The Impact of Microbial Composition on Postprandial Glycaemia and Lipidaemia: A Systematic Review of Current Evidence
Abstract
:1. Introduction
2. Results
2.1. Literature Search and Study Selection Process
2.2. Characteristics of the Included Studies
2.3. Modulation of the Gut Microbiome Using Dietary Intervention
2.4. Individual Intervariability of Metabolic Responses and Microbial Diversity Using a Standardised Approach
2.5. Interaction between Drugs and the Gut Microbiome
3. Discussion
4. Materials and Methods
4.1. Literature Search Strategy
4.2. Search Methods
4.3. Selection Criteria
4.4. Data Extraction and Critical Appraisal
4.5. Quality Assessment
4.6. Data Extraction and Management
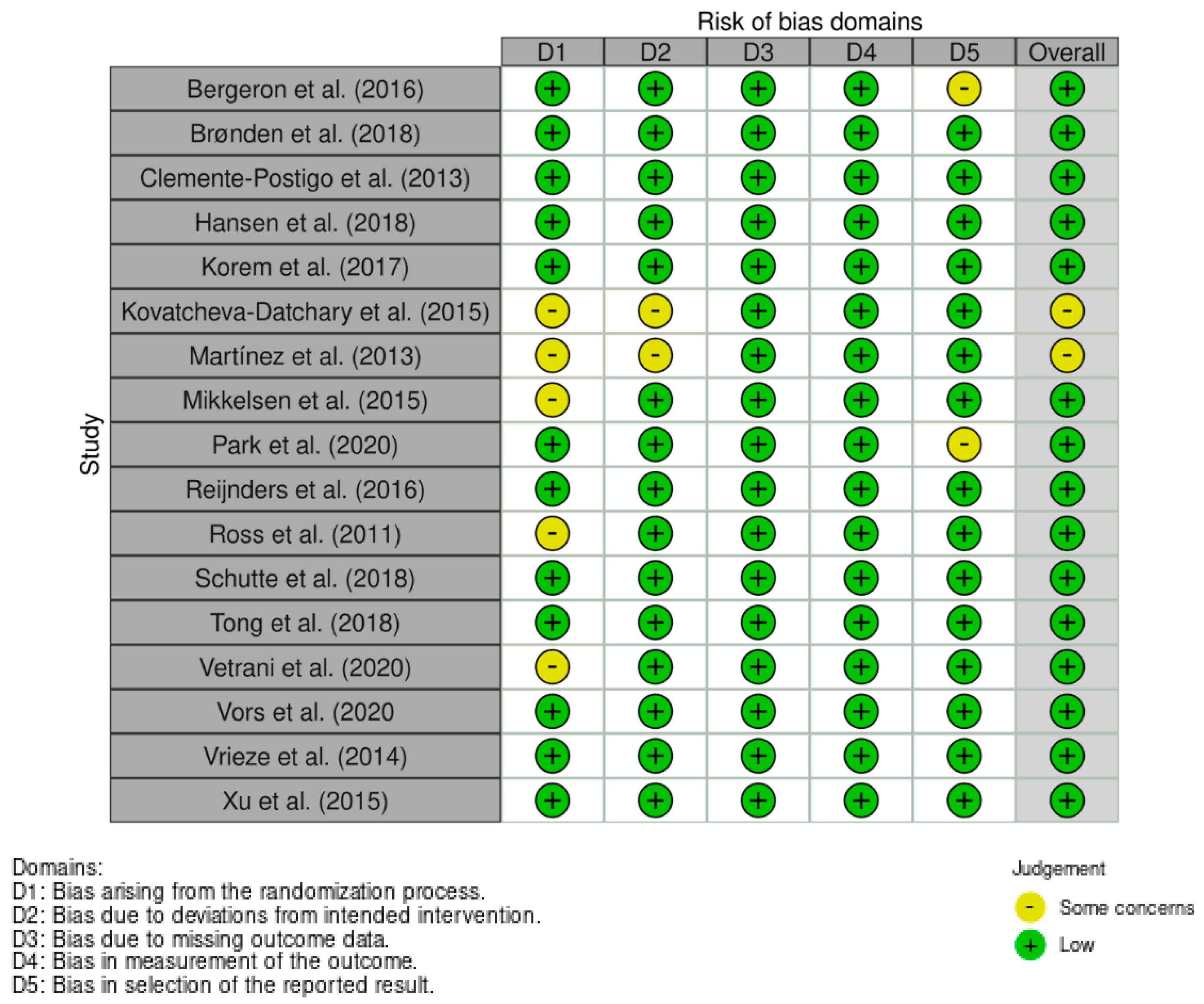
4.7. Assessment of Risk of Bias
5. Strengths and Limitations of the Current Review
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Node, K.; Inoue, T. Postprandial hyperglycemia as an etiological factor in vascular failure. Cardiovasc. Diabetol. 2009, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Sargsyan, A.; Herman, M.A. Regulation of Glucose Production in the Pathogenesis of Type 2 Diabetes. Curr. Diabetes Rep. 2019, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.B.; Moughan, P.J.; Wood, L.G.; Singh, H.; Garg, M.L. Postprandial lipemia: Factoring in lipemic response for ranking foods for their healthiness. Lipids Health Dis. 2017, 16, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmarchelier, C.; Borel, P.; Lairon, D.; Maraninchi, M.; Valéro, R. Effect of Nutrient and Micronutrient Intake on Chylomicron Production and Postprandial Lipemia. Nutrients 2019, 11, 1299. [Google Scholar] [CrossRef] [Green Version]
- Bozzetto, L.; Pepa, G.D.; Vetrani, C.; Rivellese, A.A. Dietary Impact on Postprandial Lipemia. Front. Endocrinol. 2020, 11, 337. [Google Scholar] [CrossRef]
- Panagiotakos, D.B.; Kouvari, M. Behavioural healthy nutrition and physical activity counselling in cardiovascular disease prevention: Where we are now? Hepatobiliary Surg. Nutr. 2019, 8, 534. [Google Scholar] [CrossRef]
- De Roos, B.; Brennan, L. Personalised interventions—A precision approach for the next generation of dietary intervention studies. Nutrients 2017, 2017, 847. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, L.R.; De Caterina, R.; Görman, U.; Allayee, H.; Kohlmeier, M.; Prasad, C.; Kang, J.X. Guide and position of the international society of nutrigenetics/nutrigenomics on personalised nutrition: Part 1-fields of precision nutrition. Lifestyle Genom. 2016, 9, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Bush, C.L.; Blumberg, J.B.; El-Sohemy, A.; Minich, D.M.; Ordovás, J.M.; Reed, D.G.; Behm, V.A.Y. Toward the definition of personalized nutrition: A proposal by The American Nutrition Association. J. Am. Coll. Nutr. 2020, 39, 5–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betts, J.A.; Gonzalez, J.T. Personalised nutrition: What makes you so special? Nutr. Bull. 2016, 41, 353–359. [Google Scholar] [CrossRef]
