The Emulsifier Carboxymethylcellulose Induces More Aggressive Colitis in Humanized Mice with Inflammatory Bowel Disease Microbiota Than Polysorbate-80
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Materials
2.3. Colonization with Human Fecal Bacteria
2.4. Emulsifier Treatment
2.5. Lipocalin 2 Assay (Lcn-2)
2.6. Colonic RNAs Extraction and qRT-PCR Analysis
2.7. Cytokine Measurement for Interferon Gamma Protein in Unstimulated Colonic Strip Cultures and Serum
2.8. Myeloperoxidase Assay
2.9. Assessment of Histologic Inflammation
2.10. Library Preparation and Shotgun Metagenomic DNA Sequencing
2.11. Microbial Analysis
2.12. Statistical Analysis of Non-Microbial Data
3. Results
3.1. The Effect of CMC and P80 on Body Weight and Inflammatory Biomarkers
3.2. CMC Enhanced Histologic Colonic Inflammation
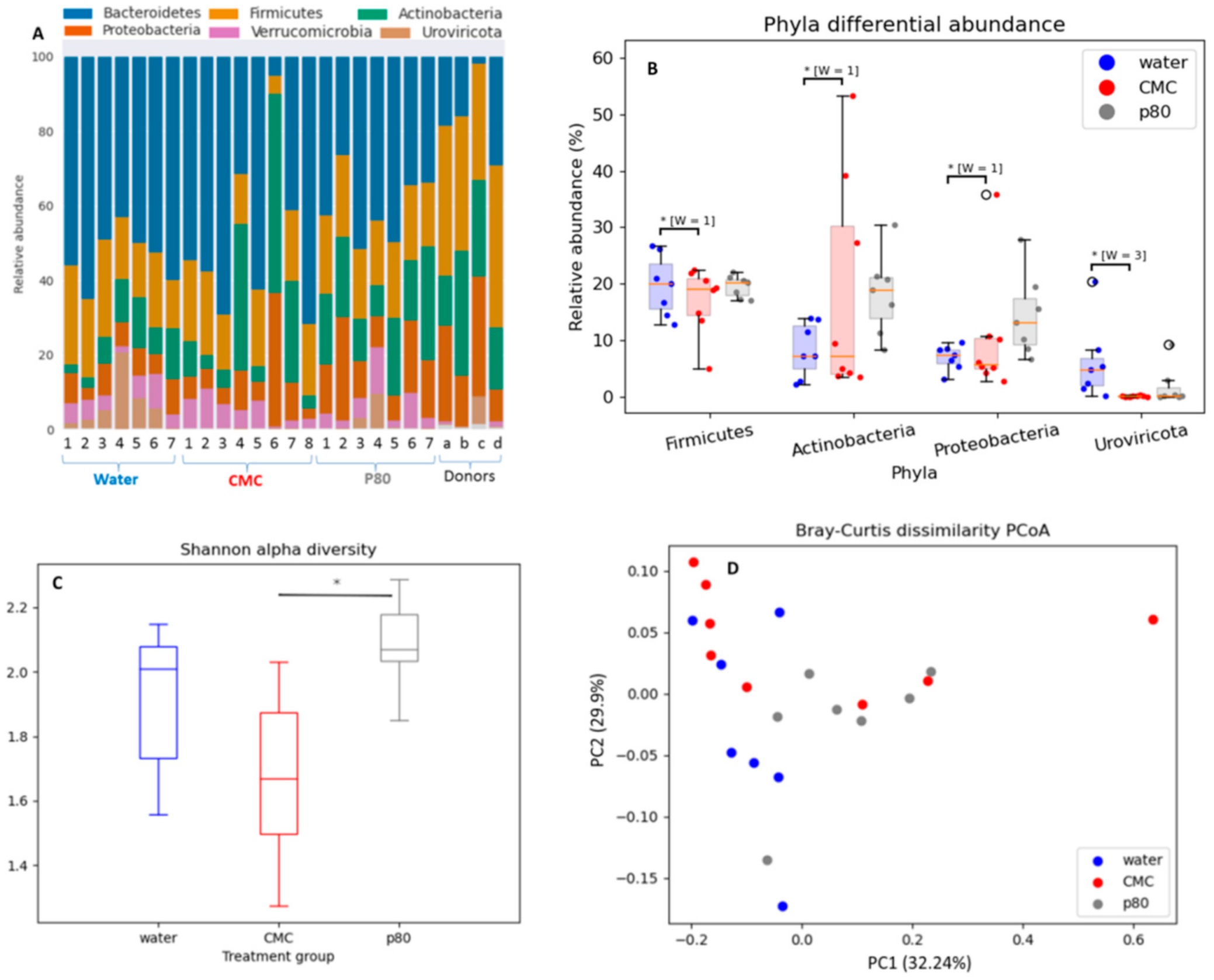
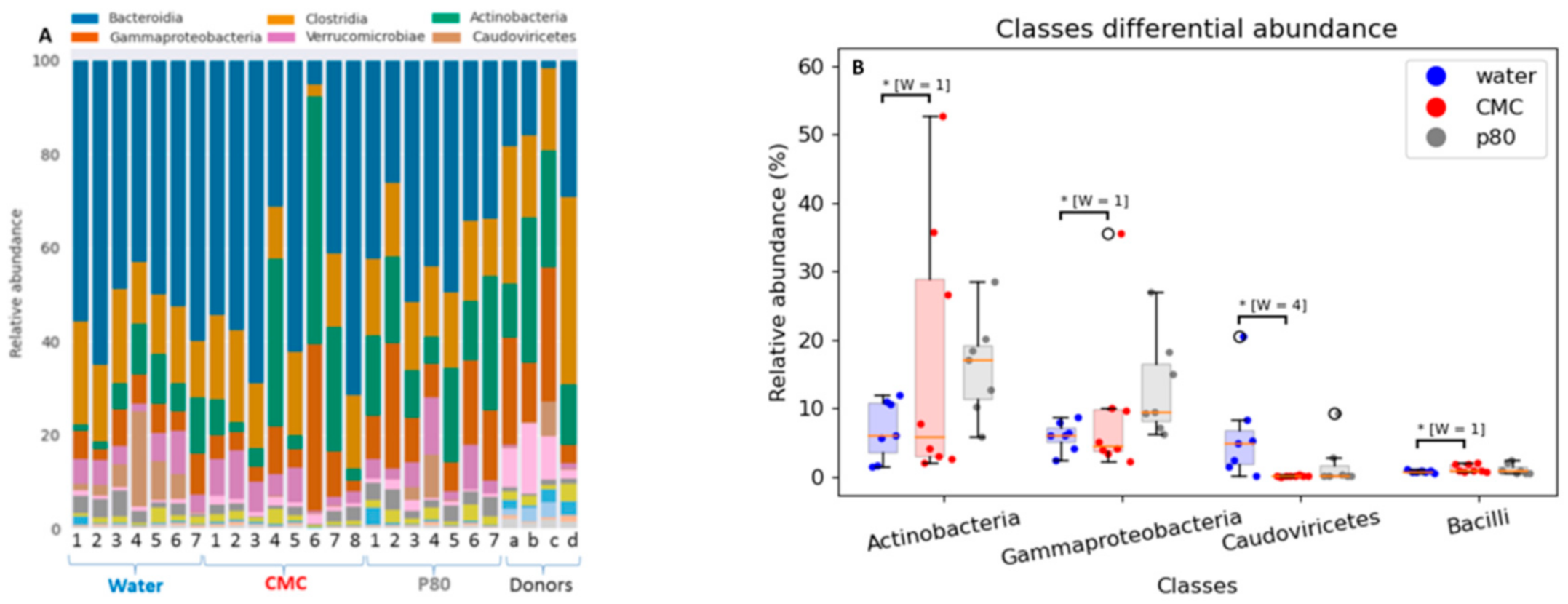
3.3. Differential Effects of CMC and P80 on Gut Microbiota Abundance and Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef]
- Rogala, A.R.; Oka, A.; Sartor, R.B. Strategies to dissect host-microbial immune interactions that determine mucosal homeostasis vs. Intestinal inflammation in Gnotobiotic mice. Front. Immunol. 