Impact of Fetal Growth Restriction on the Neonatal Microglial Proteome in the Rat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Magnetic Sorting of Microglial Cells
2.3. Primary Microglial Cell Culture and Immunocytochemistry
2.4. Measurements of Reactive Oxygen Species (ROS) Production by Luminometry
2.5. Sample Preparation for Proteomic Analysis
2.6. Liquid Chromatography–Coupled Mass Spectrometry Analysis (LC-MS)
2.7. Protein Quantification and Comparison
2.8. Bioinformatic Analysis
2.9. Statistical Analysis
3. Results
3.1. Evaluation of Purity after Cell Sorting and Sample Clustering
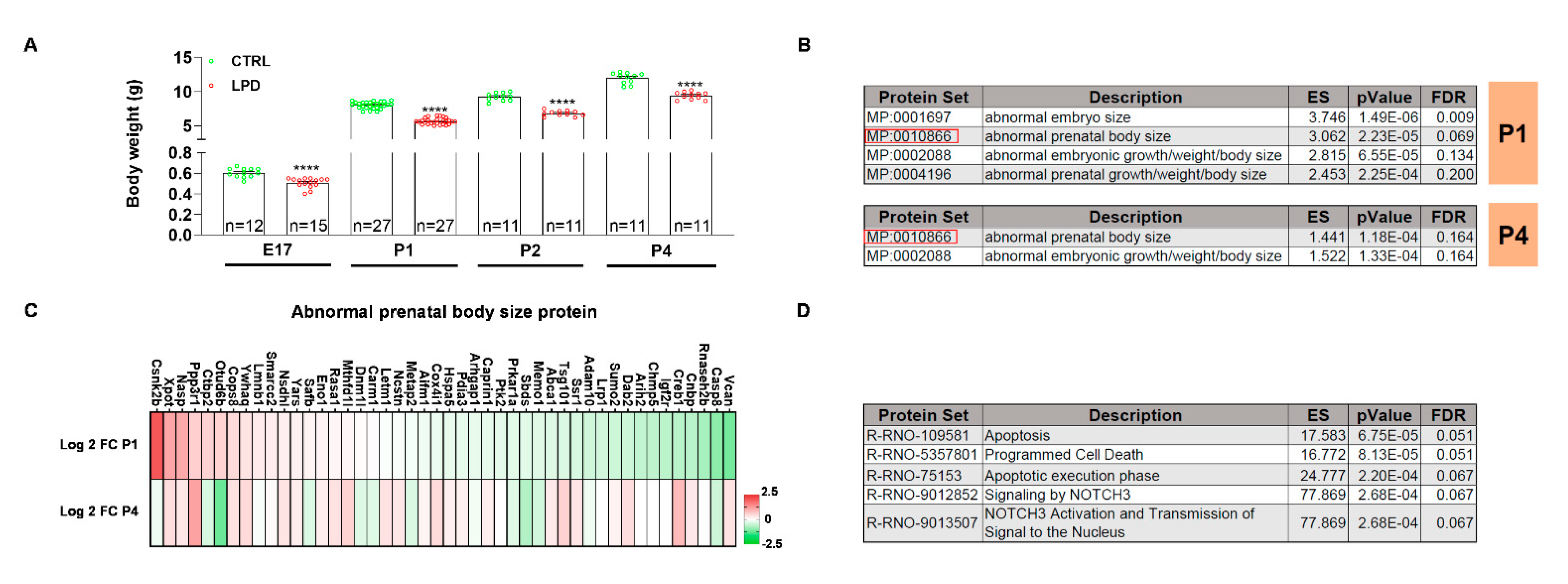
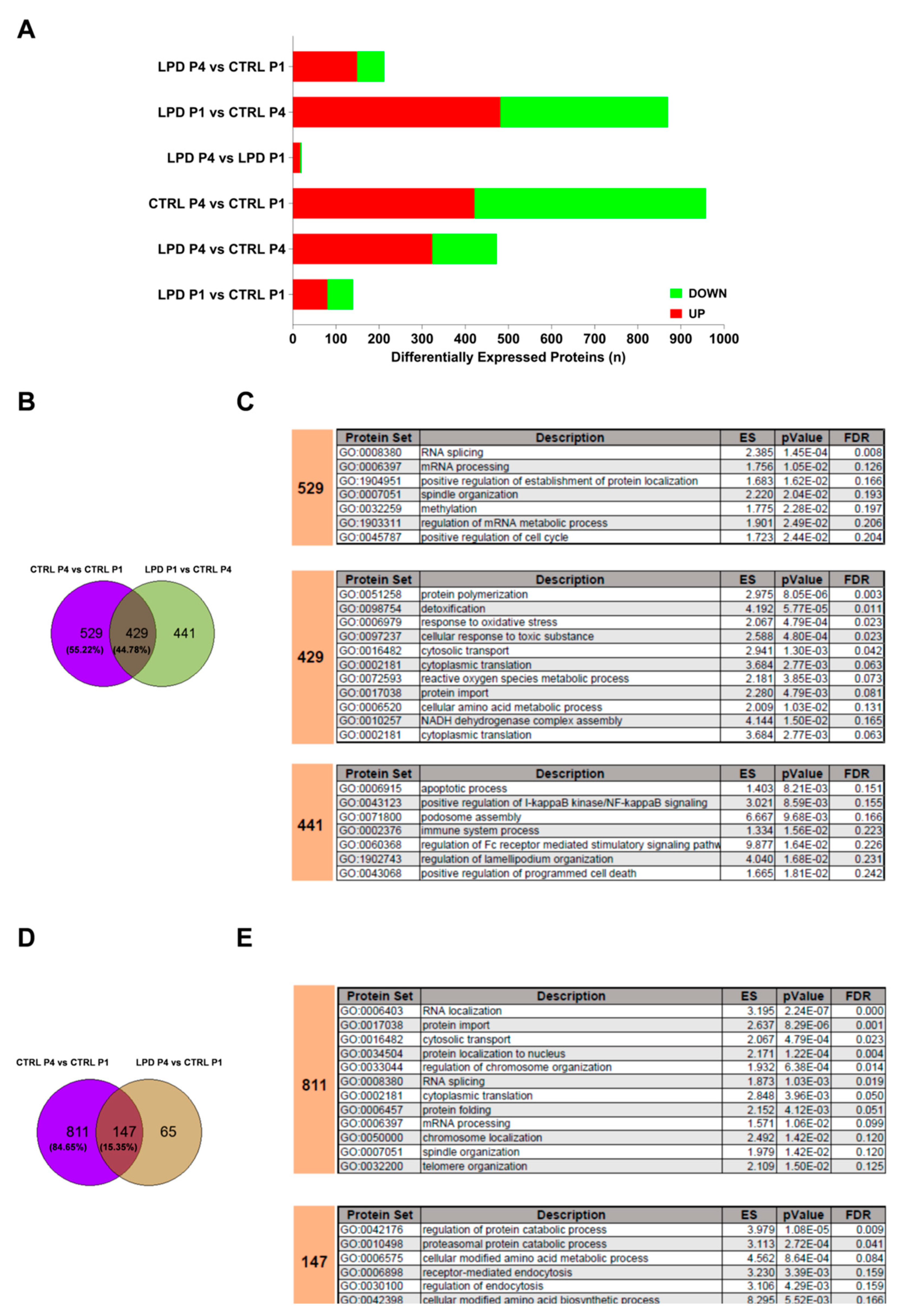
3.2. The Microglial Proteome Early after Birth Reflects the Growth Restriction following LPD Exposure
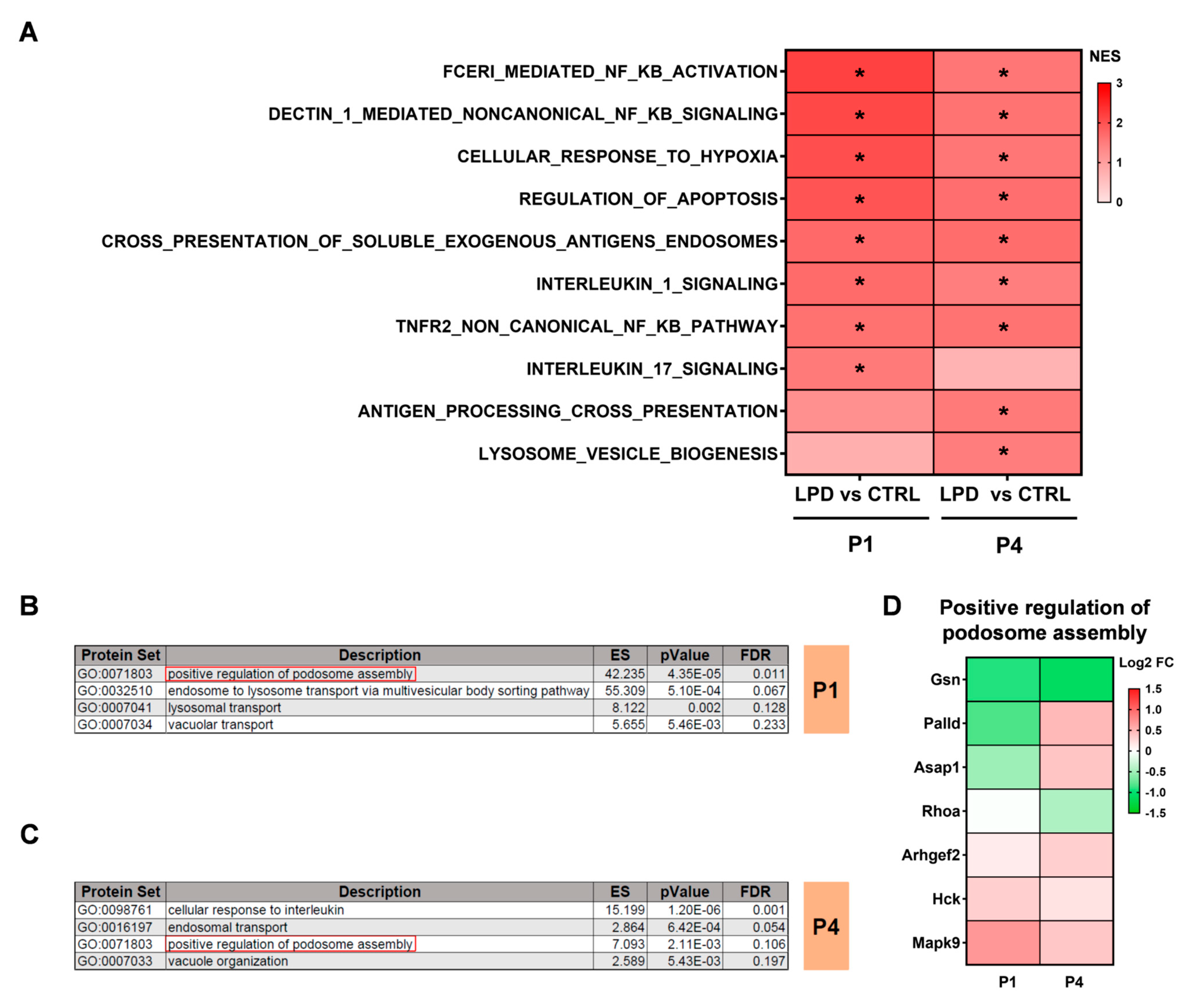
3.3. Protein Sets Associated with Inflammation Are Deregulated in the LPD Microglial Proteome
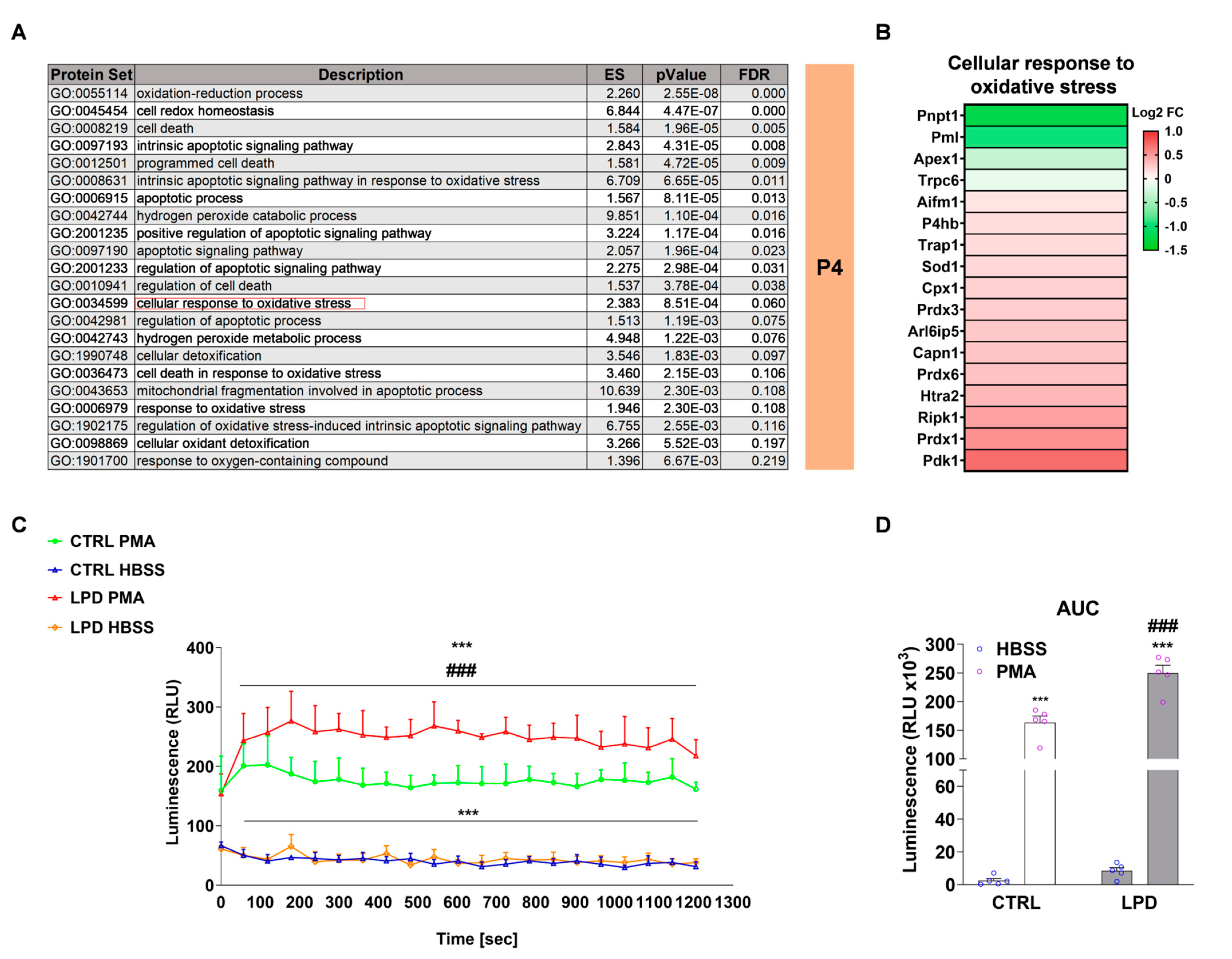
3.4. Protein Sets Associated with Oxidative Stress and Cellular Response to Oxidative Stress Are Enriched in LPD Microglia
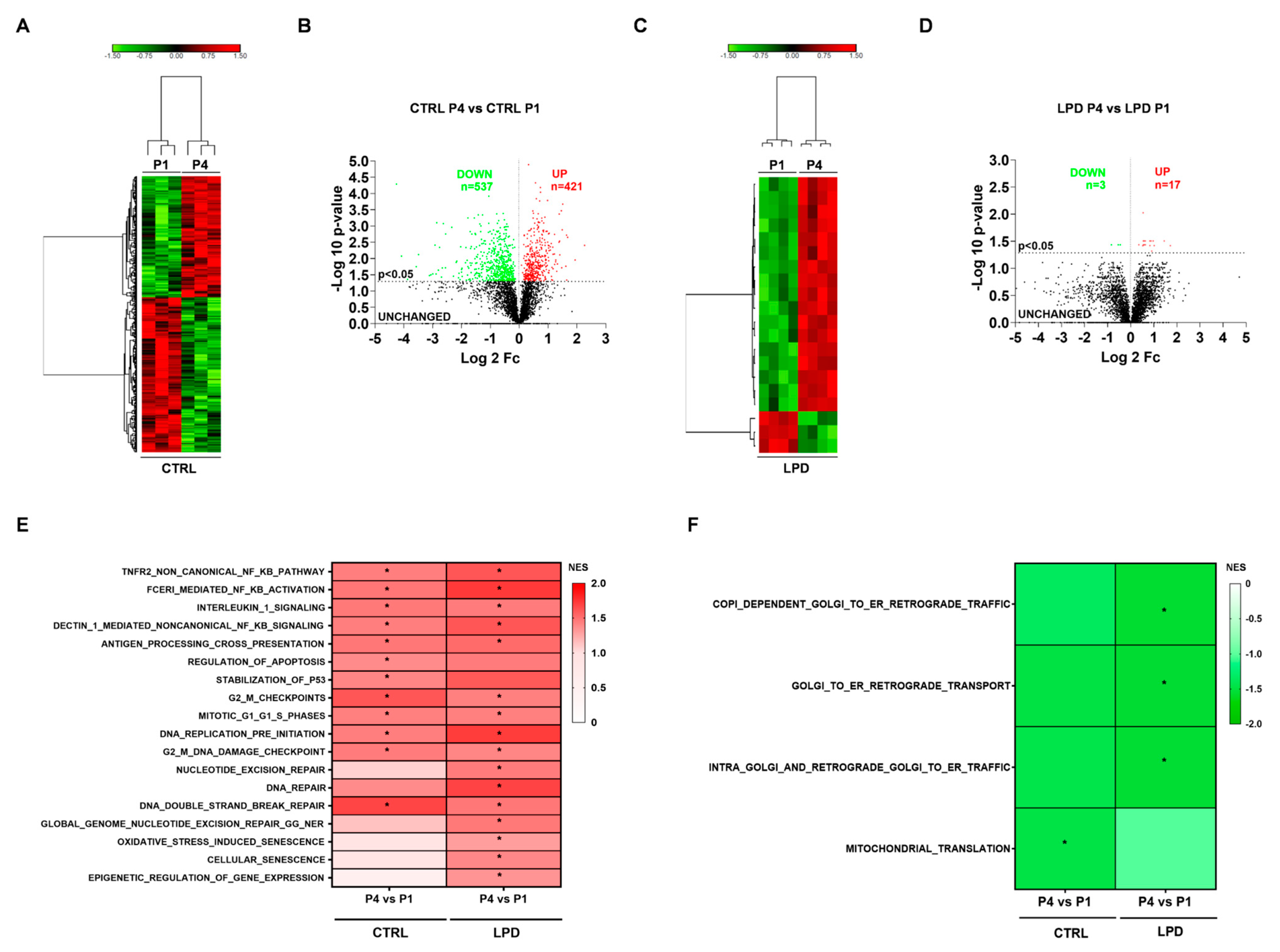
3.5. The Early Developmental Changes of the Microglial Proteome Are Modified by Antenatal LPD Exposure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miller, L.S.; Huppi, P.S.; Mallard, C. The Consequences of Fetal Growth Restriction on Brain Structure and Neurodevelopmental Outcome. J. Physiol. 2016, 594, 807–823. [Google Scholar] [CrossRef] [Green Version]
- Wolke, D. Preterm Birth: High Vulnerability and No Resiliency? Reflections on Van Lieshout et al. J. Child Psychol. Psychiatry 2018, 59, 1201–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacchi, C.J.; O’Muircheartaigh, D.; Batalle, S.J.; Counsell, A.; Simonelli, M.; Cesano, S.; Falconer, A.; Chew, N.; Kennea, P.; Nongena, M.A.; et al. Neurodevelopmental Outcomes Following Intrauterine Growth Restriction and Very Preterm Birth. J. Pediatr. 2021. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.T.; Alun, E.; Elizabeth, S.; Draper, D.J.; Field, B.N.; Manktelow, N.M.; Matthews, R.; Stavros, P.; Sarah, E.; Seaton, L.; et al. Neurodevelopmental Outcomes Following Late and Moderate Prematurity: A Population-Based Cohort Study. Arch. Dis. Child. Fetal Neonatal Ed. 2015, 100, F301. [Google Scholar] [CrossRef] [Green Version]