- Lampe, J.W.; Navarro, S.L.; Hullar, M.A.J.; Shojaie, A. Inter-individual differences in response to dietary intervention: Integrating omics platforms towards personalised dietary recommendations. Proc. Nutr. Soc. 2013, 72, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Ryan, E.P.; Heuberger, A.L.; Broeckling, C.D.; Borresen, E.C.; Tillotson, C.; Prenni, J.E. Advances in nutritional metabolomics. Curr. Metab. 2013, 1, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castaner, O.; Goday, A.; Park, Y.-M.; Lee, S.-H.; Magkos, F.; Shiow, S.-A.T.E.; Schröder, H. The Gut Microbiome Profile in Obesity: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 4095789. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Verma, M.K.; Chauhan, N.S. A review of metabolic potential of human gut microbiome in human nutrition. Arch. Microbiol. 2018, 200, 203–217. [Google Scholar] [CrossRef]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Bäckhed, F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef] [Green Version]
- Burcelin, R. Gut microbiota and immune crosstalk in metabolic disease. Mol. Metab. 2016, 5, 771–781. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [Green Version]
- Berry, S.E.; Valdes, A.M.; Drew, D.A.; Asnicar, F.; Mazidi, M.; Wolf, J.; Capdevila, J.; Hadjigeorgiou, G.; Davies, R.; Al Khatib, H.; et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 2020, 26, 964–973. [Google Scholar] [CrossRef]
- Mendes-Soares, H.; Raveh-Sadka, T.; Azulay, S.; Edens, K.; Ben-Shlomo, Y.; Cohen, Y.; Ofek, T.; Bachrach, D.; Stevens, J.; Colibaseanu, D.; et al. Assessment of a Personalized Approach to Predicting Postprandial Glycemic Responses to Food Among Individuals Without Diabetes. JAMA Netw. Open 2019, 2, e188102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Raka, F.; Adeli, K. The Role of the Gut Microbiota in Lipid and Lipoprotein Metabolism. J. Clin. Med. 2019, 8, 2227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961.e17. [Google Scholar] [CrossRef] [Green Version]
- Mendes-Soares, H.; Raveh-Sadka, T.; Azulay, S.; Ben-Shlomo, Y.; Cohen, Y.; Ofek, T.; Stevens, J.; Bachrach, D.; Kashyap, P.; Segal, L.; et al. Model of personalized postprandial glycemic response to food developed for an Israeli cohort predicts responses in Midwestern American individuals. Am. J. Clin. Nutr. 2019, 110, 63–75. [Google Scholar] [CrossRef] [Green Version]
- Brønden, A.; Mikkelsen, K.; Sonne, D.P.; Hansen, M.; Våben, C.; Gabe, M.N.; Rosenkilde, M.; Tremaroli, V.; Wu, H.; Bäckhed, F.; et al. Glucose-lowering effects and mechanisms of the bile acid-sequestering resin sevelamer. Diabetes Obes. Metab. 2018, 20, 1623–1631. [Google Scholar] [CrossRef]
- Mikkelsen, K.H.; Frost, M.; Bahl, M.I.; Licht, T.R.; Jensen, U.S.; Rosenberg, J.; Pedersen, O.; Hansen, T.; Rehfeld, J.F.; Holst, J.J.; et al. Effect of Antibiotics on Gut Microbiota, Gut Hormones and Glucose Metabolism. PLoS ONE 2015, 10, e0142352. [Google Scholar] [CrossRef] [Green Version]
