Brain-Restricted Inhibition of IL-6 Trans-Signaling Mildly Affects Metabolic Consequences of Maternal Obesity in Male Offspring
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model
2.2. Analytical Procedures
2.3. Food Intake (FI) and Food Preference (FP) Test
2.4. Intraperitoneal Glucose Tolerance Test (ipGTT) and Intraperitoneal Insulin Tolerance Test (ipITT)
2.5. Proteomics Analysis
2.6. Quantitative PCR
2.7. ELISA Analysis
2.8. µ CTanalysis
2.9. Statistical Analysis
3. Results
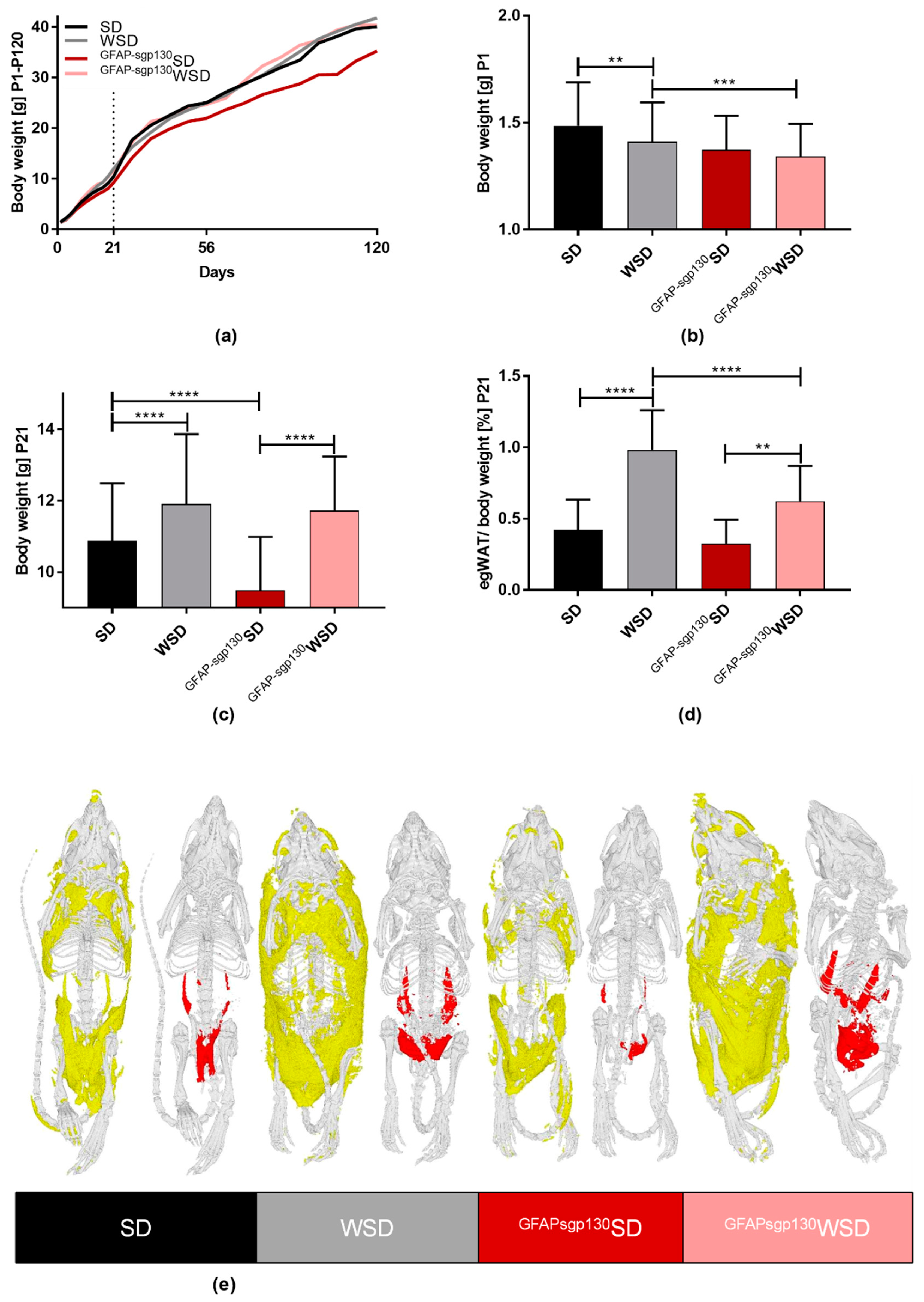
3.1. Brain-Restricted Inhibition of IL-6tS Mildly Attenuates WSD Impact on Dams
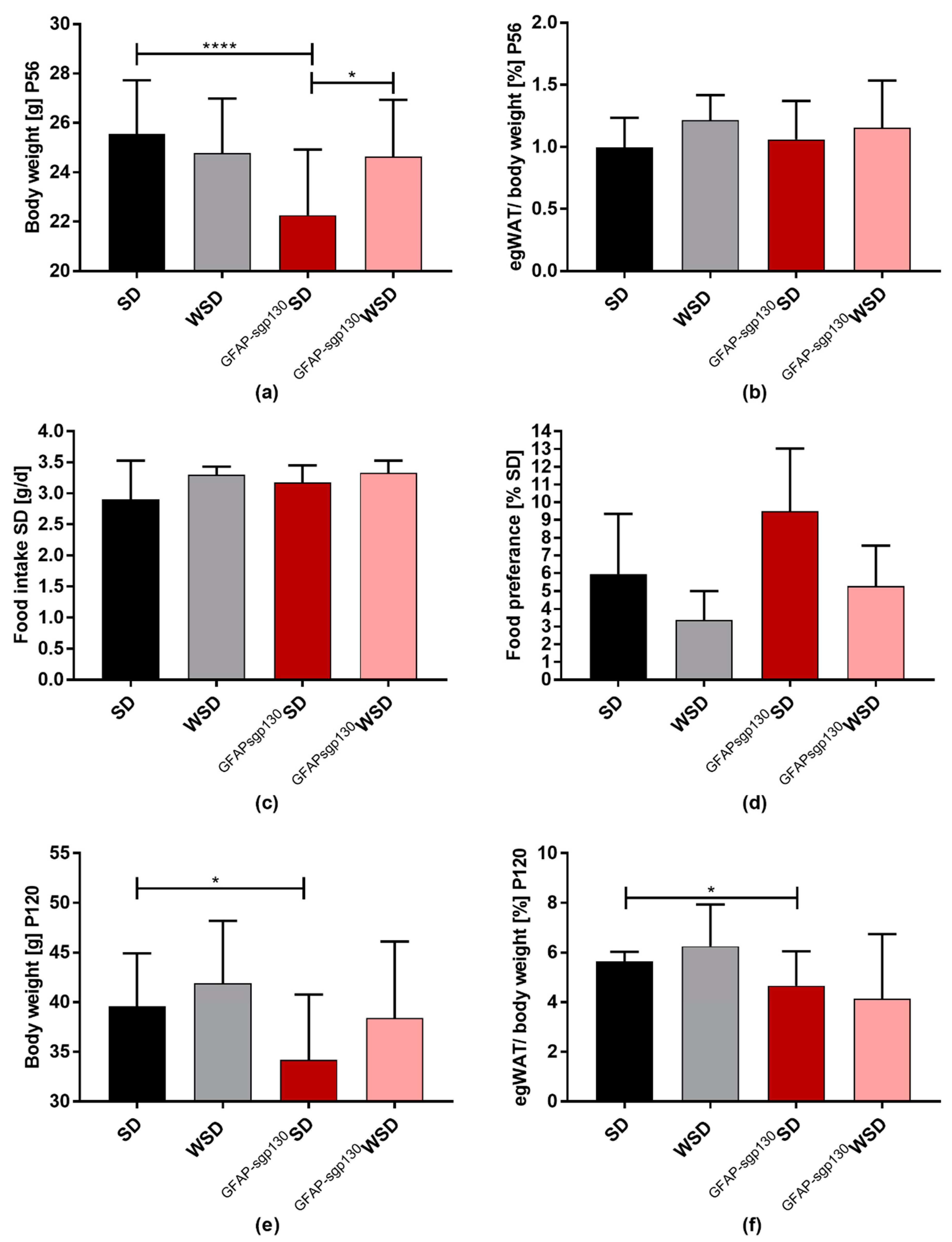
3.2. Maternal Diet Shows Effects on Offspring’s Body Weight and egWAT Percentage with Only Partial Effects of Brain-Restricted Inhibition of IL-6tS
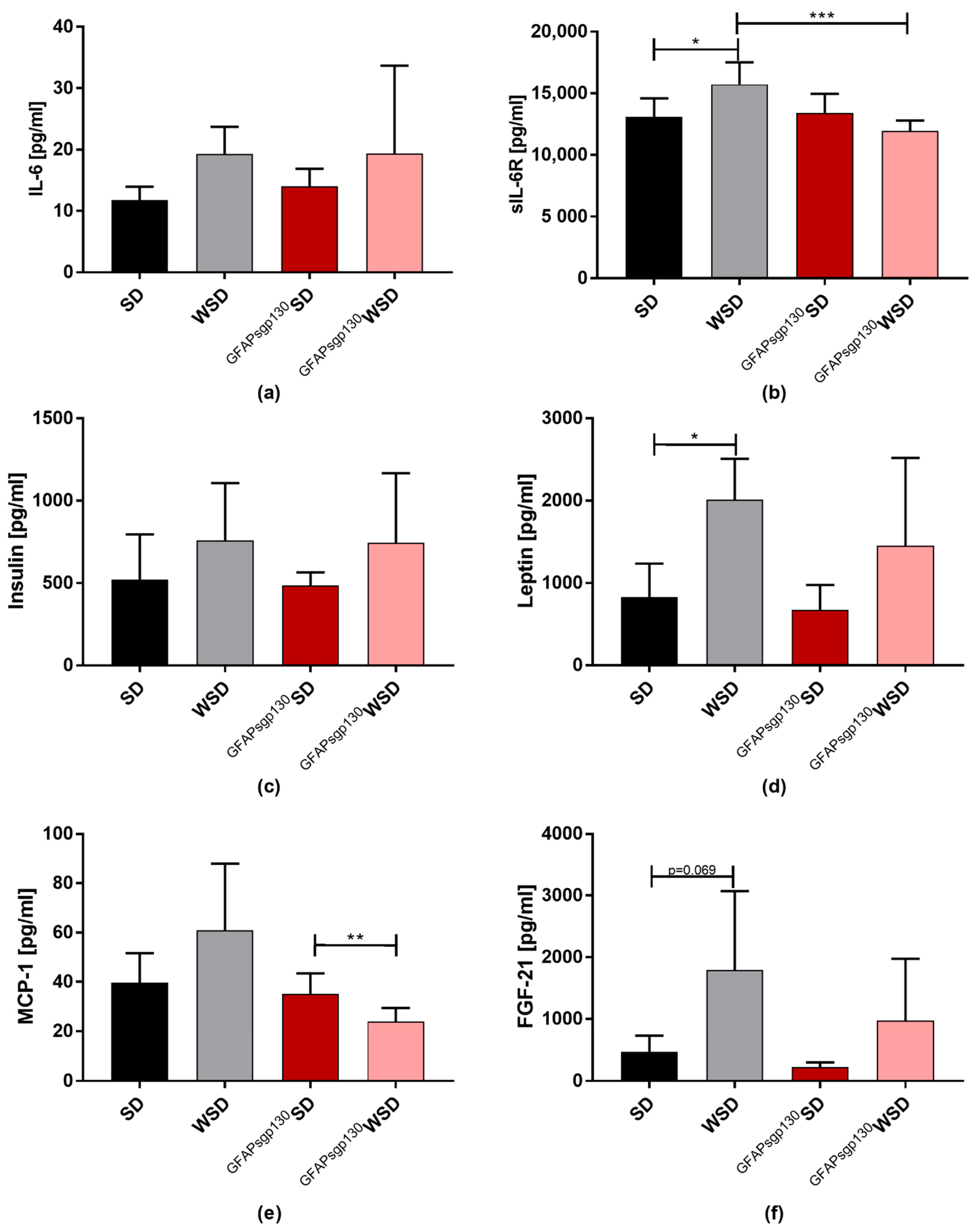
3.3. Effects of Maternal Diet Outweight Effects of Brain Restricted Inhibition of IL-6tS on Metabolic Serum Parameters and ipITT in the Offspring at P21
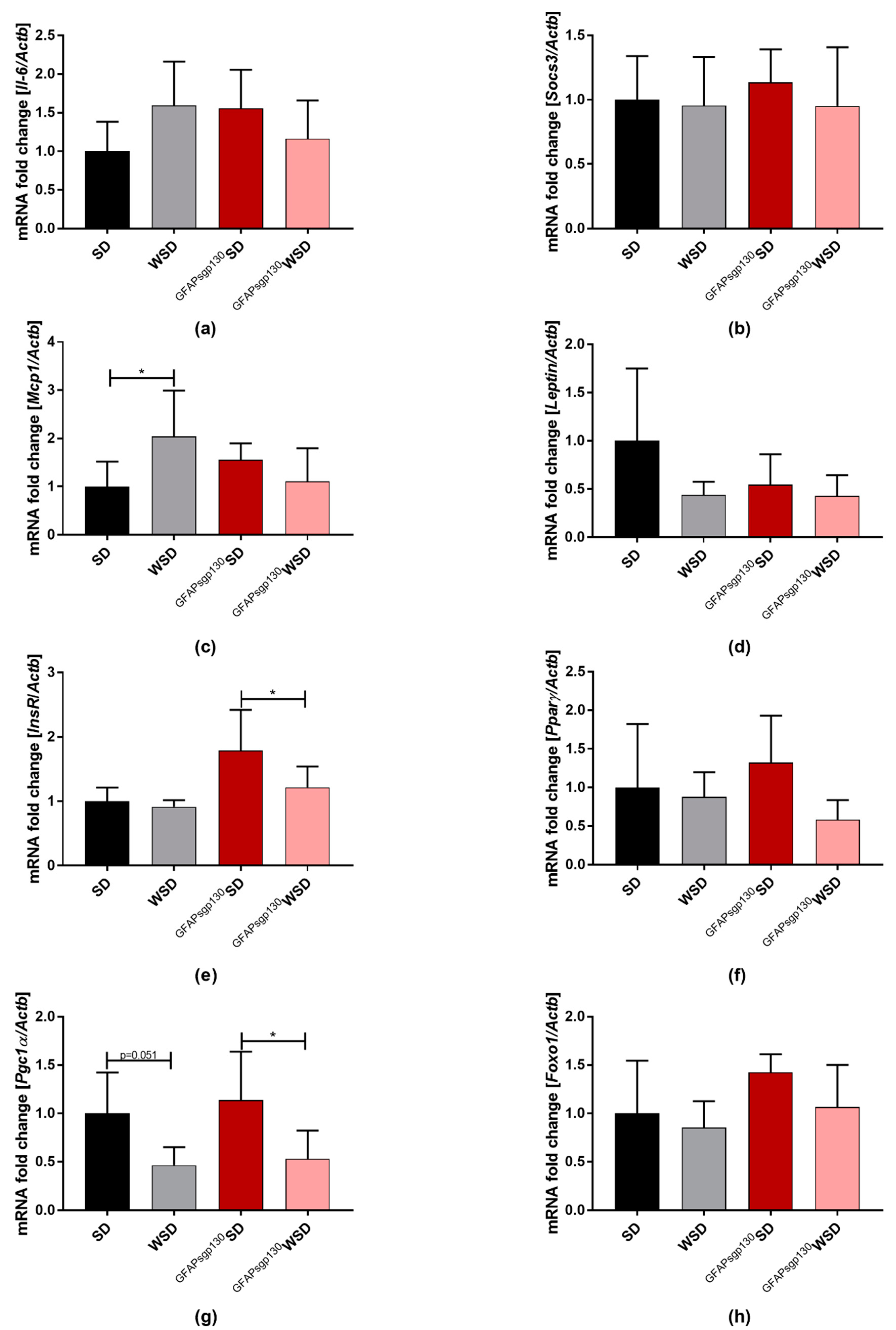
3.4. Effects of Maternal Obesity and Brain Restricted Inhibition of IL-6tS on Offspring egWAT mRNA Expression of Adipokines and egWAT Metabolism
3.5. Effects of Maternal Obesity and Brain-Restricted Inhibition of IL-6tS on the Offspring’s egWAT Proteome at P21
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catalano, P.M.; Ehrenberg, H.M. The short- and long-term implications of maternal obesity on the mother and her offspring. BJOG 2006, 113, 1126–1133. [Google Scholar] [CrossRef]
- Boney, C.M.; Verma, A.; Tucker, R.; Vohr, B.R. Metabolic syndrome in childhood: Association with birth weight, maternal obesit and gestational diabetes mellitus. Pediatrics 2005, 115, e290–e296. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, J.; Heslehurst, N.; Hall, J.; Schoenaker, D.A.J.M.; Hutchinson, J.; Cade, J.E.; Poston, L.; Barrett, G.; Crozier, S.R.; Barker, M.; et al. Before the beginning: Nutrition and lifestyle in the preconception period and its importance for future health. Lancet 2018, 391, 1830–1841. [Google Scholar] [CrossRef]
- Calcaterra, V.; Cena, H.; Verduci, E.; Bosetti, A.; Pelizzo, G.; Zuccotti, G.V. Nutritional surveillance for the best start in life, promoting health for neonates, infants and children. Nutrients 2020, 12, 3386. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Hernández, A.; Beneit, N.; Díaz-Castroverde, S.; Escribano, Ó. Differential Role of Adipose Tissues in Obesity and Related Metabolic and Vascular Complications. Int. J. Endocrinol. 2016, 2016. [Google Scholar] [CrossRef]
- Pereira, S.S.; Alvarez-Leite, J.I. Low-Grade Inflammation, Obesity, and Diabetes. Curr. Obes. Rep. 2014, 3, 422–431. [Google Scholar] [CrossRef]
