya, S.; Wu, W.; Strom, A.M.; Warner, M.; Gustafsson, J. Estrogen receptor β regulates AKT activity through up-regulation of INPP4B and inhibits migration of prostate cancer cell line PC-3. Proc. Natl. Acad. Sci. USA 2020, 117, 26347–26355. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yu, W.; Liu, B.; Yang, M.; Tao, H. Estrogen receptor β induces autophagy of osteosarcoma through the mTOR signaling pathway. J. Orthop. Surg. Res. 2020, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.D.; Han, C.C.; Wan, H.F.; He, F.; Xu, H.Y.; Wei, S.H.; Du, X.H.; Xu, F. Effects of inhibiting PI3K-Akt-mTOR pathway on lipid metabolism homeostasis in goose primary hepatocytes. Anim. Int. J. Anim. Biosci. 2016, 10, 1319–1327. [Google Scholar] [CrossRef] [Green Version]
- Ogo, Y.; Taniuchi, S.; Ojima, F.; Hayashi, S.; Murakami, I.; Saito, Y.; Takeuchi, S.; Kudo, T.; Takahashi, S. IGF-1 gene expression is differentially regulated by estrogen receptors α and β in mouse endometrial stromal cells and ovarian granulosa cells. J. Reprod. Dev. 2014, 60, 216–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Rogoschin, J.; Niehoff, A.; Oden, K.; Kulling, S.E.; Xie, M.; Diel, P. Combinatory effects of phytoestrogens and exercise on body fat mass and lipid metabolism in ovariectomized female rats. J. Steroid Biochem. Mol. Biol. 2018, 178, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.J.; Song, W.R.; Gao, L.Y.; Nie, S.P.; Eisenbrand, G.; Xie, M.Y. Assessment of dietary phytoestrogen intake via plant-derived foods in China. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2014, 31, 1325–1335. [Google Scholar] [CrossRef]
- Kim, J.; Kwon, C. Estimated dietary isoflavone intake of Korean population based on National Nutrition Survey. Nutr. Res. 2001, 21, 947–953. [Google Scholar] [CrossRef]
- Boker, L.K.; Van der Schouw, Y.T.; De Kleijn, M.J.; Jacques, P.F.; Grobbee, D.E.; Peeters, P.H. Intake of dietary phytoestrogens by Dutch women. J. Nutr. 2002, 132, 1319–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taxvig, C.; Specht, I.O.; Boberg, J.; Vinggaard, A.M.; Nellemann, C. Dietary relevant mixtures of phytoestrogens inhibit adipocyte differentiation in vitro. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2013, 55, 265–271. [Google Scholar] [CrossRef]
- Shin, E.S.; Lee, H.H.; Cho, S.Y.; Park, H.W.; Lee, S.J.; Lee, T.R. Genistein downregulates SREBP-1 regulated gene expression by inhibiting site-1 protease expression in HepG2 cells. J. Nutr. 2007, 137, 1127–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Shin, H.J.; Kim, S.Y.; Kim, J.H.; Lee, Y.S.; Kim, D.H.; Lee, M.O. Genistein enhances expression of genes involved in fatty acid catabolism through activation of PPARalpha. Mol. Cell. Endocrinol. 2004, 220, 51–58. [Google Scholar] [CrossRef]
- Fu, W.; Gao, X.P.; Zhang, S.; Dai, Y.P.; Zou, W.J.; Yue, L.M. 17β-Estradiol Inhibits PCSK9-Mediated LDLR Degradation Through GPER/PLC Activation in HepG2 Cells. Front. Endocrinol. 2019, 10, 930. [Google Scholar] [CrossRef] [Green Version]
- Sibbons, C.M.; Brenna, J.T.; Lawrence, P.; Hoile, S.P.; Clarke-Harris, R.; Lillycrop, K.A.; Burdge, G.C. Effect of sex hormones on n-3 polyunsaturated fatty acid biosynthesis in HepG2 cells and in human primary hepatocytes. Prostaglandins Leukot. Essent. Fat. Acids 2014, 90, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Luo, T.; Snyder, S.M.; Zhao, B.; Sullivan, D.K.; Hamilton-Reeves, J.; Guthrie, G.; Ricketts, M.L.; Shiverick, K.T.; Shay, N. Gene Expression Patterns Are Altered in Athymic Mice and Metabolic Syndrome Factors Are Reduced in C57BL/6J Mice Fed High-Fat Diets Supplemented with Soy Isoflavones. J. Agric. Food Chem. 2016, 64, 7492–7501. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zheng, Z.; Yin, Y.; Jiang, Z. Effect of Genistein on Cholesterol Metabolism-Related Genes in HepG2 Cell. J. Food Sci. 2019, 84, 2330–2336. [Google Scholar] [CrossRef] [PubMed]