- Tong, X.; Xu, J.; Lian, F.; Yu, X.; Zhao, Y.; Xu, L.; Zhang, M.; Zhao, X.; Shen, J.; Wu, S.; et al. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: A Multicenter, Randomized, Open Label Clinical Trial. mBio 2018, 9, e02392-17. [Google Scholar] [CrossRef] [Green Version]
- Bergeron, N.; Williams, P.T.; Lamendella, R.; Faghihnia, N.; Grube, A.; Li, X.; Wang, Z.; Knight, R.; Jansson, J.; Hazen, S.L.; et al. Diets high in resistant starch increase plasma levels of trimethylamine-N-oxide, a gut microbiome metabolite associated with CVD risk. Br. J. Nutr. 2016, 116, 2020–2029. [Google Scholar] [CrossRef] [Green Version]
- Clemente-Postigo, M.; Ortuño, M.I.Q.; Boto-Ordoñez, M.; Coin-Aragüez, L.; Roca-Rodriguez, M.D.M.; Delgado-Lista, J.; Cardona, F.; Andres-Lacueva, C.; Tinahones, F.J. Effect of acute and chronic red wine consumption on lipopolysaccharide concentrations. Am. J. Clin. Nutr. 2013, 97, 1053–1061. [Google Scholar] [CrossRef] [Green Version]
- Hansen, L.B.S.; Roager, H.M.; Søndertoft, N.B.; Gøbel, R.J.; Kristensen, M.; Vallès-Colomer, M.; Vieira-Silva, S.; Ibrügger, S.; Lind, M.V.; Mærkedahl, R.B.; et al. A low-gluten diet induces changes in the intestinal microbiome of healthy Danish adults. Nat. Commun. 2018, 9, 4360. [Google Scholar] [CrossRef] [Green Version]
- Korem, T.; Zeevi, D.; Zmora, N.; Weissbrod, O.; Bar, N.; Lotan-Pompan, M.; Avnit-Sagi, T.; Kosower, N.; Malka, G.; Rein, M.; et al. Bread Affects Clinical Parameters and Induces Gut Microbiome-Associated Personal Glycemic Responses. Cell Metab. 2017, 25, 1243–1253.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Bäckhed, F. Dietary fibre-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [Green Version]
- Martínez, I.; Lattimer, J.; Hubach, K.L.; A Case, J.; Yang, J.; Weber, C.G.; A Louk, J.; Rose, D.J.; Kyureghian, G.; A Peterson, D.; et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2012, 7, 269–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.; Zenger, K.R. Methods for normalizing microbiome data: An ecological perspective. Methods Ecol. Evol. 2019, 10, 389–400. [Google Scholar] [CrossRef]
- Park, Y.E.; Kim, M.S.; Shim, K.W.; Kim, Y.-I.; Chu, J.; Kim, B.-K.; Choi, I.S.; Kim, J.Y. Effects of Lactobacillus plantarum Q180 on Postprandial Lipid Levels and Intestinal Environment: A Double-Blind, Randomized, Placebo-Controlled, Parallel Trial. Nutrients 2020, 12, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reijnders, D.; Goossens, G.H.; Hermes, G.D.A.; Neis, E.P.; van der Beek, C.M.; Most, J.; Holst, J.J.; Lenaerts, K.; Kootte, R.S.; Nieuwdorp, M.; et al. Effects of Gut Microbiota Manipulation by Antibiotics on Host Metabolism in Obese Humans: A Randomized Double-Blind Placebo-Controlled Trial. Cell Metab. 2016, 24, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Ross, A.B.; Bruce, S.J.; Blondel-Lubrano, A.; Oguey-Araymon, S.; Beaumont, M.; Bourgeois, A.; Nielsen-Moennoz, C.; Vigo, M.; Fay, L.