Serum Untargeted Metabolism Reveals the Mechanism of L. plantarum ZDY2013 in Alleviating Kidney Injury Induced by High-Salt Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. DNA Extraction from Mouse Feces and 16S rDNA Sequencing Analysis
2.3. H&E and Masson’s Staining
2.4. Protein and Genes Expression
2.5. Determination of the Albumin Level of Fecal Bacteria
2.6. Serum Metabolomic Analysis
2.7. Statistical Analysis
3. Results
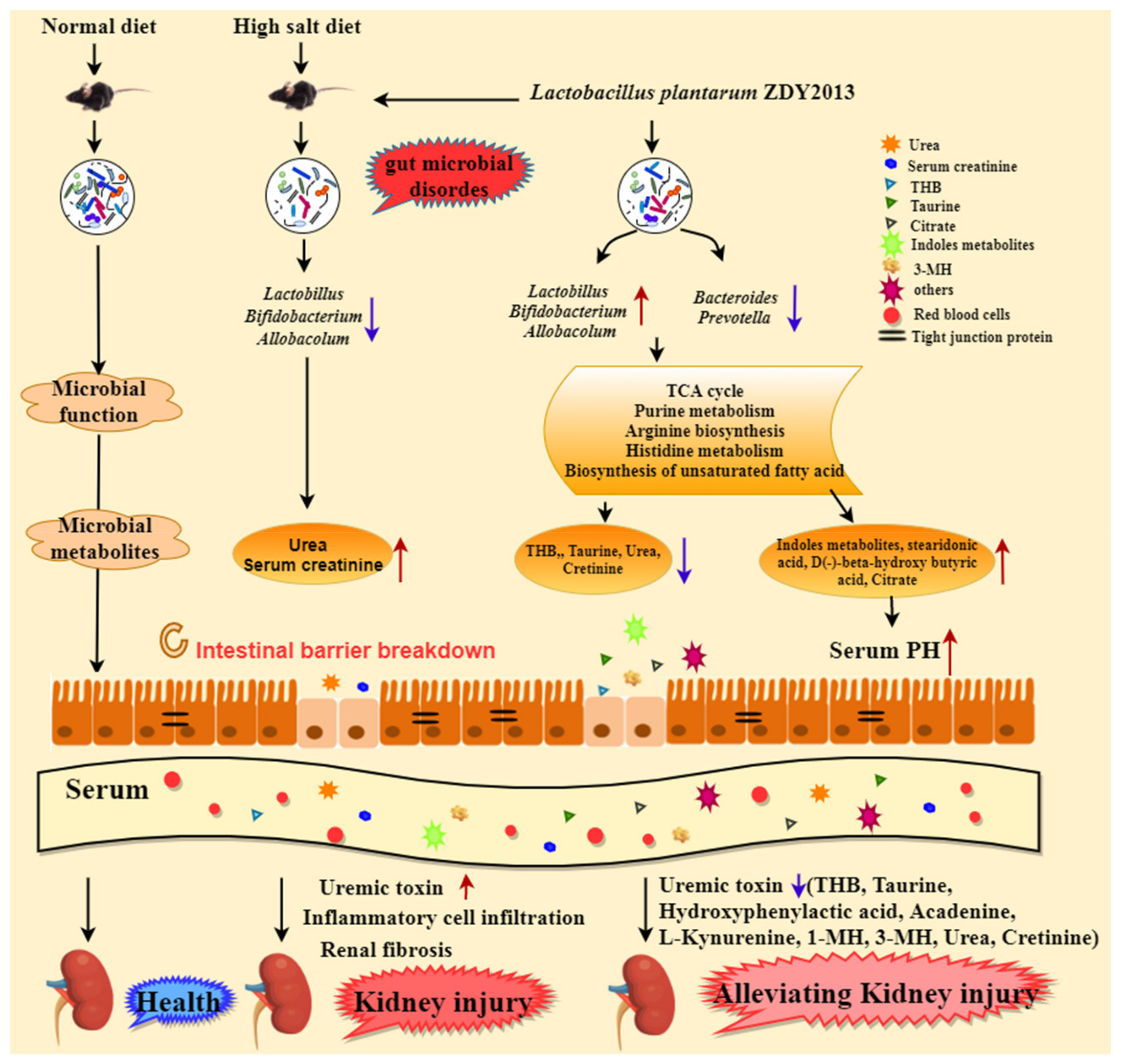
3.1. HSD Broke the Balance of Intestinal Flora, while L. plantarum ZDY2013 Alleviated It
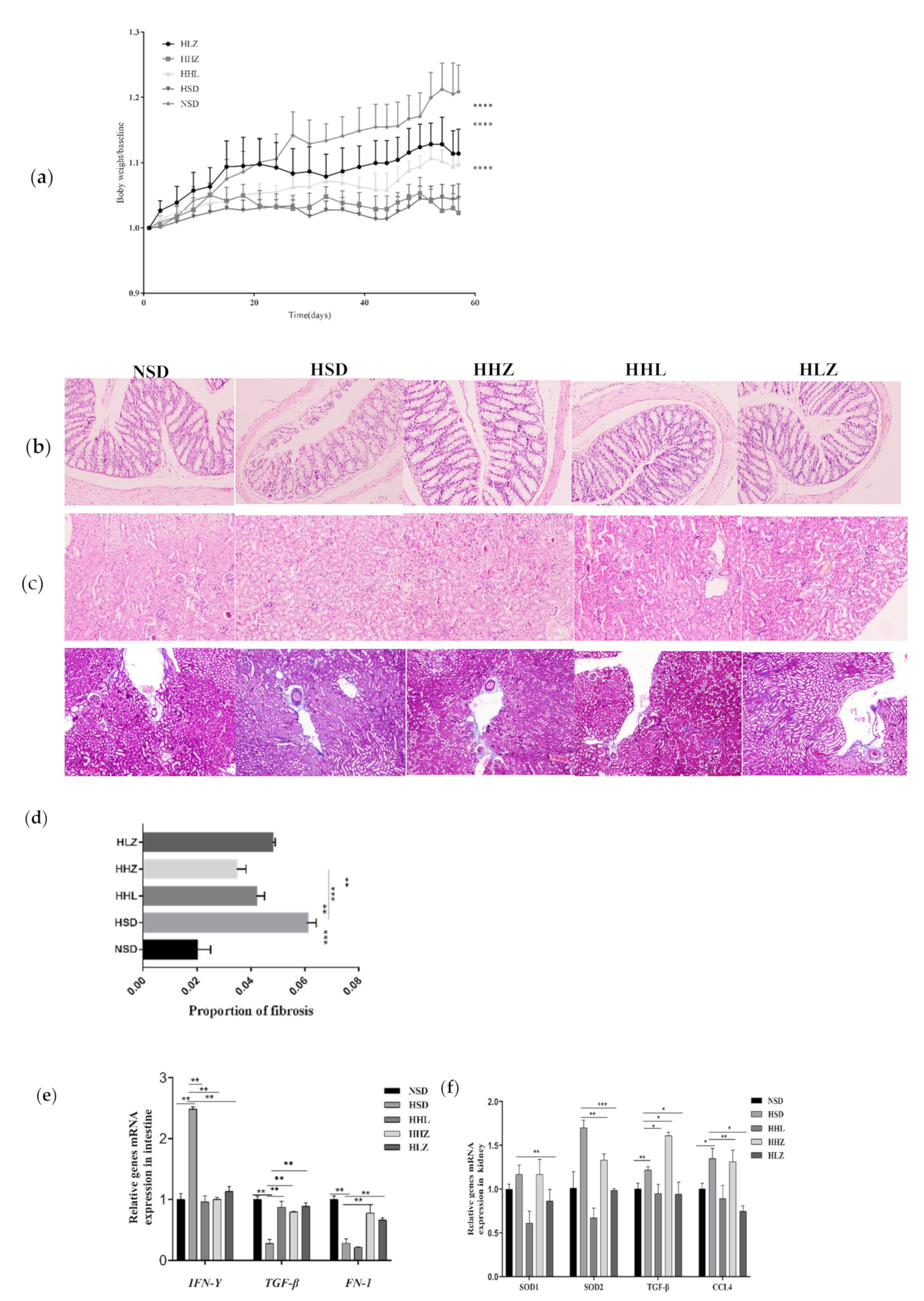
3.2. L. plantarum ZDY2013 Alleviated Body Damage Caused by Long-Term High-Salt Diet
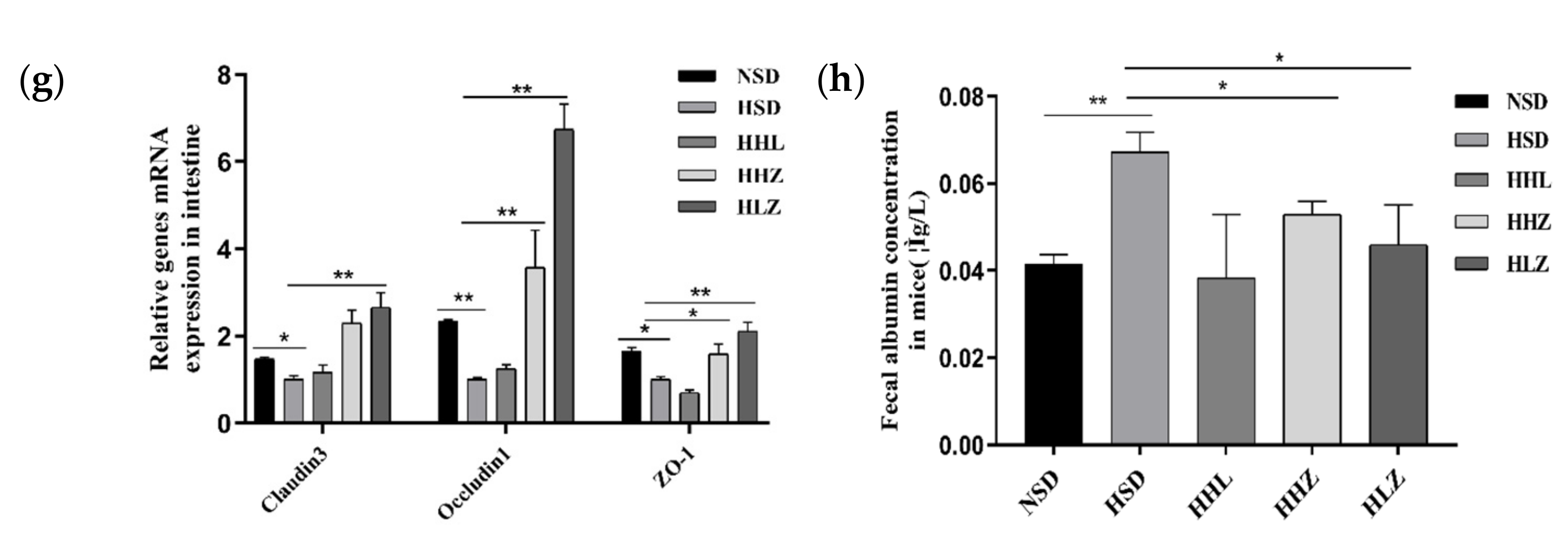
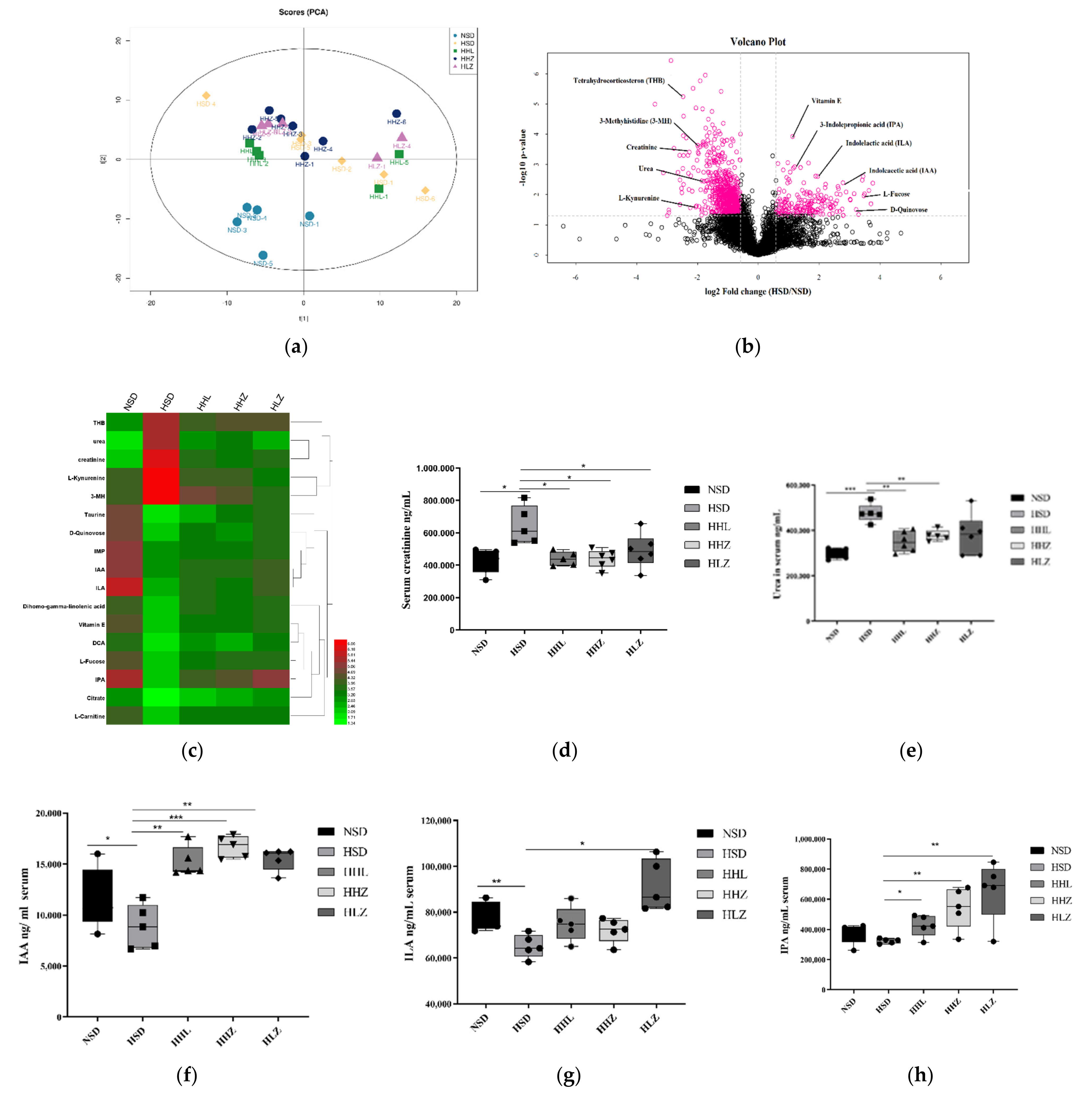
3.3. L. plantarum ZDY2013 Relieved Kidney Injury by Reducing Uremic Toxin
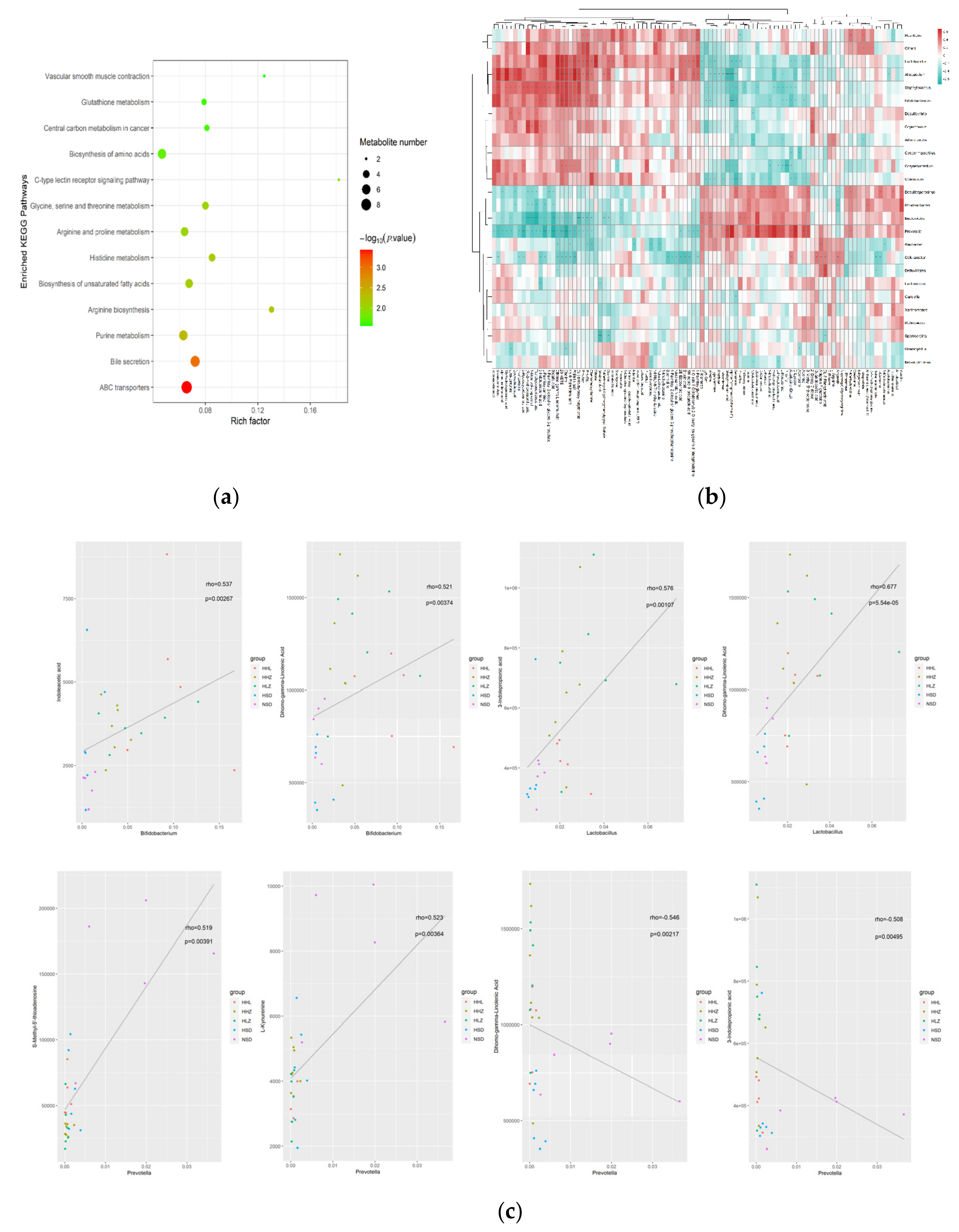
3.4. L. plantarum ZDY2013 Alleviates Kidney Injury via Regulating Intestinal Microorganisms to Reduce Urinary Toxin Concentration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bragulat, E.; de la Sierra, A. Salt intake, endothelial dysfunction, and salt-sensitive hypertension. J. Clin. Hypertens. 2002, 4, 41–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuda, N.; Stamler, J.; Brown, I.; Ueshima, H.; Miura, K.; Okayama, A.; Saitoh, S.; Nakagawa, H.; Sakata, K.; Yoshita, K.; et al. Individual efforts to reduce salt intake in China, Japan, UK, USA: What did people achieve? The INTERMAP Population Study. J. Hypertens. 2014, 32, 2385–2392. [Google Scholar] [CrossRef] [PubMed]
- Meneton, P.; Jeunemaitre, X.; de Wardener, H.; MacGregor, G. Links between dietary salt intake, renal salt handling, blood pressure, and cardiovascular diseases. Physiol. Rev. 2005, 85, 679–715. [Google Scholar] [CrossRef] [PubMed]