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartor, R.B.; Wu, G.D. Roles for intestinal bacteria, viruses, and fungi in the pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017, 152, 327–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Lelie, D.; Oka, A.; Taghavi, S.; Umeno, J.; Fan, T.J.; Merrell, K.E.; Watson, S.D.; Ouellette, L.; Liu, B.; Awoniyi, M.; et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat. Commun. 2021, 12, 3105. [Google Scholar] [CrossRef]
- van Wijk, F.; Cheroutre, H. Mucosal T cells in gut homeostasis and inflammation. Expert Rev. Clin. Immunol. 2011, 6, 559–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, T.; Kamm, M.A.; Colombel, J.F.; Ng, S.C. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Abreu, M.T. Diet as a trigger or therapy for inflammatory bowel diseases. Gastroenterology 2017, 152, 398–414. [Google Scholar] [CrossRef]
- Laudisi, F.; Stolfi, C.; Monteleone, G. Impact of food additives on gut homeostasis. Nutrients 2019, 11, 2334. [Google Scholar] [CrossRef] [Green Version]
- Bancil, A.S.; Sandall, A.M.; Rossi, M.; Chassaing, B.; Lindsay, J.O.; Whelan, K. Food additive emulsifiers and their impact on gut microbiome, permeability and inflammation: Mechanistic insights in inflammatory bowel disease. J. Crohn’s Colitis 2021, 15, 1068–1079. [Google Scholar] [CrossRef]