-B.; Kochhar, S.; et al. A whole-grain cereal-rich diet increases plasma betaine, and tends to decrease total and LDL-cholesterol compared with a refined-grain diet in healthy subjects. Br. J. Nutr. 2011, 105, 1492–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schutte, S.; Esser, D.; Hoevenaars, F.P.M.; Hooiveld, G.; Priebe, M.G.; Vonk, R.J.; Wopereis, S.; A Afman, L. A 12-wk whole-grain wheat intervention protects against hepatic fat: The Graandioos study, a randomized trial in overweight subjects. Am. J. Clin. Nutr. 2018, 108, 1264–1274. [Google Scholar] [CrossRef] [Green Version]
- Tily, H.; Perlina, A.; Patridge, E.; Gline, S.; Genkin, M.; Gopu, V.; Banavar, G. Gut microbiome activity contributes to individual variation in glycemic response in adults. bioRxiv 2019, 641019. [Google Scholar] [CrossRef]
- Vetrani, C.; Maukonen, J.; Bozzetto, L.; Pepa, G.D.; Vitale, M.; Costabile, G.; Annuzzi, G. Diets naturally rich in poly-phenols and/or long-chain n-3 polyunsaturated fatty acids differently affect microbiota composition in high-cardiometabolic-risk individuals. Acta Diabetol. 2020, 57, 853–860. [Google Scholar] [CrossRef]
- Vors, C.; Joumard-Cubizolles, L.; Lecomte, M.; Combe, E.; Ouchchane, L.; Drai, J.; Michalski, M.C. Milk polar lipids reduce lipid cardiovascular risk factors in overweight postmenopausal women: Towards a gut sphingomyelin-cholesterol interplay. Gut 2020, 69, 487–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef]
- Xu, J.; Lian, F.; Zhao, L.; Zhao, Y.; Chen, X.; Zhang, X.; Guo, Y.; Zhang, C.; Zhou, Q.; Xue, Z.; et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015, 9, 552–562. [Google Scholar] [CrossRef]
- Fiedorová, K.; Radvanský, M.; Němcová, E.; Grombiříková, H.; Bosák, J.; Černochová, M.; Freiberger, T. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front. Microbiol. 2019, 10, 821. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Gill, K.D. Estimation of Blood Glucose Levels by Glucose Oxidase Method. In Basic Concepts in Clinical Biochemistry: A Practical Guide; Springer: Singapore, 2018; pp. 57–60. [Google Scholar]
- Akintola, A.A.; Noordam, R.; Jansen, S.W.; De Craen, A.J.; Ballieux, B.E.; Cobbaert, C.; Mooijaart, S.; Pijl, H.; Westendorp, R.G.; Van Heemst, D. Accuracy of Continuous Glucose Monitoring Measurements in Normo-Glycemic Individuals. PLoS ONE 2015, 10, e0139973. [Google Scholar] [CrossRef] [Green Version]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 579–588. [Google Scholar] [CrossRef]
- Miyagi, S.; Takamura, T.; Nguyen, T.T.T.; Tsujiguchi, H.; Hara, A.; Nakamura, H.; Nakamura, H. Moderate alcohol consumption is associated with impaired insulin secretion and fasting glucose in non-obese non-diabetic men. J. Diabetes Investig. 2020, 12, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, S.; Joseph, P.; Rangarajan, S.; Islam, S.; Mente, A.; Hystad, P.; Brauer, M.; Kutty, V.R.; Gupta, R.; Wielgosz, A.; et al. Modifiable risk factors, cardiovascular disease, and mortality in 155,722 individuals from 21 high-income, middle-income, and low-income countries (PURE): A prospective cohort study. Lancet 2020, 395, 795–808. [Google Scholar] [CrossRef] [Green Version]