- Borradaile, N.M.; de Dreu, L.E.; Wilcox, L.J.; Edwards, J.Y.; Huff, M.W. Soya phytoestrogens, genistein and daidzein, decrease apolipoprotein B secretion from HepG2 cells through multiple mechanisms. Biochem. J. 2002, 366, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Guillou, H.; Martin, P.G.; Pineau, T. Transcriptional regulation of hepatic fatty acid metabolism. Sub-Cell. Biochem. 2008, 49, 3–47. [Google Scholar]
- Han, J.; Li, E.; Chen, L.; Zhang, Y.; Wei, F.; Liu, J.; Deng, H.; Wang, Y. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 2015, 524, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.S.; Qi, W.T.; Guo, W.; Wang, C.L.; Hu, Z.B.; Li, A.K. Genistein and daidzein induce apoptosis of colon cancer cells by inhibiting the accumulation of lipid droplets. Food Nutr. Res. 2018, 62, 1384. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Pang, D.; Luo, Q.; Chen, X.; Gao, Q.; Shi, L.; Liu, W.; Zou, Y.; Li, L.; Chen, Z. Soy Isoflavones Regulate Lipid Metabolism through an AKT/mTORC1 Pathway in Diet-Induced Obesity (DIO) Male Rats. Molecules 2016, 21, 586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, J.D.; Jones, M.E.; Prelle, K.; Simpson, E.R.; Boon, W.C. A selective estrogen receptor α agonist ameliorates hepatic steatosis in the male aromatase knockout mouse. J. Endocrinol. 2011, 210, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, G. Localization of androgen and estrogen receptors in rat and primate tissues. Histol. Histopathol. 2000, 15, 1261–1270. [Google Scholar]
- Inoue, H.; Shimizu, I.; Lu, G.; Itonaga, M.; Cui, X.; Okamura, Y.; Shono, M.; Honda, H.; Inoue, S.; Muramatsu, M.; et al. Idoxifene and estradiol enhance antiapoptotic activity through estrogen receptor-beta in cultured rat hepatocytes. Dig. Dis. Sci. 2003, 48, 570–580. [Google Scholar] [CrossRef]
- Maruyama, K.; Endoh, H.; Sasaki-Iwaoka, H.; Kanou, H.; Shimaya, E.; Hashimoto, S.; Kato, S.; Kawashima, H. A novel isoform of rat estrogen receptor beta with 18 amino acid insertion in the ligand binding domain as a putative dominant negative regular of estrogen action. Biochem. Biophys. Res. Commun. 1998, 246, 142–147. [Google Scholar] [CrossRef]
- Yepuru, M.; Eswaraka, J.; Kearbey, J.D.; Barrett, C.M.; Raghow, S.; Veverka, K.A.; Miller, D.D.; Dalton, J.T.; Narayanan, R. Estrogen receptor-{beta}-selective ligands alleviate high-fat diet- and ovariectomy-induced obesity in mice. J. Biol. Chem. 2010, 285, 31292–31303. [Google Scholar] [CrossRef] [Green Version]
- Zidon, T.M.; Padilla, J.; Fritsche, K.L.; Welly, R.J.; McCabe, L.T.; Stricklin, O.E.; Frank, A.; Park, Y.; Clegg, D.J.; Lubahn, D.B.; et al. Effects of ERβ and ERα on OVX-induced changes in adiposity and insulin resistance. J. Endocrinol. 2020, 245, 165–178. [Google Scholar] [CrossRef]