- Roberts, C.L.; Keita, V.; Duncan, S.H.; O’Kennedy, N.; Söderholm, J.D.; Rhodes, J.M.; Campbell, B.J. Translocation of Crohn’s disease Escherichia coli across M-cells: Contrasting effects of soluble plant fibres and emulsifiers. Gut 2010, 59, 1331–1339. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Koren, O.; Goodrich, J.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 404–413. [Google Scholar] [CrossRef] [Green Version]
- Holder, M.K.; Peters, N.V.; Whylings, J.; Fields, C.T.; Gewirtz, A.T.; Chassaing, B.; de Vries, G.J. Dietary emulsifiers consumption alters anxiety-like and social-related behaviors in mice in a sex-dependent manner. Sci. Rep. 2019, 172, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Ishikawa, S. Food additive P-80 impacts mouse gut microbiota promoting intestinal inflammation, obesity and liver dysfunction. SOJ Microbiol. Infect. Dis. 2016, 4, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viennois, E.; Merlin, D.; Gewirtz, A.T.; Chassaing, B. Dietary emulsifier-induced low-grade inflammation promotes colon carcinogenesis. Cancer Res. 2017, 77, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viennois, E.; Chassaing, B. First victim, later aggressor: How the intestinal microbiota drives the pro-inflammatory effects of dietary emulsifiers? Gut Microbes 2018, 9, 288–291. [Google Scholar] [CrossRef] [Green Version]
- Vo, T.D.; Lynch, B.S.; Roberts, A. Dietary exposures to common emulsifiers and their impact on the gut microbiota: Is there a cause for concern? Compr. Rev. Food Sci. Saf. 2019, 18, 31–47. [Google Scholar] [CrossRef] [Green Version]