- Cancello, R.; Clément, K. Review article: Is obesity an inflammatory illness? Role of low-grade inflammation and macrophage infiltration in human white adipose tissue. BJOG Int. J. Obstet. Gynaecol. 2006, 113, 1141–1147. [Google Scholar] [CrossRef]
- Bae-Gartz, I.; Janoschek, R.; Breuer, S.; Schmitz, L.; Hoffmann, T.; Ferrari, N.; Branik, L.; Oberthuer, A.; Kloppe, C.S.; Appel, S.; et al. Maternal Obesity Alters Neurotrophin-Associated MAPK Signaling in the Hypothalamus of Male Mouse Offspring. Front. Neurosci. 2019, 13, 1–17. [Google Scholar] [CrossRef]
- Burhans, M.S.; Hagman, D.K.; Kuzma, J.N.; Schmidt, K.A.; Kratz, M. Contribution of adipose tissue inflammation to the development of type 2 diabetes mellitus. Compr. Physiol. 2019, 9, 1–58. [Google Scholar] [CrossRef]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose tissue remodeling: Its role in energy metabolism and metabolic disorders. Front. Endocrinol. 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Funcke, J.B.; Scherer, P.E. Beyond adiponectin and leptin: Adipose tissue-derived mediators of inter-organ communication. J. Lipid Res. 2019, 60, 1648–1697. [Google Scholar] [CrossRef] [PubMed]
- Crichton, G.E.; Howe, P.R.C.; Buckley, J.D.; Coates, A.M.; Murphy, K.J.; Bryan, J. Long-term dietary intervention trials: Critical issues and challenges. Trials 2012, 13. [Google Scholar] [CrossRef]
- Kern, L.; Mittenbühler, M.J.; Vesting, A.J.; Ostermann, A.L.; Wunderlich, C.M.; Wunderlich, F.T. Obesity-induced TNFα and IL-6 signaling: The missing link between obesity and inflammation- driven liver and colorectal cancers. Cancers 2019, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.L.; Erta, M.; Lim, S.L.; Frausto, R.; May, U.; Rose-John, S.; Scheller, J.; Hidalgo, J. Trans-signaling is a dominant mechanism for the pathogenic actions of interleukin-6 in the brain. J. Neurosci. 2014, 34, 2503–2513. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 2011, 1813, 878–888. [Google Scholar] [CrossRef]
- Jostock, T.; Müllberg, J.; Özbek, S.; Atreya, R.; Blinn, G.; Voltz, N.; Fischer, M.; Neurath, M.F.; Rose-John, S. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur. J. Biochem. 2001, 268, 160–167. [Google Scholar] [CrossRef]
- Eulenfeld, R.; Dittrich, A.; Khouri, C.; Müller, P.J.; Mütze, B.; Wolf, A.; Schaper, F. Interleukin-6 signalling: More than Jaks and STATs. Eur. J. Cell Biol. 2012, 91, 486–495. [Google Scholar] [CrossRef]
- Kraakman, M.J.; Kammoun, H.L.; Allen, T.L.; Deswaerte, V.; Henstridge, D.C.; Estevez, E.; Matthews, V.B.; Neill, B.; White, D.A.; Murphy, A.J.; et al. Blocking IL-6 trans-signaling prevents high-fat diet-induced adipose tissue macrophage recruitment but does not improve insulin resistance. Cell Metab. 2015, 21, 403–416. [Google Scholar] [CrossRef]