- Abdoli, A.; Jahromi, M.; Roustazadeh, A. Role of diet and gut microbiota in multiple sclerosis: New findings on the role of high-salt intake in induction of neuroinflammation. Clin. Exp. Neuroimmunol. 2019, 10, 149–151. [Google Scholar] [CrossRef]
- Kassem, S.; Howard, A.; Yehuda, S.; MaACR. The role of dietary sodium in autoimmune diseases: The salty truth. Autoimmun. Rev. 2018, 17, S1568997218302040. [Google Scholar]
- Jca, D.; Xl, B.; Hha, D.; Fza, C.; Yl, B.; Hai, H. High salt diet may promote progression of breast tumor through eliciting immune response. Int. Immunopharmacol. 2020, 87, 106816. [Google Scholar]
- Yoshida, Y.; Kosaki, K.; Sugasawa, T.; Matsui, M.; Takekoshi, K. High Salt Diet Impacts the Risk of Sarcopenia Associated with Reduction of Skeletal Muscle Performance in the Japanese Population. Nutrients 2020, 12, 3474. [Google Scholar] [CrossRef]
- Hosohata, K.; Yoshioka, D.; Tanaka, A.; Ando, H.; Fujimura, A. Early urinary biomarkers for renal tubular damage in spontaneously hypertensive rats on a high salt intake. Hypertens. Res. 2016, 39, 19–26. [Google Scholar] [CrossRef]
- Healy, E. Role of tubule epithelial cells in the pathogenesis of tubulointerstitial fibrosis induced by glomerular disease. Curr. Opin. Nephrol. Hypertens. 1998, 7, 525–530. [Google Scholar]
- Banday, A.; Lokhandwala, M. Renal Dopamine Oxidation and Inflammation in High Salt Fed Rats. J. Am. Heart Assoc. 2020, 9, e014977. [Google Scholar] [CrossRef]
- Hu, J.; Luo, H.; Wang, J.; Tang, W.; Lu, J.; Wu, S.; Xiong, Z.; Yang, G.; Chen, Z.; Lan, T.; et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp. Mol. Med. 2017, 49, e370. [Google Scholar] [CrossRef] [Green Version]
- Pahmi, K. The Effect of Telmisartan on Bone Morphogenetic Protein-7 (BMP-7) Expression in the Kidney of 8% Sodium Chloride-Treated Rats. Biol. Med. 2018, 10, 1–3. [Google Scholar] [CrossRef]
- Moonen, L.; Geryl, H.; D’Haese, P.; Vervaet, B.A. Short-term dexamethasone treatment transiently, but not permanently, attenuates fibrosis after acute-to-chronic kidney injury. BMC Nephrol. 2018, 19, 343. [Google Scholar] [CrossRef]
