PPARGC1A Gene Promoter Methylation as a Biomarker of Insulin Secretion and Sensitivity in Response to Glucose Challenges
Abstract
1. Introduction
2. Subjects and Methods
2.1. Design, Subjects and Anthropometric Measurements
2.2. Insulin Secretion/Sensitivity Indexes Derived from the Oral Glucose Tolerance Test (OGTT)
2.3. Insulin Secretion/Sensitivity Indexes Derived from the Intravenous Glucose Tolerance Test (IVGTT)
2.4. Hemogram and Differential Leukocyte Counts
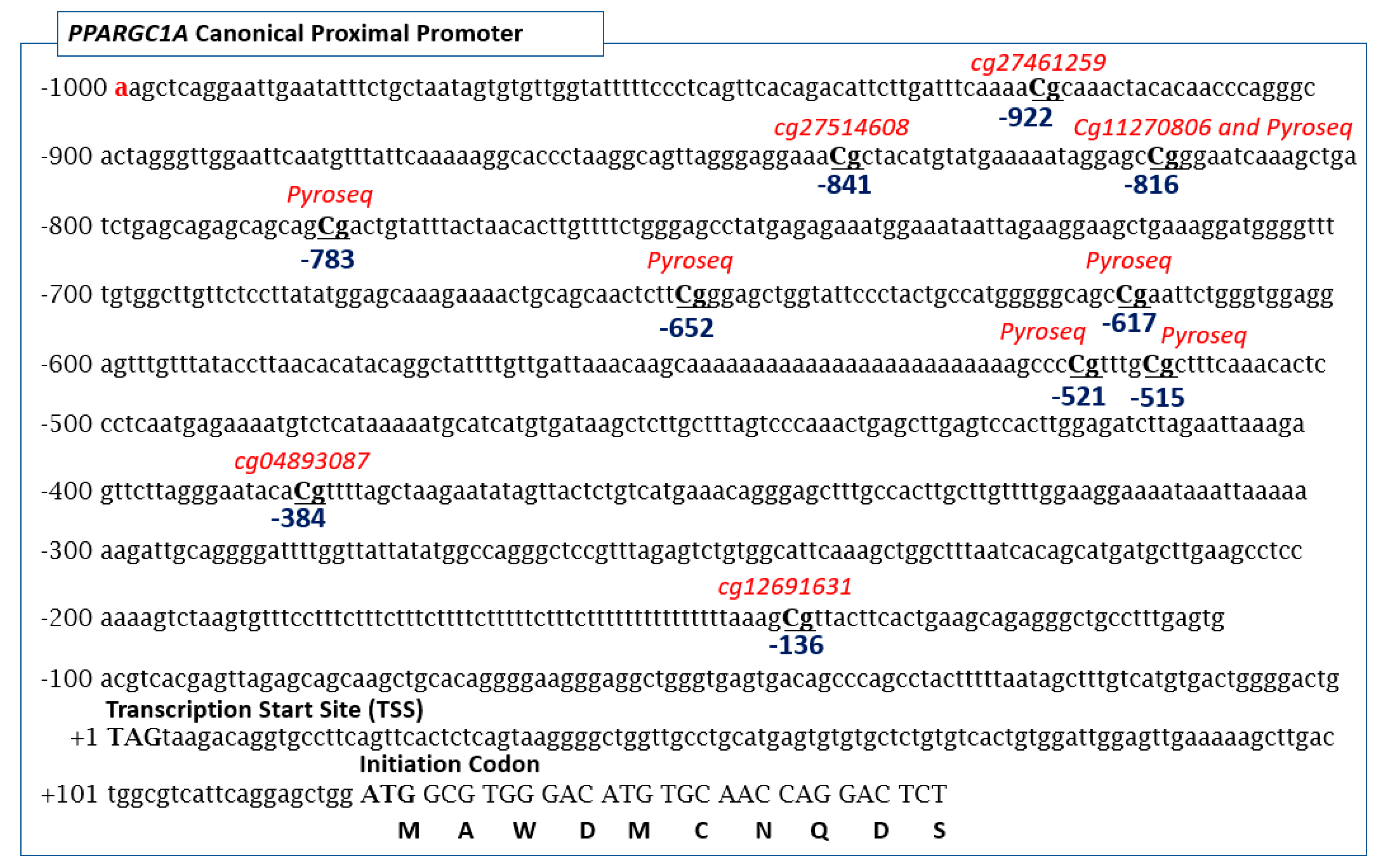
2.5. Pyrosequencing and Array-Based Methylation Analysis of the Promoter Region of PPARGC1A Gene
2.6. Determination of PPARGC1A Gly482Ser Genotypes
2.7. Statistical Methods
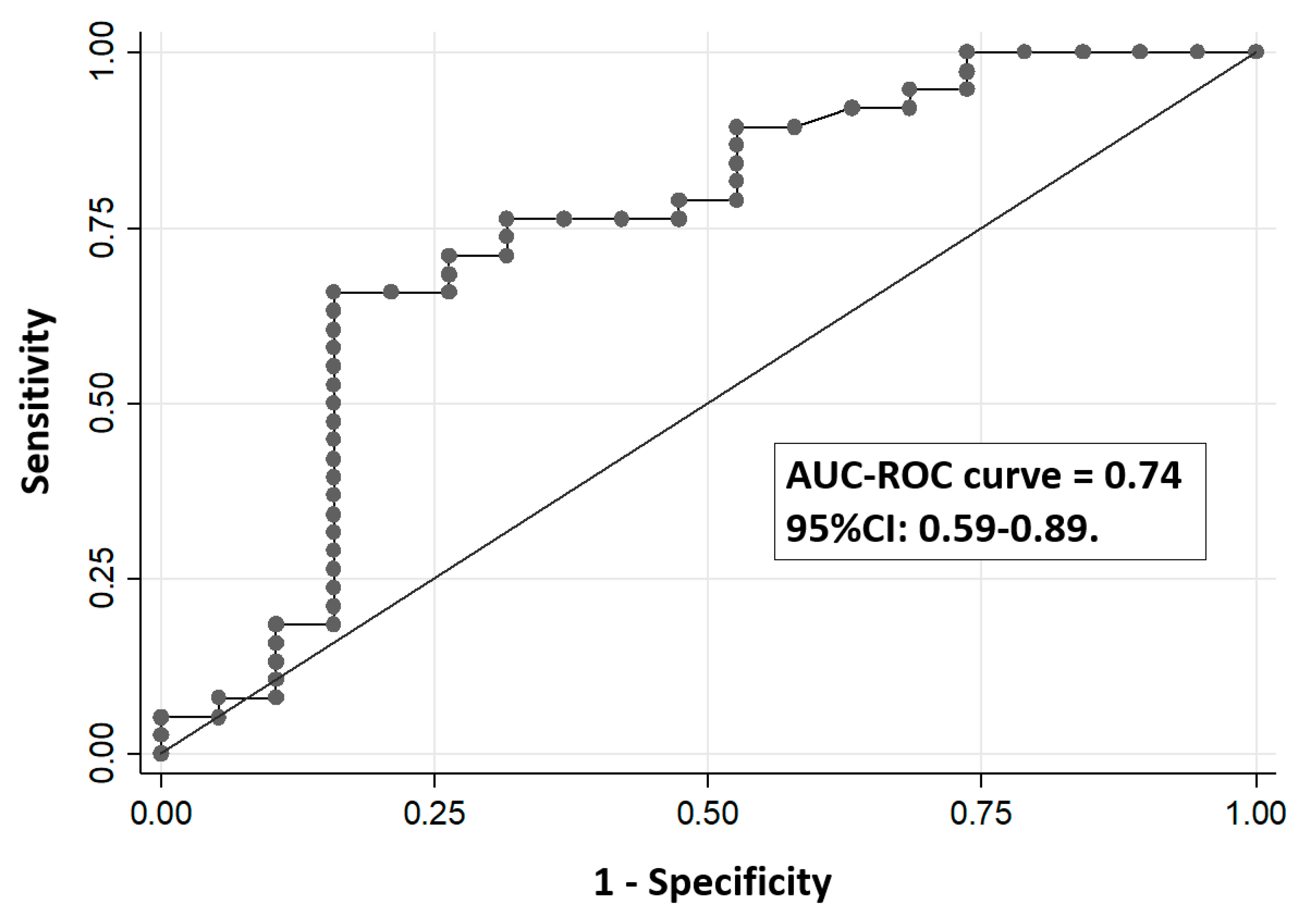
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Tondt, J.; Yancy, W.S.; Westman, E.C. Application of nutrient essentiality criteria to dietary carbohydrates. Nutr. Res. Rev. 2020, 27, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cust, A.E.; Skilton, M.R.; van Bakel, M.M.E.; Halkjaer, J.; Olsen, A.; Agnoli, C.; Psaltopoulou, T.; Buurma, E.; Sonestedt, E.; Chirlaque, M.D.; et al. Total dietary carbohydrate, sugar, starch and fibre intakes in the European prospective investigation into cancer and nutrition. Eur. J. Clin. Nutr. 2009, 63, S37–S60. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, A.; Bingham, S.A.; Cummings, J.H. Starch intake and colorectal cancer risk: An international comparison. Br. J. Cancer 1994, 69, 937–942. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A. From the triumvirate to the ominous octet: A new paradigm for the treatment of type 2 diabetes mellitus. Diabetes 2009, 58, 773–795. [Google Scholar] [CrossRef]