- Musso, G.; Gambino, R.; Cassader, M. Interactions Between Gut Microbiota and Host Metabolism Predisposing to Obesity and Diabetes. Annu. Rev. Med. 2011, 62, 361–380. [Google Scholar] [CrossRef]
- Søndertoft, N.B.; Vogt, J.K.; Arumugam, M.; Kristensen, M.; Gøbel, R.J.; Fan, Y.; Lyu, L.; Bahl, M.I.; Eriksen, C.; Ängquist, L.; et al. The intestinal microbiome is a co-determinant of the postprandial plasma glucose response. PLoS ONE 2020, 15, e0238648. [Google Scholar] [CrossRef]
- Nordestgaard, B.G.; Langsted, A.; Freiberg, J.J. Nonfasting hyperlipidemia and cardiovascular disease. Curr. Drug Targets 2009, 10, 328–335. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Li, J.V.; Zhou, N.; Tang, H.; Wang, Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J. Proteome Res. 2013, 12, 2987–2999. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Leshem, A.; Elinav, E. Breakthroughs and Bottlenecks in Microbiome Research. Trends Mol. Med. 2021, 27, 298–301. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, Z. Impact of trimethylamine N-oxide (TMAO) metaorganismal pathway on cardiovascular disease. J. Lab. Precis. Med. 2020, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Marventano, S.; Vetrani, C.; Vitale, M.; Godos, J.; Riccardi, G.; Grosso, G. Whole grain intake and glycaemic control in health subjects: A systematic review and meta-analysis of randomised controlled trials. Nutrients 2017, 9, 769. [Google Scholar] [CrossRef]
- Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; Bertalan, M.; Borruel, N.; Casellas, F.; Fernandez, L.; Gautier, L.; Hansen, T.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Hjorth, M.F.; Christensen, L.; Kjølbæk, L.; Larsen, L.H.; Roager, H.M.; Kiilerich, P.; Kristansen, K.; Astrup, A. Pretreatment Prevotella-to-Bacteroides ratio and markers of glucose metabolism as prognostic markers for dietary weight loss maintenance. Eur. J. Clin. Nutr. 2020, 74, 338–347. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Jiang, Q.; Yin, Y. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol. Metab. 2021, 32, 159–169. [Google Scholar] [CrossRef]
- Lavelle, A.; Hoffmann, T.W.; Pham, H.-P.; Langella, P.; Guédon, E.; Sokol, H. Baseline microbiota composition modulates antibiotic-mediated effects on the gut microbiota and host. Microbiome 2019, 7, 111. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Saxen, H.; Nikkonen, A.; Peltola, V.; Jaakkola, T.; De Vos, W.; Kolho, K.-L. Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatr. Res. 2020, 88, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Chng, K.R.; Ghosh, T.; Tan, Y.H.; Nandi, T.; Lee, I.R.; Ng, A.H.Q.; Li, C.; Ravikrishnan, A.; Lim, K.M.; Lye, D.; et al. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat. Ecol. Evol. 2020, 4, 1256–1267. [Google Scholar] [CrossRef]