- Alzueta, N.; Echeverría, A.; Sanz, L.; Fontela, C.; Acín, T.; Montenegro, L.; Garjón, J. Telmisartan-induced sprue-like enteropathy: A case report. Eur. J. Hosp. Pharm. Sci. Pract. 2020, 27, 49–51. [Google Scholar] [CrossRef]
- Malkawi, A.; Alzoubi, K.; Jacob, M.; Matic, G.; Ali, A.; Al Faraj, A.; Almuhanna, F.; Dasouki, M.; Abdel Rahman, A. Metabolomics Based Profiling of Dexamethasone Side Effects in Rats. Front. Pharmacol. 2018, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Jeung, W.; Choi, I.D.; Jeong, J.W.; Ahn, Y.T. Lactic Acid Bacteria Improves Peyers Patch Cell-Mediated Immunoglobulin A and Tight-Junction Expression in a Destructed Gut Microbial Environment. J. Microbiol. Biotechnol. 2016, 26, 1035–1045. [Google Scholar] [CrossRef] [Green Version]
- Ku, S.; Yang, S.; Lee, H.H.; Choe, D.; Park, M.S. Biosafety assessment of Bifidobacterium animalis subsp. lactis AD011 used for human consumption as a probiotic microorganism. Food Control 2020, 117, 106985. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, H.; Jeong, D.; Kang, I.B.; Chon, J.W.; Kim, H.S.; Song, K.Y.; Seo, K.H. Kefir alleviates obesity and hepatic steatosis in high-fat diet-fed mice by modulation of gut microbiota and mycobiota: Targeted and untargeted community analysis with correlation of biomarkers. J. Nutr. Biochem. 2017, 44, 35–43. [Google Scholar] [CrossRef]
- Park, J.; Joe, I.; Rhee, P.; Jeong, C.; Jeong, G. A lactic acid bacterium isolated from kimchi ameliorates intestinal inflammation in DSS-induced colitis. J. Microbiol. 2017, 55, 304–310. [Google Scholar] [CrossRef]