- Krentz, A.J.; Hompesch, M. Glucose: Archetypal biomarker in diabetes diagnosis, clinical management and research. Biomark. Med. 2016, 10, 1153–1166. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37, S81–S90. [Google Scholar] [CrossRef]
- Lorenzo, C.; Wagenknecht, L.E.; D’Agostino, R.B., Jr.; Rewers, M.J.; Karter, A.J.; Haffner, S.M. Insulin resistance, β-cell dysfunction, and conversion to type 2 diabetes in a multiethnic population: The Insulin Resistance Atherosclerosis Study. Diabetes Care 2010, 33, 67–72. [Google Scholar] [CrossRef]
- Utzschneider, K.M.; Prigeon, R.L.; Faulenbach, M.V.; Tong, J.; Carr, D.B.; Boyko, E.J.; Leonetti, D.L.; McNeely, M.J.; Fujimoto, W.Y.; Kahn, S.E. Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care 2009, 32, 335–341. [Google Scholar] [CrossRef]
- Prasad, R.B.; Groop, L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes 2015, 6, 87–123. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Davegårdh, C.; García-Calzón, S.; Bacos, K.; Ling, C. DNA methylation in the pathogenesis of type 2 diabetes in humans. Mol. Metab. 2018, 14, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Ling, C. Epigenetic regulation of insulin action and secretion—Role in the pathogenesis of type 2 diabetes. J. Intern. Med. 2020, 288, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Cardona, A.; Day, F.R.; Perry, J.R.B.; Loh, M.; Chu, A.Y.; Lehne, B.; Paul, D.S.; Lotta, L.A.; Stewart, I.D.; Kerrison, N.D.; et al. Epigenome-wide association study of incident type 2 diabetes in a British population: EPIC-Norfolk study. Diabetes 2019, 68, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Deng, X.; Shi, Y.; Su, Y.; Wei, J.; Duan, H. PGC-1α, glucose metabolism and type 2 diabetes mellitus. J. Endocrinol. 2016, 229, R99–R115. [Google Scholar] [CrossRef] [PubMed]