- Das, B.; Nair, G.B. Homeostasis and dysbiosis of the gut microbiome in health and disease. J. Biosci. 2019, 44, 117. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Fei, N.; Pang, X.; Shen, J.; Wang, L.; Zhang, B.; Zhang, M.; Zhang, X.; Zhang, C.; Li, M.; et al. A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol. Ecol. 2014, 87, 357–367. [Google Scholar] [CrossRef]
- Hall, A.B.; Tolonen, A.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Patumcharoenpol, P.; Nakphaichit, M.; Panagiotou, G.; Senavonge, A.; Suratannon, N.; Vongsangnak, W. MetGEMs Toolbox: Metagenome-scale models as integrative toolbox for uncovering metabolic functions and routes of human gut microbiome. PLoS Comput. Biol. 2021, 17, e1008487. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [Green Version]
- Methley, A.M.; Campbell, S.; Chew-Graham, C.; McNally, R.; Cheraghi-Sohi, S. PICO, PICOS and SPIDER: A comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv. Res. 2014, 14, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgins, J.P.; Savović, J.; Page, M.J.; Elbers, R.G.; Sterne, J.A. Assessing risk of bias in a randomized trial. In Cochrane Handbook for Systematic Reviews of Interventions; Wiley Blackwell: Hoboken, NJ, USA, 2019; pp. 205–228. [Google Scholar] [CrossRef]
- Sterne, J.A.; A Hernán, M.; McAleenan, A.; Reeves, B.C.; Higgins, J.P. Assessing risk of bias in a non-randomized study. In Cochrane Handbook for Systematic Reviews of Interventions; Wiley Blackwell: Hoboken, NJ, USA, 2019; pp. 621–641. [Google Scholar] [CrossRef]

| First Author, Year, Country | Study Design | Age Range (year) | Male (%) | Background Disease | Sample Size (n) | Study Duration | Intervention Type | Summary of Key Findings |
|---|---|---|---|---|---|---|---|---|
| Bergeron et al. (2016), USA [29] | RCT | ≥20 y | 38.5 | Healthy men and post-menopausal women with no history of CVD or other chronic diseases. | 52 | 8 weeks | Diet: High and low total CHO intake compared to high vs. low resistant starch intake. |
|
| Berry et al. (2020), UK [20] | Series of acute experimental studies | 18–65 y | 27.8 | Healthy subjects with no history of chronic diseases. | 1002 | 2 weeks | Diet: Standardised meal testing to measure and predict individual metabolic responses. |
|
| Brønden et al. (2018), Denmark [26] | RCT | 35–80 y | 70 | Healthy subjects and patients with T2D for at least > 3 months. | 30 | 7 days | Drug: Treatment of 1600 mg of sevelamer or placebo. |
|
| Clemente-Postigo et al. (2013), Spain [30] | RCT | 45–50 y | 100 | Healthy subjects with no history of chronic diseases. | 5 | 12 days | Diet: 4-arm crossover intervention: 50 g fat overload, red wine/fat overload, dealcoholised red wine/fat overload, and 100 mL gin/fat overload. |
|
| Hansen et al. (2018), Denmark [31] | RCT | 18–65 y | 42.6 | Obesity/Risk of MetS. | 60 | 16 weeks | Diet: Low or high-gluten diet in comparison to habitual intake. |
|