- Korzeniewski, S.J.; Elizabeth, N.; Allred, R.M.; Joseph, T.H.; Karl, C.K.; Kuban, T.; O’Shea, M.; Leviton, A. ELGAN Study Investigators. Neurodevelopment at Age 10 Years of Children Born <28 Weeks with Fetal Growth Restriction. Pediatrics 2017, 140, e20170697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadar, O.; Sheiner, E.; Wainstock, T. The Association between Delivery of Small-for-Gestational-Age Neonate and Their Risk for Long-Term Neurological Morbidity. J. Clin. Med. 2020, 9, 3199. [Google Scholar] [CrossRef]
- Morsing, E.; Åsard, M.; Ley, D.; Stjernqvist, K.; Maršál, K. Cognitive Function after Intrauterine Growth Restriction and Very Preterm Birth. Pediatrics 2011, 127, e874–e882. [Google Scholar] [CrossRef]
- Bruno, C.J.; Bengani, S.; Gomes, W.A.; Brewer, M.; Vega, M.; Xie, X.; Kim, M.; Fuloria, M. Mri Differences Associated with Intrauterine Growth Restriction in Preterm Infants. Neonatology 2017, 111, 317–323. [Google Scholar] [CrossRef]
- Hagberg, H.; Gressens, P.; Mallard, C. Inflammation During Fetal and Neonatal Life: Implications for Neurologic and Neuropsychiatric Disease in Children and Adults. Ann. Neurol. 2012, 71, 444–457. [Google Scholar] [CrossRef]
- Fleiss, B.; Wong, F.; Brownfoot, F.; Shearer, I.K.; Baud, O.; Walker, D.W.; Gressens, P.; Tolcos, M. Knowledge Gaps and Emerging Research Areas in Intrauterine Growth Restriction-Associated Brain Injury. Front. Endocrinol. 2019, 10, 188. [Google Scholar] [CrossRef] [Green Version]
- Paolicelli, R.; Bolasco, G.C.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; et al. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science 2011, 333, 1456–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.; Wang, J.; Huang, M.; Chen, Z.; Liu, J.; Zhang, Q.; Zhang, C.; Xiang, Y.; Zen, K.; Li, L. Loss of Microglial Sirpα Promotes Synaptic Pruning in Preclinical Models of Neurodegeneration. Nat. Commun. 2021, 12, 2030. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Ann. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ben, A. Barres. Microglia and Macrophages in Brain Homeostasis and Disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Willis, E.; Kelli, F.; MacDonald, P.A.; Quan Nguyen, H.; Garrido, A.L.; Gillespie, R.E.; Harley, B.R.S.; Bartlett, F.P.; Schroder, A.W.; Yates, G.A.; et al. Repopulating Microglia Promote Brain Repair in an Il-6-Dependent Manner. Cell 2020, 180, 833–846. [Google Scholar] [CrossRef]
- Neumann, H.; Kotter, M.R.; Franklin, R.J.M. Debris Clearance by Microglia: An Essential Link between Degeneration and Regeneration. Brain 2008, 132, 288–295. [Google Scholar] [CrossRef]
- Dong, R.; Huang, R.; Wang, J.; Liu, H.; Xu, Z. Effects of Microglial Activation and Polarization on Brain Injury after Stroke. Front. Neurol. 2021, 12, 620948. [Google Scholar] [CrossRef]
- Edmonson, C.; Mark, A.; Ziats, N.; Rennert, O.M. A Non-Inflammatory Role for Microglia in Autism Spectrum Disorders. Front. Neurol. 2016, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Morgan, J.T.; Chana, G.; Abramson, I.; Semendeferi, K.; Courchesne, E.; Everall, I.P. Abnormal Microglial–Neuronal Spatial Organization in the Dorsolateral Prefrontal Cortex in Autism. Brain Res. 2012, 1456, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.X.; Kim, G.H.; Tan, J.W.; Riso, A.E.; Sun, Y.; Xu, E.Y.; Liao, G.Y.; Xu, H.; Lee, S.H.; Do, N.Y.; et al. Elevated Protein Synthesis in Microglia Causes Autism-Like Synaptic and Behavioral Aberrations. Nat. Commun. 2020, 11, 1797. [Google Scholar] [CrossRef]
- Mallard, C.; Davidson, J.O.; Tan, S.; Green, C.R.; Bennet, L.; Robertson, N.J.; Gunn, A.J. Astrocytes and Microglia in Acute Cerebral Injury Underlying Cerebral Palsy Associated with Preterm Birth. Pediatric Res. 2014, 75, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Vuguin, P.M. Animal Models for Small for Gestational Age and Fetal Programming of Adult Disease. Horm. Res. 2007, 68, 113–123. [Google Scholar] [PubMed] [Green Version]
- Felicioni, F.; Santos, T.G.; Domingues, T.d.; Paula, E.; Chiarini-Garcia, H.; de Almeida, F.R.C.L. Intrauterine Growth Restriction: Screening and Diagnosis Using Animal Models. Anim. Reprod. 2020, 16, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Colella, M.; Zinni, M.; Pansiot, J.; Cassanello, M.; Mairesse, J.; Ramenghi, L.; Baud, O. Modulation of Microglial Activation by Adenosine A2a Receptor in Animal Models of Perinatal Brain Injury. Front. Neurol. 2018, 9, 605. [Google Scholar] [CrossRef]