- Bereswill, S.; Ekmekciu, I.; Escher, U.; Fiebiger, U.; Stingl, K.; Heimesaat, M. Lactobacillus johnsonii ameliorates intestinal, extra-intestinal and systemic pro-inflammatory immune responses following murine Campylobacter jejuni infection. Sci. Rep. 2017, 7, 2138. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Tao, X.; Wan, C.; Li, S.; Xu, H.; Xu, F.; Shah, N.; Wei, H. In vitro probiotic characteristics of Lactobacillus plantarum ZDY 2013 and its modulatory effect on gut microbiota of mice. J. Dairy Sci. 2015, 98, 5850–5861. [Google Scholar] [CrossRef]
- Huang, R.; Pan, M.; Wan, C.; Shah, N.P.; Tao, X.; Hua, W. Physiological and transcriptional responses and cross protection of Lactobacillus plantarum ZDY2013 under acid stress. J. Dairy Sci. 2016, 99, 1002–1010. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Lozupone, C.; Turnbaugh, P.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Erben, U.; Loddenkemper, C.; Doerfel, K.; Spieckermann, S.; Haller, D.; Heimesaat, M.; Zeitz, M.; Siegmund, B.; Kühl, A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 2014, 7, 4557–4576. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [Green Version]
- Zelante, T.; Iannitti, R.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Batlle, D.; Sharma, A.; Alsheikha, M.; Sobrero, M.; Saleh, A.; Gutterman, C. Renal acid excretion and intracellular pH in salt-sensitive genetic hypertension. J. Clin. Investig. 1993, 91, 2178–2184. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, J.; Aden, L.; Barbaro, N.; Van Beusecum, J.; Xiao, L.; Simmons, A.; Warden, C.; Pasic, L.; Himmel, L.; Washington, M.; et al. High dietary salt-induced dendritic cell activation underlies microbial dysbiosis-associated hypertension. JCI Insight 2019, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Oudot, C.; Lajoix, A.D.; Jover, B.; Rugale, C. Dietary sodium restriction prevents kidney damage in high fructose-fed rats. Kidney Int. 2013, 83, 674–683. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Hu, X.; Zhang, X.; Chen, Z.; Wang, J.; Jin, K.; Cao, F.; Sun, B.; Bellanti, J.; Olsen, N.; et al. High salt diet accelerates the progression of murine lupus through dendritic cells via the p38 MAPK and STAT1 signaling pathways. Signal Transduct. Target. Ther. 2020, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- De Miguel, C.; Sedaka, R.; Kasztan, M.; Lever, J.M.; Sonnenberger, M.; Abad, A.; Jin, C.; Carmines, P.K.; Pollock, D.M.; Pollock, J.S. Tauroursodeoxycholic acid (TUDCA) abolishes chronic high salt-induced renal injury and inflammation. Acta Physiol. 2019, 226, e13227. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Nikolic-Paterson, D.; Lan, H. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Pardali, E.; Sanchez-Duffhues, G.; Gomez-Puerto, M.; Ten Dijke, P. TGF-β-Induced Endothelial-Mesenchymal Transition in Fibrotic Diseases. Int. J. Mol. Sci. 2017, 18, 2157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Kitai, T.; Hazen, S.L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhao, J.; Tian, C.; Chen, X.; Li, H.; Wei, X.; Lin, W.; Zheng, N.; Jiang, A.; Feng, R.; et al. Targeting the Gut Microbiota to Investigate the Mechanism of Lactulose in Negating the Effects of a High-Salt Diet on Hypertension. Mol. Nutr. Food Res. 2019, 63, e1800941. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Chen, M.; Zhang, X.; Ji, Q.; Wang, Y.; Zheng, Q.; Tan, S.; Gao, X.; Lu, Y. Salted and Unsalted Zhàcài (Brassica juncea var. tumida) Alleviated High-Fat Diet-Induced Dyslipidemia by Regulating Gut Microbiota: A Multiomics Study. Mol. Nutr. Food Res. 2020, e2000798. [Google Scholar] [CrossRef]
- Fu, L.; Xie, M.; Wang, C.; Qian, Y.; Huang, J.; Sun, Z.; Zhang, H.; Wang, Y. Lactobacillus Casei Zhang Alleviates Shrimp Tropomyosin-Induced Food Allergy by Switching Antibody Isotypes through the NF-κB-Dependent Immune Tolerance. Mol. Nutr. Food Res. 2020, 64, e1900496. [Google Scholar] [CrossRef]