| Korem et al. (2017), Israel [32] | Randomised crossover trial | 18–70 y | 55 | Healthy subjects with no history of chronic diseases. | 20 | 2 weeks | Diet: Consumption of sourdough bread compared to industrially made white bread. |
|
| Kovatcheva-Datchary et al. (2015), Sweden [33] | Randomised crossover trial | 50–70 y | 15.4 | Healthy subjects with no history of chronic diseases. | 39 | 3 weeks | Diet: Consumption of barley kernel-based bread (BKB) or white wheat flour bread (WWB). |
|
| Martínez et al. (2013), USA [34] | Randomised crossover trial | 18–65 y | 39.3 | Healthy subjects with no history of chronic diseases. | 28 | 17 weeks | Diet: Consumption of 60 g of whole-grain barley, brown rice, or an equal mixture of the two. |
|
| Mendes-Soares et al. (2019), USA [25] | Series of acute experimental studies | ≥18 y | 23 | Healthy subjects with no history of chronic diseases. | 297 | 6 days | Diet: Standardised meal testing to measure and predict individual metabolic responses. |
|
| Mendes-Soares et al. (2019), USA [21] | Series of acute experimental studies | ≥18 y | 22 | Healthy subjects with no history of chronic diseases. | 327 | 6 days | Diet: Consumption of two different standardised test meals (plain bagel with cream cheese and cereal with or without milk) to measure and predict individual metabolic responses. |
|
| Mikkelsen et al. (2015), Denmark [27] | Clinical trial | 18–40 y | 100 | Healthy subjects with no history of chronic diseases. | 12 | 180 days | Drug: 4-day treatment of antibiotics (500 mg vancomycin, 40 mg gentamycin, and 500 mg of meropenem) daily. |
|
| Park et al. (2020), Korea [36] | RCT | >20 y | 34.3 | Subjects with healthy and slightly elevated fasting TG levels (<200 mg/dL). | 62 | 14 weeks | Diet: supplementation of probiotic Lactobacillus plantarum Q180 (LPQ180) or placebo daily. |
|
| Reijnders et al. (2016), Netherlands [37] | RCT | 35–70 y | 100 | Obesity and impaired fasting glucose and/or impaired glucose tolerance | 57 | 7 days | Drug: Comparison of treatment with 1500 mg amoxicillin, 1500 mg vancomycin, or placebo. |
|
| Ross et al. (2011), Switzerland [38] | Randomised crossover trial | 20–50 y | 35.3 | Healthy subjects with no history of chronic diseases. | 17 | 2 weeks | Diet: Whole grain rich foods (WG) vs. refined grains (RG). |
|
| Schutte et al. (2018), Netherlands [39] | Randomised parallel trial | 45–70 y | 62 | Subjects with increased risk of CVD; overweight males and postmenopausal females with mildly elevated levels of plasma total cholesterol (>5 mmol/L). | 50 | 12 weeks | Diet: Whole grain wheat diet (WGW) vs. refined wheat (RW) diet. |
|
| Tily et al. (2019), USA [40] | Series of acute experimental studies | ≥18 y | ~34 | Healthy subjects with no history of chronic diseases. | 550 | 2 weeks | Diet: Standardised meal testing to measure individual glycaemic responses. |
|
| Tong et al. (2018), China [28] | RCT | 30–65 y | 50 | Untreated subjects that meet diagnostic criteria for T2D with an elevated waist circumference.) | 100 | 12 weeks | Drug: Comparison of treatment with Chinese herbal formula (AMC) or metformin as a positive control. |