- Novais, R.B.; Pham, H.A.; van de Looij, Y.; Bernal, M.; Mairesse, J.; Zana-Taieb, E.; Colella, M.; Jarreau, P.H.; Pansiot, J.; Dumont, F.; et al. Transcriptomic Regulations in Oligodendroglial and Microglial Cells Related to Brain Damage Following Fetal Growth Restriction. Glia 2016, 64, 2306–2320. [Google Scholar] [CrossRef] [Green Version]
- Mairesse, J.; Zinni, M.; Pansiot, J.; Hassan-Abdi, R.; Demene, C.; Colella, M.; Charriaut-Marlangue, C.; Novais, A.R.B.; Tanter, M.; Maccari, S.; et al. Oxytocin Receptor Agonist Reduces Perinatal Brain Damage by Targeting Microglia. Glia 2019, 67, 345–359. [Google Scholar] [CrossRef]
- Zinni, M.; Mairesse, J.; Pansiot, J.; Fazio, F.; Iacovelli, L.; Antenucci, N.; Orlando, R.; Nicoletti, F.; Vaiman, D.; Baud, O. Mglu3 Receptor Regulates Microglial Cell Reactivity in Neonatal Rats. J. NeuroInflamm. 2021, 18, 13. [Google Scholar] [CrossRef]
- Pan, J.; Ma, N.; Yu, B.; Zhang, W.; Wan, J. Transcriptomic Profiling of Microglia and Astrocytes Throughout Aging. J. Neuroinflamm. 2020, 17, 97. [Google Scholar] [CrossRef] [Green Version]
- Qingyun, L.; Cheng, Z.; Zhou, L.; Darmanis, S.; Neff, N.F.; Okamoto, J.; Gulati, G.; Bennett, M.L.; Lu Sun, O.; Clarke, L.E.; et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell Rna Sequencing. Neuron 2019, 101, 207–223. [Google Scholar]
- Hammond, T.R.; Dufort, C.; Dissing-Olesen, L.; Giera, S.; Young, A.; Wysoker, A.; Walker, A.J.; Gergits, F.; Segel, M.; Nemesh, J.; et al. Single-Cell Rna Sequencing of Microglia Throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 2019, 50, 253–271. [Google Scholar] [CrossRef] [Green Version]
- Antwoine, F.; Bell-Temin, H.; Jalloh, A.; Stanley, M.; Stevens, P.; Bickford, C. Proteomic Analysis of Aged Microglia: Shifts in Transcription, Bioenergetics, and Nutrient Response. J. Neuroinflamm. 2017, 14, 96. [Google Scholar]
- Monasor, S.; Stephan, L.; Müller, A.; Colombo, A.V.; Tanrioever, G.; König, J.; Roth, S.; Liesz, A.; Berghofer, A.; Piechotta, A.; et al. Fibrillar Aβ Triggers Microglial Proteome Alterations and Dysfunction in Alzheimer Mouse Models. eLife 2020, 9, e54083. [Google Scholar] [CrossRef]