- Zhang, X.; Tong, Y.; Lyu, X.; Wang, J.; Wang, Y.; Yang, R. Bacillus subtilisPrevention and Alleviation of Dextran Sulfate Sodium Salt-Induced Inflammatory Bowel Disease in Mice With -Fermented Milk Inhibition of the Inflammatory Responses and Regulation of the Intestinal Flora. Front. Microbiol. 2020, 11, 622354. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, K.; Wu, W.; Lv, L.; Bian, X.; Yang, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; et al. Administration of Bifidobacterium bifidum CGMCC 15068 modulates gut microbiota and metabolome in azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced colitis-associated colon cancer (CAC) in mice. Appl. Microbiol. Biotechnol. 2020, 104, 5915–5928. [Google Scholar] [CrossRef]
- Panpetch, W.; Hiengrach, P.; Nilgate, S.; Tumwasorn, S.; Somboonna, N.; Wilantho, A.; Chatthanathon, P.; Prueksapanich, P.; Leelahavanichkul, A. Candida albicansAdditional administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by L34. Gut Microbes 2020, 11, 465–480. [Google Scholar] [CrossRef]
- Alicia, B.; Nicole, R.; Adrian, C.; Warren, M.N. Metabolism of Caprine Milk Carbohydrates by Probiotic Bacteria and Caco-2:HT29–MTX Epithelial Co-Cultures and Their Impact on Intestinal Barrier Integrity. Nutrients 2018, 10, 949. [Google Scholar]
- Krumbeck, J.; Rasmussen, H.; Hutkins, R.; Clarke, J.; Shawron, K.; Keshavarzian, A.; Walter, J. Probiotic Bifidobacterium strains and galactooligosaccharides improve intestinal barrier function in obese adults but show no synergism when used together as synbiotics. Microbiome 2018, 6, 121. [Google Scholar] [CrossRef]
- Wang, W.; Liu, F.; Xu, C.; Liu, Z.; Ma, J.; Gu, L.; Jiang, Z.; Hou, J. Lactobacillus plantarum 69-2 Combined with Galacto-Oligosaccharides Alleviates d-Galactose-Induced Aging by Regulating the AMPK/SIRT1 Signaling Pathway and Gut Microbiota in Mice. J. Agric. Food Chem. 2021, 69, 2745–2757. [Google Scholar] [CrossRef]
- Evenepoel, P.; Meijers, B.; Bammens, B.; Verbeke, K. Uremic toxins originating from colonic microbial metabolism. Kidney Int. 2009, 76, S12–S19. [Google Scholar] [CrossRef] [Green Version]