- McClements, D.J.; Decker, E.A.; Weiss, J. Emulsion-based delivery systems for lipophilic bioactive components. J. Food Sci. 2007, 72, 109–124. [Google Scholar] [CrossRef]
- Partridge, D.; Lloyd, K.A.; Rhodes, J.M.; Walker, A.W.; Johnstone, A.M.; Campbell, B.J. Food additives: Assessing the impact of exposure to permitted emulsifiers on the bowel and metabolic health—introducing the FADiets study. Nutr. Bull. 2019, 44, 329–349. [Google Scholar] [CrossRef] [Green Version]
- Viennois, E.; Bretin, A.; Barnich, N.; Maue, A.C.; Dauriat, C.J.G.; Barnich, N.; Gewirtz, A.T.; Chassaing, B. Dietary emulsifiers directly impact adherent- invasive E. coli gene expression to drive chronic intestinal inflammation. Cell. Rep. 2020, 33, 108229. [Google Scholar] [CrossRef]
- Levine, A.A.; Wine, E.; Assa, A.; Sigall, B.R.; Shaoul, R.K.M.; Cohen, S.; Peleg, S.; Shamaly, H.; On, A.; Millman, P.; et al. Crohn’s disease exclusion diet plus partial enteral nutrition induces sustained remission in a randomized controlled trial. Gastroenterology 2019, 157, 440–450. [Google Scholar] [CrossRef] [Green Version]
- Sellon, R.K.; Tonkonogy, S.; Schultz, M.; Dieleman, L.A.; Grenther, W.; Balish, E.; Rennick, D.M.; Sartor, R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998, 66, 5224–5231. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.C.; Tonkonogy, S.L.; Albright, C.A.; Tsang, J.; Balish, E.J.; Braun, J.; Huycke, M.M.; Sartor, R.B. Variable phenotypes of enterocolitis in interleukin 10–deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005, 128, 891–906. [Google Scholar] [CrossRef]
- Mishima, Y.; Oka, A.; Liu, B.; Herzog, J.W.; Eun, C.S.; Fan, T.J.; Bulik-Sullivan, E.; Carroll, I.M.; Hansen, J.J.; Chen, L.; et al. Microbiota maintain colonic homeostasis by activating TLR2/MyD88/PI3K signaling in IL-10—Producing regulatory B cells. J. Clin. Investig. 2019, 129, 3702–3716. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Tonkonogy, S.N.; Sartor, R.B. IL-10 produced by antigen presenting cells inhibits bacterial- responsive TH1/TH17 cells and suppresses colitis in mice. Gastroenterology 2011, 141, 653–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, S.; van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, S.K.; Knight, R.; Caporaso, G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Moschen, A.R.; Gerner, R.R.; Wang, J.; Klepsch, V.; Adolph, T.E.; Reider, S.J.; Hackl, H.; Pfister, A.; Schilling, J.; Moser, P.L.; et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 2016, 19, 455–469. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Foncea, R.; Deis, J.A.; Guo, H.; Bernlohr, D.A.; Chen, X. Lipocalin 2 expression and secretion is highly regulated by metabolic stress, cytokines, and nutrients in adipocytes. PLoS ONE 2014, 9, e96997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chassaing, B.; Van de Wiele, T.; De Bodt, J.; Marzorati, M.; Gewirtz, A.T. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2018, 66, 1414–1427. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, R. Humanizing the gut microbiota of mice: Opportunities and challenges. Lab. Anim. 2019, 53, 244–251. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Lloyd-Price, J.; Arez, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, A. Upregulation of T-bet and tight junction molecules by Bifidobactrium longum improves colonic inflammation of ulcerative colitis. Inflamm. Bowel Dis. 2009, 15, 1617–1618. [Google Scholar] [CrossRef]
- Zhuang, X.; Tian, Z.; Feng, R.; Li, M.; Li, T.; Zhou, G.; Qiu, Y.; Chen, B.; He, Y.; Chen, M.; et al. Fecal microbiota alterations associated with clinical and endoscopic response to infliximab therapy in Crohn’s disease. Inflamm. Bowel Dis. 2020, 26, 1636–1647. [Google Scholar] [CrossRef]
- Wang, W.; Jovel, J.; Halloran, B.; Wine, E.; Patterson, J.; Ford, G.; O’Keefe, S.; Meng, B.; Song, D.; Zhang, Y.; et al. Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory nowel diseases reveals interplay of viruses and bacteria. Inflamm. Bowel Dis. 2015, 21, 1419–1427. [Google Scholar] [CrossRef]