- Jones, S.A.; Scheller, J.; Rose-John, S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Investig. 2011, 121, 3375–3383. [Google Scholar] [CrossRef] [PubMed]
- Bae-Gartz, I.; Janoschek, R.; Kloppe, C.S.; Vohlen, C.; Roels, F.; Oberthür, A.; Alejandre Alcazar, M.A.; Lippach, G.; Muether, P.S.; Dinger, K.; et al. Running exercise in obese pregnancies prevents IL-6 trans-signaling in male offspring. Med. Sci. Sports Exerc. 2016. [Google Scholar] [CrossRef]
- Timper, K.; Brüning, J.C. Hypothalamic circuits regulating appetite and energy homeostasis: Pathways to obesity. Dis. Model. Mech. 2017, 10, 679–689. [Google Scholar] [CrossRef]
- Wang, B.; Li, A.; Li, X.; Ho, P.W.; Wu, D.; Wang, X.; Liu, Z.; Wu, K.K.; Yau, S.S.; Xu, A.; et al. Activation of hypothalamic RIP -Cre neurons promotes beiging of WAT via sympathetic nervous system. EMBO Rep. 2018, 19, 1–17. [Google Scholar] [CrossRef]
- Thaler, J.P.; Yi, C.-X.; Schur, E.A.; Guyenet, S.J.; Hwang, B.H.; Dietrich, M.; Zhao, X.; Sarruf, D.A.; Izgur, V.; Maravilla, K.R.; et al. Obesity is associated with hypothalamic injury in rodents and humans. J. Clin. Investig. 2012, 122, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Horvath, T.L.; Sarman, B.; García-Cáceres, C.; Enriori, P.J.; Sotonyi, P.; Shanabrough, M.; Borok, E.; Argente, J.; Chowen, J.A.; Perez-Tilve, D.; et al. Synaptic input organization of the melanocortin system predicts diet-induced hypothalamic reactive gliosis and obesity. Proc. Natl. Acad. Sci. USA 2010, 107, 14875–14880. [Google Scholar] [CrossRef] [PubMed]
- Rother, E.; Kuschewski, R.; Alcazar, M.A.A.; Oberthuer, A.; Bae-Gartz, I.; Vohlen, C.; Roth, B.; Dötsch, J. Hypothalamic JNK1 and IKKβ activation and impaired early postnatal glucose metabolism after maternal perinatal high-fat feeding. Endocrinology 2012, 153, 770–781. [Google Scholar] [CrossRef]
- Dearden, L.; Buller, S.; Furigo, I.; Fernandez-Twinn, D.; Ozanne, S. Maternal obesity causes fetal hypothalamic insulin resistance and disrupts development of hypothalamic feeding pathways. Mol. Metab. 2020, 42. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, L.; Kuglin, R.; Bae-Gartz, I.; Janoschek, R.; Appel, S.; Mesaros, A.; Jakovcevski, I.; Vohlen, C.; Handwerk, M.; Ensenauer, R.; et al. Hippocampal insulin resistance links maternal obesity with impaired neuronal plasticity in adult offspring. Psychoneuroendocrinology 2018, 89, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Masuzaki, H.; Paterson, J.; Shinyama, H.; Morton, N.M.; Mullins, J.J.; Seckl, J.R.; Flier, J.S. A Transgenic Model of Visceral Obesity and the Metabolic Syndrome. Science 2001, 294, 2166–2170. [Google Scholar] [CrossRef]