- Zanella, I.; Marrazzo, E.; Biasiotto, G.; Penza, M.; Romani, A.; Vignolini, P.; Caimi, L.; Di Lorenzo, D. Soy and the soy isoflavone genistein promote adipose tissue development in male mice on a low-fat diet. Eur. J. Nutr. 2015, 54, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Al-Alem, U.; Ali, M.M.; Bosland, M.C. Genistein increases estrogen receptor beta expression in prostate cancer via reducing its promoter methylation. J. Steroid Biochem. Mol. Biol. 2015, 152, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Luo, F.; Guo, H.; Yu, H.; Li, Y.; Feng, Y.; Wang, Y. PM2.5 organic extract mediates inflammation through the ERβ pathway to contribute to lung carcinogenesis in vitro and vivo. Chemosphere 2021, 263, 127867. [Google Scholar] [CrossRef]
- Osborne, C.K.; Wakeling, A.; Nicholson, R.I. Fulvestrant: An oestrogen receptor antagonist with a novel mechanism of action. Br. J. Cancer 2004, 90 (Suppl. S1), S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Kahraman, M.; Govek, S.; Nagasawa, J.; Bonnefous, C.; Julien, J.; Douglas, K.; Sensintaffar, J.; Lu, N.; Lee, K.J.; et al. Identification of GDC-0810 (ARN-810), an Orally Bioavailable Selective Estrogen Receptor Degrader (SERD) that Demonstrates Robust Activity in Tamoxifen-Resistant Breast Cancer Xenografts. J. Med. Chem. 2015, 58, 4888–4904. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Yu, X.; Chen, Q.; Wu, X.; Kong, X.; Wang, S.; Cai, D.; Cheng, P.; Li, Y.; Bian, H. Liuwei Dihuang soft capsules inhibits the phenotypic conversion of VSMC to prevent the menopausal atherosclerosis by up-regulating the expression of myocardin. J. Ethnopharmacol. 2020, 246, 112207. [Google Scholar] [CrossRef]
- Kumar, S.; Patel, R.; Moore, S.; Crawford, D.K.; Suwanna, N.; Mangiardi, M.; Tiwari-Woodruff, S.K. Estrogen receptor β ligand therapy activates PI3K/Akt/mTOR signaling in oligodendrocytes and promotes remyelination in a mouse model of multiple sclerosis. Neurobiol. Dis. 2013, 56, 131–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guido, C.; Panza, S.; Santoro, M.; Avena, P.; Panno, M.L.; Perrotta, I.; Giordano, F.; Casaburi, I.; Catalano, S.; De Amicis, F.; et al. Estrogen receptor beta (ERβ) produces autophagy and necroptosis in human seminoma cell line through the binding of the Sp1 on the phosphatase and tensin homolog deleted from chromosome 10 (PTEN) promoter gene. Cell Cycle 2012, 11, 2911–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Name | Primer Sequence (5′ to 3′) |
|---|---|
| Srebp1c | forward: GCAACACAGCAACCAGAA |
| reserve: GAAAGGTGAGCCAGCATC | |
| fasn | forward: GCCCAAGGGAAGCACATT |
| reserve: CGAAGCCACCCAGACCAC | |
| pparα | forward: TAGGGACAGACTGACACC |
| reserve: CATAACAAAAGATACGGG | |
| cpt1α | forward: CTACTTCCAGACTTGCCC |
| reserve: ACACCATTTCCATTCCAC | |
| mttp | forward: GGAAATGGTCGCTCACAA |
| reserve: TGCCAGAACCCGAGTAGAGA | |
| β-actin | forward: TTGCGTTACACCCTTTCT |
| reserve: ACCTTCACCGTTCCAGTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, H.; Song, Z.; Shaukat, H.; Zheng, W. Genistein Regulates Lipid Metabolism via Estrogen Receptor β and Its Downstream Signal Akt/mTOR in HepG2 Cells. Nutrients 2021, 13, 4015. https://doi.org/10.3390/nu13114015
Qin H, Song Z, Shaukat H, Zheng W. Genistein Regulates Lipid Metabolism via Estrogen Receptor β and Its Downstream Signal Akt/mTOR in HepG2 Cells. Nutrients. 2021; 13(11):4015. https://doi.org/10.3390/nu13114015
Chicago/Turabian StyleQin, Hong, Ziyu Song, Horia Shaukat, and Wenya Zheng. 2021. "Genistein Regulates Lipid Metabolism via Estrogen Receptor β and Its Downstream Signal Akt/mTOR in HepG2 Cells" Nutrients 13, no. 11: 4015. https://doi.org/10.3390/nu13114015
APA StyleQin, H., Song, Z., Shaukat, H., & Zheng, W. (2021). Genistein Regulates Lipid Metabolism via Estrogen Receptor β and Its Downstream Signal Akt/mTOR in HepG2 Cells. Nutrients, 13(11), 4015. https://doi.org/10.3390/nu13114015