- Boneh, R.S.; Sarbagili-Shabat, C.; Yanai, H.; Chermesh, I.; Ben Avraham, S.; Boaz, M.; Levine, A. Dietary therapy with the Crohn’s disease exclusion diet is a successful strategy for induction of remission in children and adults failing biological therapy. J. Crohn’s Colitis 2017, 11, 1205–1212. [Google Scholar] [CrossRef] [Green Version]
- Sandall, A.M.; Cox, S.R.; Lindsay, J.O.; Gewirtz, A.T.; Chassaing, B.; Rossi, M.; Whelan, K. Emulsifiers impact colonic length in mice and emulsifier restriction is feasible in people with Crohn’s disease. Nutrients 2020, 12, 2827. [Google Scholar] [CrossRef] [PubMed]
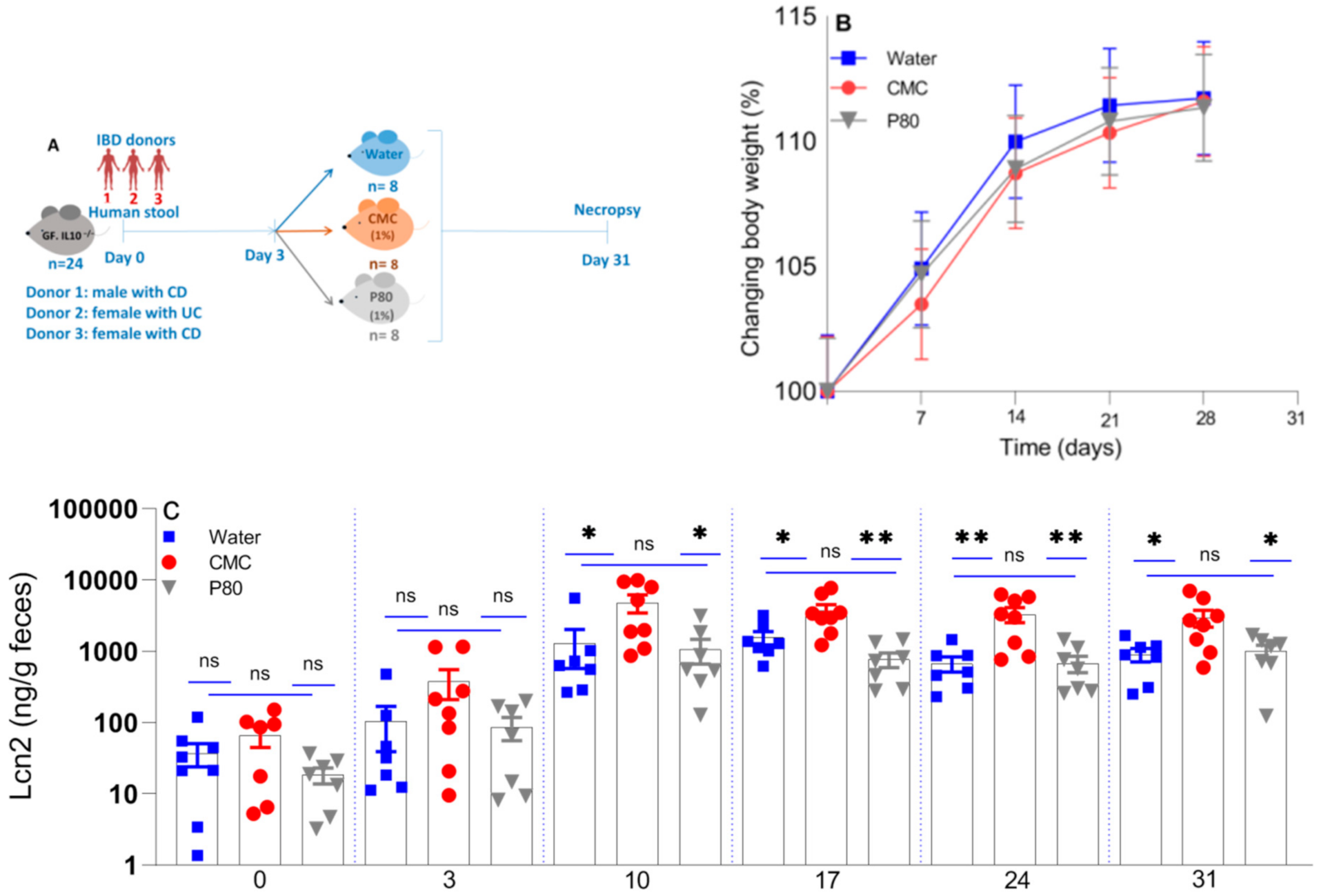
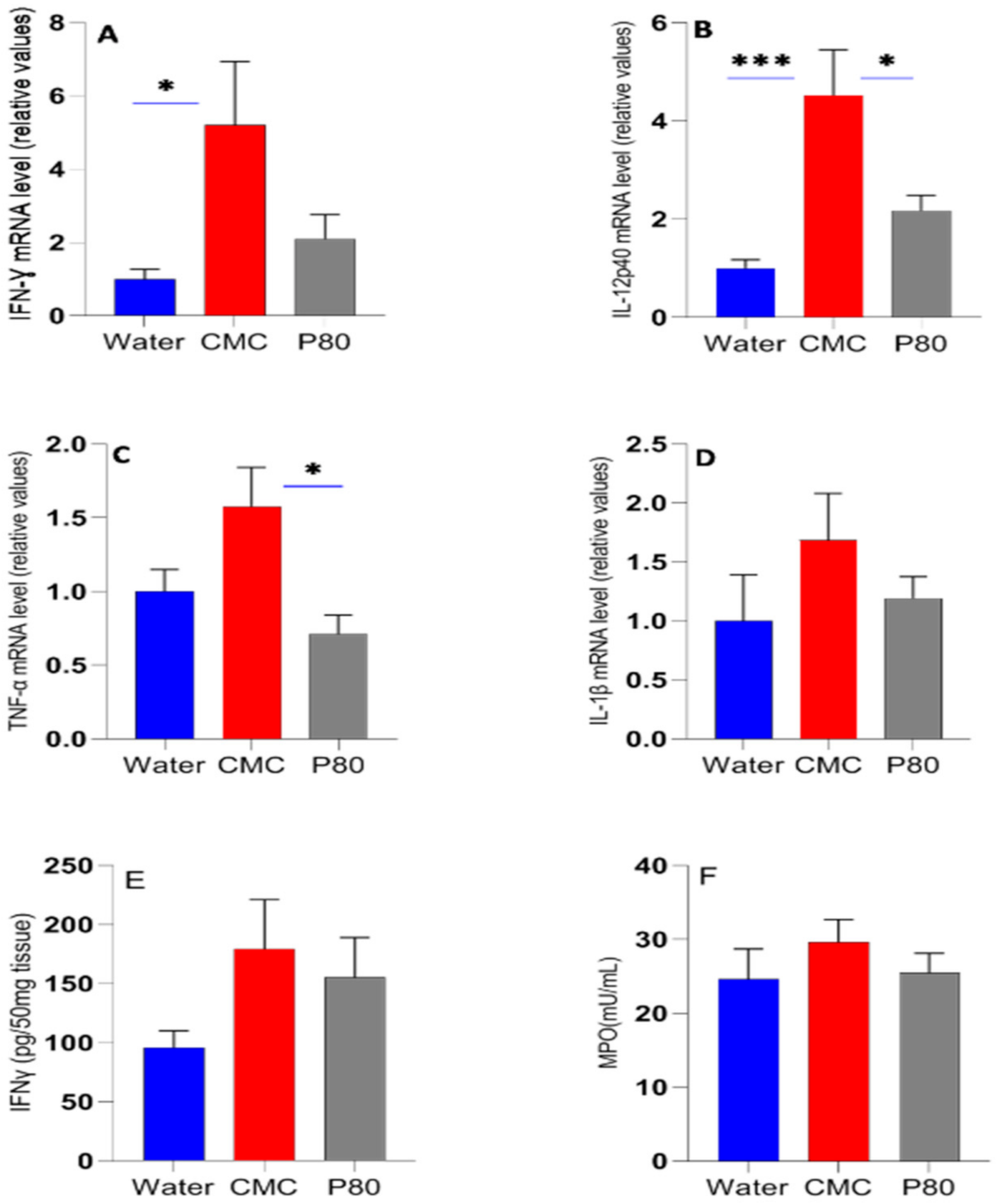
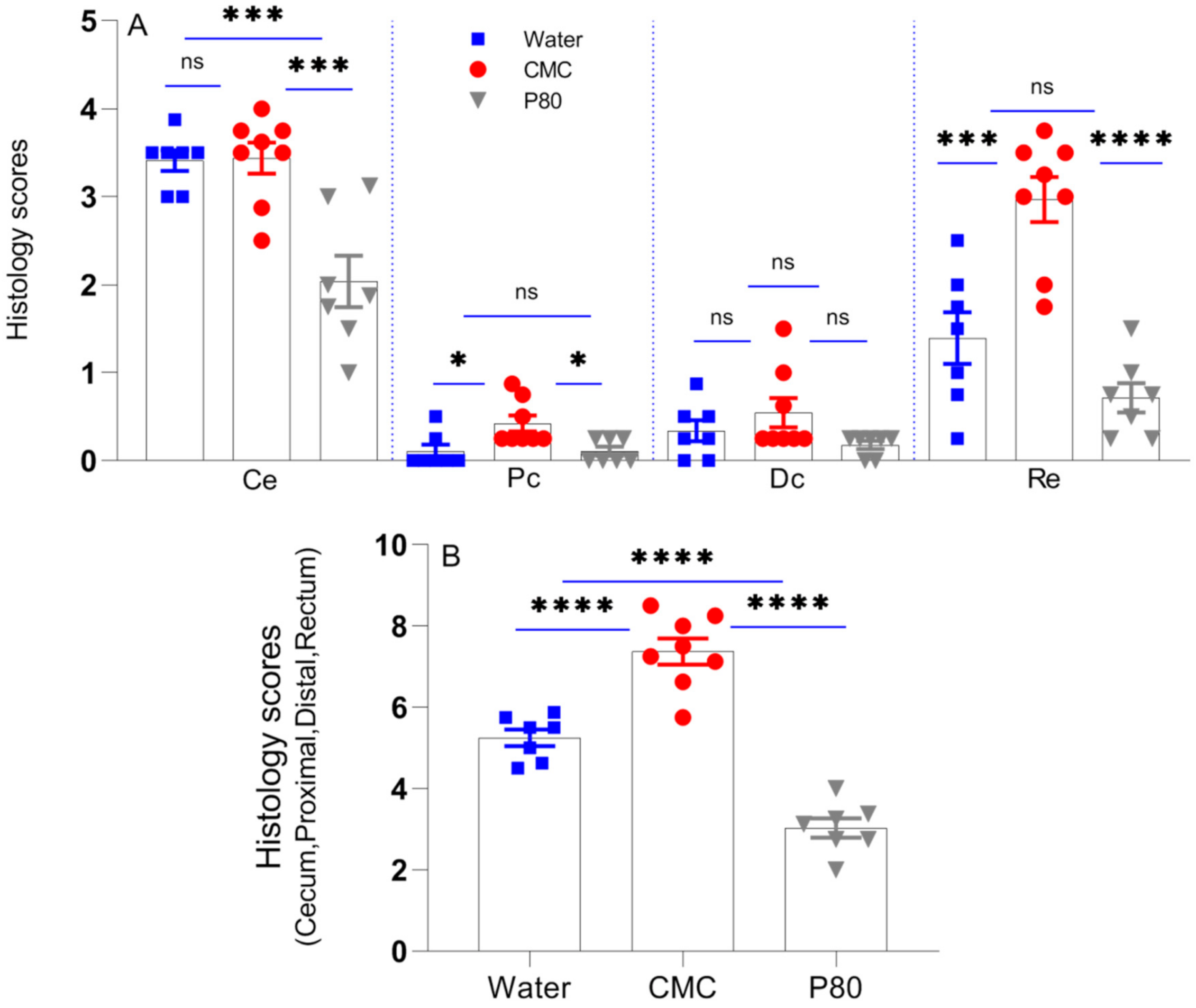
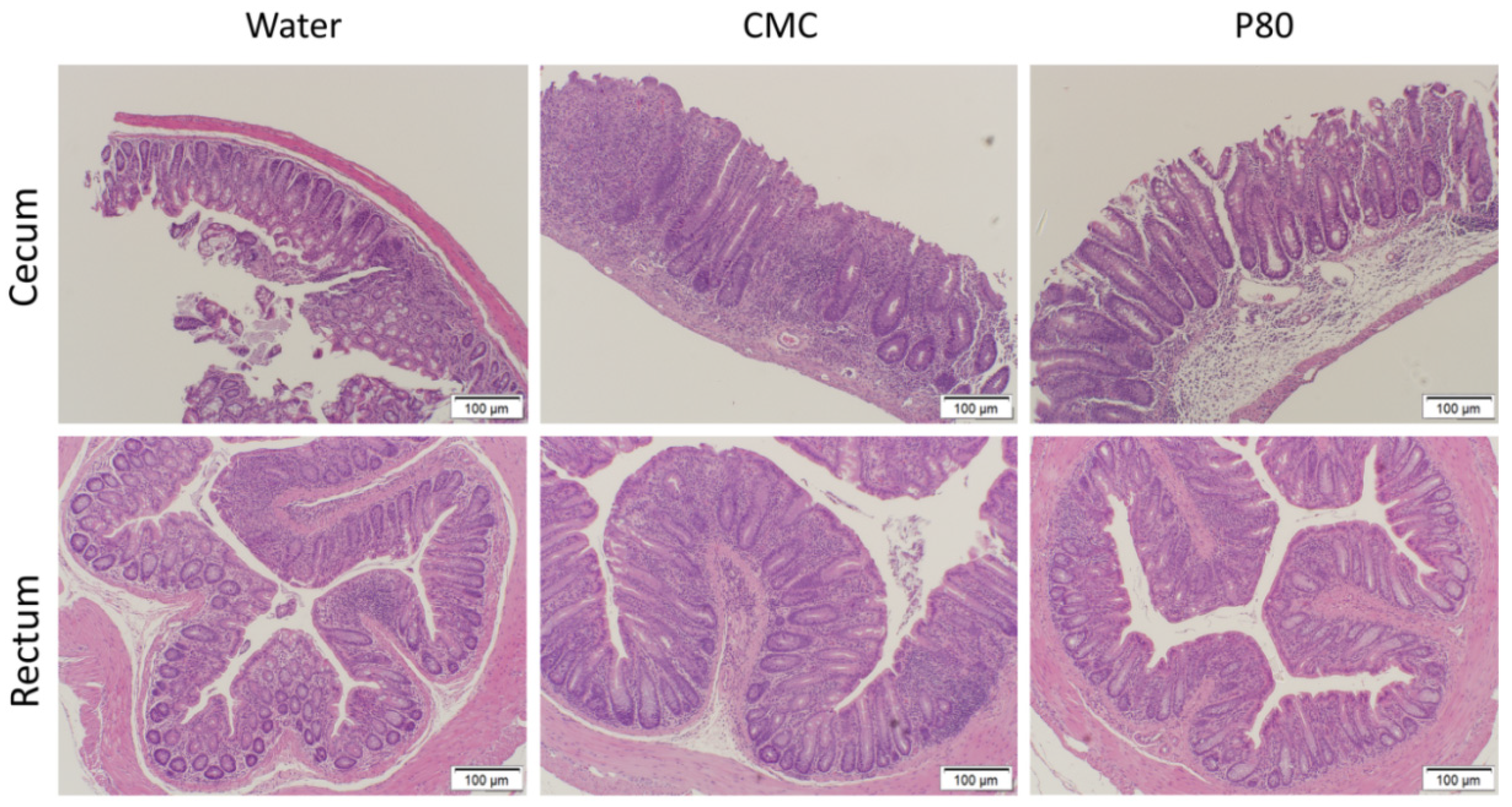

| IBD Donor 1 | Race | Sex | Disease |
|---|---|---|---|
| 1 | Caucasian | M | CD |
| 2 | Caucasian | F | UC |
| 3 | African-American | F | CD |
| Gene | Type | Sequence (5’-3’) |
|---|---|---|
| β Actin | Forward Reverse | AGCCATGTACGTAGCCATCCAG TGGCGTGAGGGAGAGCATAG |
| IFN-γ | Forward Reverse | CTTCCTCATGGCTGTTTCTGG ACGCTTATGTTGTTGCTGATGG |
| IL-12p40 | Forward Reverse | CGCAAGAAAGAAAAGATGAAGGAG TTGCATTGGACTTCGGTAGATG |
| TNF-α | Forward Reverse | ACCCTCACACACATCAGATCATCTTCTC TGAGATCCATGCCGTTGG |