- Rangaraju Srikant, E.; Dammer, B.; Raza, S.A.; Gao, T.; Xiao, H.; Betarbet, R.; Duong, M.D.; Webster, J.A.; Hales, C.M.; Lah, J.; et al. Quantitative Proteomics of Acutely-Isolated Mouse Microglia Identifies Novel Immune Alzheimer’s Disease-Related Proteins. Mol. Neurodegener. 2018, 13, 34. [Google Scholar] [CrossRef]
- Colombo, A.; Dinkel, L.; Stephan, A.; Müller, L.; Monasor, S.; Schifferer, M.; Cantuti-Castelvetri, L.; König, J.; Vidatic, L.; Bremova-Ertl, T.; et al. Loss of Npc1 Enhances Phagocytic Uptake and Impairs Lipid Trafficking in Microglia. Nat. Commun. 2021, 12, 1158. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Y.; Liu, J.; Fu, W. Proteomics Analysis of Fetal Growth Restriction and Taurine-Treated Fetal Growth Restriction Rat Brain Tissue by 2d dige and Maldi-Tof/Tof Ms Analysis. Int. J. Mol. Med. 2019, 44, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Valent, D.; Yeste, N.; Hernández-Castellano, L.E.; Arroyo, L.; Wu, W.; García-Contreras, C.; Vázquez-Gómez, M.; González-Bulnes, A.; Bendixen, E.; Bassols, A. Swath-Ms Quantitative Proteomic Investigation of Intrauterine Growth Restriction in a Porcine Model Reveals Sex Differences in Hippocampus Development. J. Proteom. 2019, 204, 103391. [Google Scholar] [CrossRef]
- Pedroso, A.P.; Souza, A.P.; Dornellas, A.P.; Oyama, L.M.; Nascimento, C.M.; Santos, G.M.; Rosa, J.C.; Bertolla, R.P.; Klawitter, J.; Christians, U.; et al. Intrauterine Growth Restriction Programs the Hypothalamus of Adult Male Rats: Integrated Analysis of Proteomic and Metabolomic Data. J. Proteome Res. 2017, 16, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Potiris, A.; Manousopoulou, A.; Zouridis, A.; Sarli, P.M.; Pervanidou, P.; Eliades, G.; Perrea, D.N.; Deligeoroglou, E.; Garbis, S.D.; Eleftheriades, M. The Effect of Prenatal Food Restriction on Brain Proteome in Appropriately Grown and Growth Restricted Male Wistar Rats. Front. Neurosci. 2021, 15, 665354. [Google Scholar] [CrossRef]