- Mohamed-Ali, V.; Goodrick, S.; Rawesh, A.; Katz, D.R.; Miles, J.M.; Yudkin, J.S.; Klein, S.; Coppack, S.W. Subcutaneous Adipose Tissue Releases Interleukin-6, But Not Tumor Necrosis Factor-α, in Vivo. J. Clin. Endocrinol. Metab. 1997, 82, 4196–4200. [Google Scholar] [CrossRef]
- Han, M.S.; White, A.; Perry, R.J.; Camporez, J.-P.; Hidalgo, J.; Shulman, G.I.; Davis, R.J. Regulation of adipose tissue inflammation by interleukin 6. Proc. Natl. Acad. Sci. USA 2020, 117, 2751–2760. [Google Scholar] [CrossRef]
- Jais, A.; Brüning, J.C. Hypothalamic inflammation in obesity and metabolic disease. J. Clin. Investig. 2017, 127, 24. [Google Scholar] [CrossRef] [PubMed]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving bioscience research reporting: The arrive guidelines for reporting animal research. PLoS Biol. 2010, 8. [Google Scholar] [CrossRef]
- Alejandre-Alcázar, M.A.; Kwapiszewska, G.; Reiss, I.; Amarie, O.V.; Marsh, L.M.; Sevilla-Pérez, J.; Wygrecka, M.; Eul, B.; Köbrich, S.; Hesse, M.; et al. Hyperoxia modulates TGF-β/BMP signaling in a mouse model of bronchopulmonary dysplasia. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2007, 292, 537–549. [Google Scholar] [CrossRef]
- Irmak, D.; Fatima, A.; Gutiérrez-Garcia, R.; Rinschen, M.M.; Wagle, P.; Altmüller, J.; Arrigoni, L.; Hummel, B.; Klein, C.; Frese, C.K.; et al. Mechanism suppressing H3K9 trimethylation in pluripotent stem cells and its demise by polyQ-expanded huntingtin mutations. Hum. Mol. Genet. 2018, 27, 4117–4134. [Google Scholar] [CrossRef]
- Bae-Gartz, I.; Kasper, P.; Großmann, N.; Breuer, S.; Janoschek, R.; Kretschmer, T.; Appel, S.; Schmitz, L.; Vohlen, C.; Quaas, A.; et al. Maternal exercise conveys protection against NAFLD in the offspring via hepatic metabolic programming. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Searle, B.C.; Swearingen, K.E.; Barnes, C.A.; Schmidt, T.; Gessulat, S.; Küster, B.; Wilhelm, M. Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Searle, B.C.; Pino, L.K.; Egertson, J.D.; Ting, Y.S.; Lawrence, R.T.; MacLean, B.X.; Villén, J.; MacCoss, M.J. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2019, 17, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Sengupta, P. Men and mice: Relating their ages. Life Sci. 2016, 152, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Escrig, A.; Molinero, A.; Méndez, B.; Giralt, M.; Comes, G.; Sanchis, P.; Fernández-Gayol, O.; Giménez-Llort, L.; Becker-Pauly, C.; Rose-John, S.; et al. IL-6 Trans-Signaling in the Brain Influences the Metabolic Phenotype of the 3xTg-AD Mouse Model of Alzheimer’s Disease. Cells 2020, 9, 1605. [Google Scholar] [CrossRef] [PubMed]