| IL-1β | Forward Reverse | GTGGACCTTCCAGGATGAGG CGGAGCCTGTAGTGCAGTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rousta, E.; Oka, A.; Liu, B.; Herzog, J.; Bhatt, A.P.; Wang, J.; Habibi Najafi, M.B.; Sartor, R.B. The Emulsifier Carboxymethylcellulose Induces More Aggressive Colitis in Humanized Mice with Inflammatory Bowel Disease Microbiota Than Polysorbate-80. Nutrients 2021, 13, 3565. https://doi.org/10.3390/nu13103565
Rousta E, Oka A, Liu B, Herzog J, Bhatt AP, Wang J, Habibi Najafi MB, Sartor RB. The Emulsifier Carboxymethylcellulose Induces More Aggressive Colitis in Humanized Mice with Inflammatory Bowel Disease Microbiota Than Polysorbate-80. Nutrients. 2021; 13(10):3565. https://doi.org/10.3390/nu13103565
Chicago/Turabian StyleRousta, Esmat, Akihiko Oka, Bo Liu, Jeremy Herzog, Aadra P. Bhatt, Jeremy Wang, Mohammad B. Habibi Najafi, and Ryan Balfour Sartor. 2021. "The Emulsifier Carboxymethylcellulose Induces More Aggressive Colitis in Humanized Mice with Inflammatory Bowel Disease Microbiota Than Polysorbate-80" Nutrients 13, no. 10: 3565. https://doi.org/10.3390/nu13103565
APA StyleRousta, E., Oka, A., Liu, B., Herzog, J., Bhatt, A. P., Wang, J., Habibi Najafi, M. B., & Sartor, R. B. (2021). The Emulsifier Carboxymethylcellulose Induces More Aggressive Colitis in Humanized Mice with Inflammatory Bowel Disease Microbiota Than Polysorbate-80. Nutrients, 13(10), 3565. https://doi.org/10.3390/nu13103565