- Buffat, C.; Mondon, F.; Rigourd, V.; Boubred, F.; Bessieres, B.; Fayol, L.; Feuerstein, J.M.; Gamerre, M.; Jammes, H.; Rebourcet, R.; et al. A Hierarchical Analysis of Transcriptome Alterations in Intrauterine Growth Restriction (Iugr) Reveals Common Pathophysiological Pathways in Mammals. J. Pathol. 2007, 213, 337–346. [Google Scholar] [CrossRef]
- Kadri-Hassani, N.; Leger, C.L.; Descomps, B. The Fatty Acid Bimodal Action on Superoxide Anion Production by Human Adherent Monocytes under Phorbol 12-Myristate 13-Acetate or Diacylglycerol Activation Can Be Explained by the Modulation of Protein Kinase C and P47phox Translocation. J. Biol. Chem. 1995, 270, 15111–15118. [Google Scholar] [CrossRef] [Green Version]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of Lc-Ms/Ms Proteomics Data in Maxquant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef] [Green Version]
- Bielow, C.; Mastrobuoni, G.; Kempa, S. Proteomics Quality Control: Quality Control Software for Maxquant Results. J. Proteome Res. 2016, 15, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.B.; Yzydorczyk, C.; Siddeek, B.; Peyter, A.C.; Simeoni, U. Intrauterine Growth Restriction: Clinical Consequences on Health and Disease at Adulthood. Reprod. Toxicol. 2021, 99, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Salam, R.; Jai, A.; Das, K.; Ali, A.; Lassi, S.Z.; Zulfiqar Bhutta, A. Maternal Undernutrition and Intrauterine Growth Restriction. Expert Rev. Obstet. Gynecol. 2013, 8, 559–567. [Google Scholar] [CrossRef]
- Butler, C.A.; Popescu, A.S.; Kitchener, E.J.A.; Allendorf, D.H.; Puigdellívol, M.; Brown, G.C. Microglial Phagocytosis of Neurons in Neurodegeneration, and Its Regulation. J. Neurochem. 2021, 158, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Deepak, S.; Shastri, S.; Pradeep, S. Intrauterine Growth Restriction: Antenatal and Postnatal Aspects. Clin. Med. Insights Pediatrics 2016, 10, 67–83. [Google Scholar]
- Gimenez da Silva-Santi, L.; Antunes, M.M.; Mori, M.A.; de Almeida-Souza, C.B.; Visentainer, J.V.; Carbonera, F.; Crisma, A.R.; Masi, L.N.; Hirabara, S.M.; Curi, R.; et al. Brain Fatty Acid Composition and Inflammation in Mice Fed with High-Carbohydrate Diet or High-Fat Diet. Nutrients 2018, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- López-López, S.; Monsalve, E.M.; Romero de Ávila, M.J.; González-Gómez, J.; Hernández de León, N.; Ruiz-Marcos, F.; Baladrón, V.; Nueda, M.L.; García-León, M.J.; Screpanti, I.; et al. Notch3 Signaling Is Essential for Nf-Κb Activation in Tlr-Activated Macrophages. Sci. Rep. 2020, 10, 14839. [Google Scholar] [CrossRef]