- Kasper, P.; Breuer, S.; Hoffmann, T.; Vohlen, C.; Janoschek, R.; Schmitz, L.; Appel, S.; Fink, G.; Hünseler, C.; Quaas, A.; et al. Maternal Exercise Mediates Hepatic Metabolic Programming via Activation of AMPK-PGC1α Axis in the Offspring of Obese Mothers. Cells 2021, 10, 1247. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.; Hasselwander, S.; Li, H.; Xia, N. Effects of different diets used in diet-induced obesity models on insulin resistance and vascular dysfunction in C57BL/6 mice. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.; Gayle, D.; Babu, J.; Ross, M.G. Programmed obesity in intrauterine growth-restricted newborns: Modulation by newborn nutrition. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288. [Google Scholar] [CrossRef]
- Kokoeva, M.V.; Yin, H.; Flier, J.S. Neurogenesis in the hypothalamus of adult mice: Potential role in energy balance. Science 2005, 310, 679–683. [Google Scholar] [CrossRef]
- Whalley, K. Hypothalamic neurogenesis regulates weight gain. Nat. Rev. Neurosci. 2012, 13, 289. [Google Scholar] [CrossRef]
- Paglia, L. Taste development and prenatal prevention. Eur. J. Paediatr. Dent. 2019, 20, 257. [Google Scholar] [CrossRef]
- Yadav, A.; Jyoti, P.; Jain, S.K.; Bhattacharjee, J. Correlation of adiponectin and leptin with insulin resistance: A pilot study in healthy north Indian population. Indian J. Clin. Biochem. 2011, 26. [Google Scholar] [CrossRef]
- Rajasekaran, M.; Sul, O.-J.; Choi, E.-K.; Kim, J.-E.; Suh, J.-H.; Choi, H.-S. MCP-1 deficiency enhances browning of adipose tissue via increased M2 polarization. J. Endocrinol. 2019, 242, 91–101. [Google Scholar] [CrossRef]
- Kado, S.; Nagase, T.; Nagata, N. Circulating levels of interleukin-6, its soluble receptor and interleukin-6/interleukin-6 receptor complexes in patients with type 2 diabetes mellitus. Acta Diabetol. 1999, 36, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Aparicio-Siegmund, S.; Garbers, Y.; Flynn, C.M.; Waetzig, G.H.; Gouni-Berthold, I.; Krone, W.; Berthold, H.K.; Laudes, M.; Rose-John, S.; Garbers, C. The IL-6-neutralizing sIL-6R-sgp130 buffer system is disturbed in patients with type 2 diabetes. Am. J. Physiol. Metab. 2019, 317, E411–E420. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Chen, S.-Y.; Sheng, L.; Jena, P.K.; Kalanetra, K.M.; Mills, D.A.; Wan, Y.-J.Y.; Slupsky, C.M. Long-term effects of western diet consumption in male and female mice. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Bastos, P.; Fontes-Villalba, M.; O’keefe, J.H.; Lindeberg, S.; Cordain, L. The western diet and lifestyle and diseases of civilization. Res. Reports Clin. Cardiol. 2011, 2, 15–35. [Google Scholar] [CrossRef]