- Schwarzer, M.; Makki, K.; Storelli, G.; Machuca-Gayet, I.; Srutkova, D.; Hermanova, P.; Martino, M.; Balmand, S.; Hudcovic, T.; Heddi, A.; et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 2016, 351, 854–857. [Google Scholar] [CrossRef]
- Nigam, S.K.; Wu, W.; Bush, K.T.; Hoenig, M.P.; Blantz, R.C.; Bhatnagar, V. Handling of Drugs, Metabolites, and Uremic Toxins by Kidney Proximal Tubule Drug Transporters. Clin. J. Am. Soc. Nephrol. 2015, 10, 2039–2049. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Klaassen, C. Gender differences in mRNA expression of ATP-binding cassette efflux and bile acid transporters in kidney, liver, and intestine of 5/6 nephrectomized rats. Drug Metab. Dispos. 2008, 36, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Barnes, S.; Gollan, J.; Billing, B. The role of tubular reabsorption in the renal excretion of bile acids. Biochem. J. 1977, 166, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Wu, I.; Gao, S.; Chou, H.; Yang, H.; Chang, L.; Kuo, Y.; Dinh, M.; Chung, W.; Yang, C.; Lai, H.; et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics 2020, 10, 5398–5411. [Google Scholar] [CrossRef]
- Curtis, M.; Hu, Z.; Klimko, C.; Narayanan, S.; Deberardinis, R.; Sperandio, V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 2014, 16, 759–769. [Google Scholar] [CrossRef] [Green Version]
- Bhattarai, Y.; Williams, B.; Battaglioli, E.; Whitaker, W.; Till, L.; Grover, M.; Linden, D.; Akiba, Y.; Kandimalla, K.; Zachos, N.; et al. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe 2018, 23, 775–785. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, A.; Destefano Shields, C.; Wu, S.; Huso, D.; Wu, X.; Murray-Stewart, T.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.; Sears, C.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [Green Version]