- Leviton, A.; Fichorova, R.N.; O’Shea, T.M.; Kuban, K.; Paneth, N.; Dammann, O.; Allred, E.N.; Elgan Study Investigators. Two-Hit Model of Brain Damage in the Very Preterm Newborn: Small for Gestational Age and Postnatal Systemic Inflammation. Pediatric Res. 2013, 73, 362–370. [Google Scholar] [CrossRef] [Green Version]
- McElrath, T.F.; Allred, E.N.; van Marter, L.; Fichorova, R.N.; Leviton, A.; ELGAN Study Investigators. Perinatal Systemic Inflammatory Responses of Growth-Restricted Preterm Newborns. Acta Paediatr. 2013, 102, e439–e442. [Google Scholar] [CrossRef]
- Guvendag Guven, E.S.; Karcaaltincaba, D.; Kandemir, O.; Kiykac, S.; Mentese, A. Cord Blood Oxidative Stress Markers Correlate with Umbilical Artery Pulsatility in Fetal Growth Restriction. J. Matern. Fetal. Neonatal. Med. 2013, 26, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Potdar, N.; Singh, R.; Mistry, V.; Evans, M.D.; Farmer, P.B.; Konje, J.C.; Cooke, M.S. First-Trimester Increase in Oxidative Stress and Risk of Small-for-Gestational-Age Fetus. Bjog 2009, 116, 637–642. [Google Scholar] [CrossRef]
- Rains, M.E.; Colin, B.; Pang, M.Y.; Lir-Wan, F.; Tien, L.-T.; Norma, B.O. Oxidative Stress and Neurodevelopmental Outcomes in Rat Offspring with Intrauterine Growth Restriction Induced by Reduced Uterine Perfusion. Brain Sci. 2021, 11, 78. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.R.; Pang, Y.; Ojeda, N.B.; Zheng, B.; Rhodes, P.G.; Alexander, B.T. Intracerebral Lipopolysaccharide Induces Neuroinflammatory Change and Augmented Brain Injury in Growth-Restricted Neonatal Rats. Pediatr. Res. 2012, 71, 645–652. [Google Scholar] [CrossRef]
- Wixey, J.A.; Lee, K.M.; Miller, S.M.; Goasdoue, K.; Colditz, P.B.; Bjorkman, S.T.; Kirat, K.C. Neuropathology in Intrauterine Growth Restricted Newborn Piglets Is Associated with Glial Activation and Proinflammatory Status in the Brain. J. Neuroinflamm. 2019, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.A.; Boddeke, H.W.; Kettenmann, H. Microglia in Physiology and Disease. Ann. Rev. Physiol. 2017, 79, 619–643. [Google Scholar] [CrossRef]