- Gao, Y.; Bielohuby, M.; Fleming, T.; Grabner, G.F.; Foppen, E.; Bernhard, W.; Guzmán-Ruiz, M.; Layritz, C.; Legutko, B.; Zinser, E.; et al. Dietary sugars, not lipids, drive hypothalamic inflammation. Mol. Metab. 2017, 6, 897–908. [Google Scholar] [CrossRef]
- Jamar, G.; Ribeiro, D.A.; Pisani, L.P. High-fat or high-sugar diets as trigger inflammation in the microbiota-gut-brain axis. Crit. Rev. Food Sci. Nutr. 2020, 61, 836–854. [Google Scholar] [CrossRef]
- Abbott, K.N.; Arnott, C.K.; Westbrook, R.F.; Tran, D.M.D. The effect of high fat, high sugar, and combined high fat-high sugar diets on spatial learning and memory in rodents: A meta-analysis. Neurosci. Biobehav. Rev. 2019, 107, 399–421. [Google Scholar] [CrossRef]
- Pendeloski, K.P.T.; Ono, E.; Torloni, M.R.; Mattar, R.; Daher, S. Maternal obesity and inflammatory mediators: A controversial association. Am. J. Reprod. Immunol. 2017, 77, e12674. [Google Scholar] [CrossRef]
- Cederbaum, A.I. CYP2E1 potentiates toxicity in obesity and after chronic ethanol treatment. Drug Metabol. Drug Interact. 2012, 27, 125–144. [Google Scholar] [CrossRef]
- Emery, M.G.; Fisher, J.M.; Chien, J.Y.; Kharasch, E.D.; Dellinger, E.P.; Kowdley, K.V.; Thummel, K.E. CYP2E1 activity before and after weight loss in morbidly obese subjects with nonalcoholic fatty liver disease. Hepatology 2003, 38, 428–435. [Google Scholar] [CrossRef]
- Yoshinari, K.; Sato, T.; Okino, N.; Sugatani, J.; Miwa, M. Expression and induction of cytochromes p450 in rat white adipose tissue. J. Pharmacol. Exp. Ther. 2004, 311, 147–154. [Google Scholar] [CrossRef]
- Abraham, N.G.; Junge, J.; Drummond, G.S. Translational Significance of Heme Oxygenase in Obesity and Metabolic Syndrome. Trends Pharmacol. Sci. 2016, 37, 17. [Google Scholar] [CrossRef]
- Kabaran, S.; Besler, H.T. Do fatty acids affect fetal programming? J. Health Popul. Nutr. 2015, 33. [Google Scholar] [CrossRef]
- Asayama, K.; Nakane, T.; Dobashi, K.; Kodera, K.; Hayashibe, H.; Uchida, N.; Nakazawa, S. Effect of obesity and troglitazone on expression of two glutathione peroxidases: Cellular and extracellular types in serum, kidney and adipose tissue. Free. Radic. Res. 2009, 34, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Galinier, A.; Carrière, A.; Fernandez, Y.; Carpéné, C.; André, M.; Caspar-Bauguil, S.; Thouvenot, J.-P.; Périquet, B.; Pénicaud, L.; Casteilla, L. Adipose Tissue Proadipogenic Redox Changes in Obesity. J. Biol. Chem. 2006, 281, 12682–12687. [Google Scholar] [CrossRef]
- Kobayashi, H.; Matsuda, M.; Fukuhara, A.; Komuro, R.; Shimomura, I. Dysregulated glutathione metabolism links to impaired insulin action in adipocytes. Am. J. Physiol.-Endocrinol. Metab. 2009, 296, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.S.; Myatt, L. Sexual dimorphism in the effect of maternal obesity on antioxidant defense mechanisms in the human placenta. Placenta 2017, 51, 64–69. [Google Scholar] [CrossRef]