|
| Vetrani et al. (2020), Italy [41] | Randomised parallel trial | 40–70 y | 42.3 | Otherwise healthy subjects at risk of MetS | 78 | 8 weeks | Diet: 4-arm intervention comparing diets of varying levels of long chain n-3 polyunsaturated fatty acids (LCn3) and/or polyphenols (PP) in subjects with MetS risk factors. |
|
| Vors et al. (2020), France [42] | RCT | <75 y | 0 | Overweight postmenopausal women. | 58 | 4 weeks | Diet: Comparison of milk polar lipid consumption (0, 3 or 5 g-PL/day) or control. |
|
| Vrieze et al. (2014), Netherlands [43] | RCT | ≥18 y | 100 | Obese subjects that meet diagnostic criteria for MetS. | 20 | 7 days | Drug: Comparison of treatment with 1500 mg amoxicillin or 1500 mg vancomycin. |
|
| Xu et al. (2015), China [44] | RCT | 30–65 y | 61.5 | Newly diagnosed but untreated T2D. | 187 | 12 weeks | Drug: Comparison of high, moderate, or low dose treatment of herbal formula GQD, or placebo. |
|
| Zeevi et al. (2015), Israel [19] | Series of acute experimental studies | 18–70 y | 40 | Healthy subjects with no previous history of chronic disease. | 800 | 7 days | Diet: Standardised meal testing to measure and predict individual metabolic responses. |
|
| Parameter | Inclusion/Exclusion Criteria |
|---|---|
| Participants | Adults aged ≥ 18 years. |
| Interventions | Diet, drug interventions. |
| Comparisons | Placebo or control group, different diet/intake. |
| Outcomes | Primary outcomes included presence of both metagenomic and postprandial plasma analysis, namely plasma glucose, lipids, and lipoproteins. |
| Study design | Randomised controlled or clinical trials with either parallel, crossover or a series of acute experimental studies. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, M.L.; Davies, I.G.; Waraksa, W.; Khayyatzadeh, S.S.; Al-Asmakh, M.; Mazidi, M. The Impact of Microbial Composition on Postprandial Glycaemia and Lipidaemia: A Systematic Review of Current Evidence. Nutrients 2021, 13, 3887. https://doi.org/10.3390/nu13113887
Wilson ML, Davies IG, Waraksa W, Khayyatzadeh SS, Al-Asmakh M, Mazidi M. The Impact of Microbial Composition on Postprandial Glycaemia and Lipidaemia: A Systematic Review of Current Evidence. Nutrients. 2021; 13(11):3887. https://doi.org/10.3390/nu13113887
Chicago/Turabian StyleWilson, Megan L., Ian G. Davies, Weronika Waraksa, Sayyed S. Khayyatzadeh, Maha Al-Asmakh, and Mohsen Mazidi. 2021. "The Impact of Microbial Composition on Postprandial Glycaemia and Lipidaemia: A Systematic Review of Current Evidence" Nutrients 13, no. 11: 3887. https://doi.org/10.3390/nu13113887
APA StyleWilson, M. L., Davies, I. G., Waraksa, W., Khayyatzadeh, S. S., Al-Asmakh, M., & Mazidi, M. (2021). The Impact of Microbial Composition on Postprandial Glycaemia and Lipidaemia: A Systematic Review of Current Evidence. Nutrients, 13(11), 3887. https://doi.org/10.3390/nu13113887