- Simpson, D.A.S.; Oliver, P.L. Ros Generation in Microglia: Understanding Oxidative Stress and Inflammation in Neurodegenerative Disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef] [PubMed]
- Matcovitch-Natan, O.; Winter, D.R.; Giladi, A.; Aguilar, S.V.; Spinrad, A.; Sarrazin, S.; Ben-Yehuda, H.; David, E.; González, F.Z.; Perrin, P.; et al. Microglia Development Follows a Stepwise Program to Regulate Brain Homeostasis. Science 2016, 353, aad8670. [Google Scholar] [CrossRef]
- Hanamsagar, R.; Alter, M.D.; Block, C.S.; Sullivan, H.; Bolton, J.L.; Bilbo, S.D. Generation of a Microglial Developmental Index in Mice and in Humans Reveals a Sex Difference in Maturation and Immune Reactivity. Glia 2017, 65, 1504–1520. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zinni, M.; Pansiot, J.; Colella, M.; Faivre, V.; Delahaye-Duriez, A.; Guillonneau, F.; Bruce, J.; Salnot, V.; Mairesse, J.; Knoop, M.; et al. Impact of Fetal Growth Restriction on the Neonatal Microglial Proteome in the Rat. Nutrients 2021, 13, 3719. https://doi.org/10.3390/nu13113719
Zinni M, Pansiot J, Colella M, Faivre V, Delahaye-Duriez A, Guillonneau F, Bruce J, Salnot V, Mairesse J, Knoop M, et al. Impact of Fetal Growth Restriction on the Neonatal Microglial Proteome in the Rat. Nutrients. 2021; 13(11):3719. https://doi.org/10.3390/nu13113719
Chicago/Turabian StyleZinni, Manuela, Julien Pansiot, Marina Colella, Valérie Faivre, Andrée Delahaye-Duriez, François Guillonneau, Johanna Bruce, Virginie Salnot, Jérôme Mairesse, Marit Knoop, and et al. 2021. "Impact of Fetal Growth Restriction on the Neonatal Microglial Proteome in the Rat" Nutrients 13, no. 11: 3719. https://doi.org/10.3390/nu13113719
APA StyleZinni, M., Pansiot, J., Colella, M., Faivre, V., Delahaye-Duriez, A., Guillonneau, F., Bruce, J., Salnot, V., Mairesse, J., Knoop, M., Possovre, M.-L., Vaiman, D., & Baud, O. (2021). Impact of Fetal Growth Restriction on the Neonatal Microglial Proteome in the Rat. Nutrients, 13(11), 3719. https://doi.org/10.3390/nu13113719