- Cantile, M.; Procino, A.; D’armiento, M.; Cindolo, L.; Cillo, C. HOX gene network is involved in the transcriptional regulation of in vivo human adipogenesis. J. Cell. Physiol. 2003, 194, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Procino, A.; Cillo, C. The HOX genes network in metabolic diseases. Cell Biol. Int. 2013, 37, 1145–1148. [Google Scholar] [CrossRef]
- Hellmann, J.; Zhang, M.J.; Tang, Y.; Rane, M.; Bhatnagar, A.; Spite, M. Increased Saturated Fatty Acids in Obesity Alter Resolution of Inflammation in Part by Stimulating Prostaglandin Production. J. Immunol. 2013, 191, 1383–1392. [Google Scholar] [CrossRef]
- Chan, P.-C.; Hsiao, F.-C.; Chang, H.-M.; Wabitsch, M.; Hsieh, P.S. Importance of adipocyte cyclooxygenase-2 and prostaglandin E2-prostaglandin E receptor 3 signaling in the development of obesity-induced adipose tissue inflammation and insulin resistance. FASEB J. 2016, 30, 2282–2297. [Google Scholar] [CrossRef]
- Lee, D.Y.; Teyssier, C.; Strahl, B.D.; Stallcup, M.R. Role of Protein Methylation in Regulation of Transcription. Endocr. Rev. 2005, 26, 147–170. [Google Scholar] [CrossRef]
- Dötsch, J. Perinatal programming-myths, fact, and future of research. Mol. Cell. Pediatr. 2014, 1. [Google Scholar] [CrossRef] [PubMed]
- Wróblewski, A.; Strycharz, J.; Świderska, E.; Drewniak, K.; Drzewoski, J.; Szemraj, J.; Kasznicki, J.; Śliwińska, A. Molecular insight into the interaction between epigenetics and leptin in metabolic disorders. Nutrients 2019, 11, 1872. [Google Scholar] [CrossRef]
- Timper, K.; Denson, J.L.; Steculorum, S.M.; Heilinger, C.; Engström-Ruud, L.; Wunderlich, C.M.; Rose-John, S.; Wunderlich, F.T.; Brüning, J.C. IL-6 Improves Energy and Glucose Homeostasis in Obesity via Enhanced Central IL-6 trans-Signaling. Cell Rep. 2017. [Google Scholar] [CrossRef]
- Mauvais-Jarvis, F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol. Sex Differ. 2015, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.G.; Agellon, L.B. Sex differences in lipid metabolism and metabolic disease risk. Biochem. Cell Biol. 2012, 90, 124–141. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, E.; Martínez-Samayoa, P.M.; Bautista, C.J.; Deás, M.; Guillén, L.; Rodríguez-González, G.L.; Guzmán, C.; Larrea, F.; Nathanielsz, P.W. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J. Physiol. 2005, 566, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Loche, E.; Blackmore, H.L.; Carpenter, A.A.; Beeson, J.H.; Pinnock, A.; Ashmore, T.J.; Aiken, C.E.; De Almeida-Faria, J.; Schoonejans, J.M.; Giussani, D.A.; et al. Maternal diet-induced obesity programmes cardiac dysfunction in male mice independently of post-weaning diet. Cardiovasc. Res. 2018, 114, 1372–1384. [Google Scholar] [CrossRef]