- Org, E.; Blum, Y.; Kasela, S.; Mehrabian, M.; Kuusisto, J.; Kangas, A.; Soininen, P.; Wang, Z.; Ala-Korpela, M.; Hazen, S.; et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017, 18, 70. [Google Scholar] [CrossRef]
- Yacoub, R.; Wyatt, C. Manipulating the gut microbiome to decrease uremic toxins. Kidney Int. 2017, 91, 521–523. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.; Heim, C.; Krastel, P.; Schmitt, E.; Omar, A.; Creasey, E.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, J.; O’Mahony, L.; O’Callaghan, L.; Sheil, B.; Vaughan, E.; Fitzsimons, N.; Fitzgibbon, J.; O’Sullivan, G.; Kiely, B.; Collins, J.; et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut 2003, 52, 975–980. [Google Scholar] [CrossRef]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Aiello, V.; Romano, B.; De Lorenzo, A.; Izzo, A.; et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, C.; Chen, S.; Zhao, K.; Ren, Z.; Peng, L.; Xia, H.; Wei, H.; Yu, B. Serum Untargeted Metabolism Reveals the Mechanism of L. plantarum ZDY2013 in Alleviating Kidney Injury Induced by High-Salt Diet. Nutrients 2021, 13, 3920. https://doi.org/10.3390/nu13113920
Wan C, Chen S, Zhao K, Ren Z, Peng L, Xia H, Wei H, Yu B. Serum Untargeted Metabolism Reveals the Mechanism of L. plantarum ZDY2013 in Alleviating Kidney Injury Induced by High-Salt Diet. Nutrients. 2021; 13(11):3920. https://doi.org/10.3390/nu13113920
Chicago/Turabian StyleWan, Cuixiang, Shufang Chen, Kui Zhao, Zhongyue Ren, Lingling Peng, Huiling Xia, Hua Wei, and Bo Yu. 2021. "Serum Untargeted Metabolism Reveals the Mechanism of L. plantarum ZDY2013 in Alleviating Kidney Injury Induced by High-Salt Diet" Nutrients 13, no. 11: 3920. https://doi.org/10.3390/nu13113920
APA StyleWan, C., Chen, S., Zhao, K., Ren, Z., Peng, L., Xia, H., Wei, H., & Yu, B. (2021). Serum Untargeted Metabolism Reveals the Mechanism of L. plantarum ZDY2013 in Alleviating Kidney Injury Induced by High-Salt Diet. Nutrients, 13(11), 3920. https://doi.org/10.3390/nu13113920