- Alfaradhi, M.Z.; Kusinski, L.C.; Fernandez-Twinn, D.S.; Pantaleão, L.C.; Carr, S.K.; Ferland-McCollough, D.; Yeo, G.S.H.; Bushell, M.; Ozanne, S.E. Maternal Obesity in Pregnancy Developmentally Programs Adipose Tissue Inflammation in Young, Lean Male Mice Offspring. Endocrinology 2016, 157, 4246–4256. [Google Scholar] [CrossRef]
- Frihauf, J.B.; Fekete, E.M.; Nagy, T.R.; Levin, B.E.; Zorrilla, E.P. Maternal Western diet increases adiposity even in male offspring of obesity-resistant rat dams: Early endocrine risk markers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R1045–R1059. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liang, X.; Yang, Q.; Fu, X.; Rogers, C.J.; Wang, B.; Pan, H.; Zhu, M.; Nathanielsz, P.W.; Du, M. Maternal obesity epigenetically alters visceral fat progenitor cell properties in male offspring mice. J. Physiol. 2016, 594, 4453. [Google Scholar] [CrossRef] [PubMed]
- Kislal, S.; Shook, L.L.; Edlow, A.G. Perinatal exposure to maternal obesity: Lasting cardiometabolic impact on offspring. Prenat. Diagn. 2020, 40, 1109–1125. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Breuer, S.; Kasper, P.; Vohlen, C.; Janoschek, R.; Hoffmann, T.; Appel, S.; Müller-Limberger, E.; Mesaros, A.; Rose-John, S.; Garbers, C.; et al. Brain-Restricted Inhibition of IL-6 Trans-Signaling Mildly Affects Metabolic Consequences of Maternal Obesity in Male Offspring. Nutrients 2021, 13, 3735. https://doi.org/10.3390/nu13113735
Breuer S, Kasper P, Vohlen C, Janoschek R, Hoffmann T, Appel S, Müller-Limberger E, Mesaros A, Rose-John S, Garbers C, et al. Brain-Restricted Inhibition of IL-6 Trans-Signaling Mildly Affects Metabolic Consequences of Maternal Obesity in Male Offspring. Nutrients. 2021; 13(11):3735. https://doi.org/10.3390/nu13113735
Chicago/Turabian StyleBreuer, Saida, Philipp Kasper, Christina Vohlen, Ruth Janoschek, Thorben Hoffmann, Sarah Appel, Elena Müller-Limberger, Andrea Mesaros, Stefan Rose-John, Christoph Garbers, and et al. 2021. "Brain-Restricted Inhibition of IL-6 Trans-Signaling Mildly Affects Metabolic Consequences of Maternal Obesity in Male Offspring" Nutrients 13, no. 11: 3735. https://doi.org/10.3390/nu13113735
APA StyleBreuer, S., Kasper, P., Vohlen, C., Janoschek, R., Hoffmann, T., Appel, S., Müller-Limberger, E., Mesaros, A., Rose-John, S., Garbers, C., Müller, S., Lackmann, J.-W., Mahabir, E., Dötsch, J., Hucklenbruch-Rother, E., & Bae-Gartz, I. (2021). Brain-Restricted Inhibition of IL-6 Trans-Signaling Mildly Affects Metabolic Consequences of Maternal Obesity in Male Offspring. Nutrients, 13(11), 3735. https://doi.org/10.3390/nu13113735