- Rohas, L.M.; St-Pierre, J.; Uldry, M.; Jäger, S.; Handschin, C.; Spiegelman, B.M. A fundamental system of cellular energy homeostasis regulated by PGC-1α. Proc. Natl. Acad. Sci. USA 2007, 104, 7933–7938. [Google Scholar] [CrossRef] [PubMed]
- Rowe, G.C.; Arany, Z. Genetic models of PGC-1 and glucose metabolism and homeostasis. Rev. Endocr. Metab. Disord. 2014, 15, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Brøns, C.; Jacobsen, S.; Nilsson, E.; Rönn, T.; Jensen, C.B.; Storgaard, H.; Poulsen, P.; Groop, L.; Ling, C.; Astrup, A.; et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J. Clin. Endocrinol. Metab. 2010, 95, 3048–3056. [Google Scholar] [CrossRef]
- Barrès, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef]
- Jørgensen, S.W.; Brøns, C.; Bluck, L.; Hjort, L.; Færch, K.; Thankamony, A.; Gillberg, L.; Friedrichsen, M.; Dunger, D.B.; Vaag, A.A. Metabolic response to 36 hours of fasting in young men born small vs. appropriate for gestational age. Diabetologia 2015, 58, 178–187. [Google Scholar] [CrossRef]
- Herzig, S.; Long, F.; Jhala, U.S.; Hedrick, S.; Quinn, R.; Bauer, A.; Rudolph, D.; Schutz, G.; Yoon, C.; Puigserver, P.; et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001, 413, 179–183. [Google Scholar] [CrossRef]
- Estall, J.L.; Kahn, M.; Cooper, M.P.; Fisher, F.M.; Wu, M.K.; Laznik, D.; Qu, L.; Cohen, D.E.; Shulman, G.I.; Spiegelman, B.M. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1α expression. Diabetes 2009, 58, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.C.; Puigserver, P.; Chen, G.; Donovan, J.; Wu, Z.; Rhee, J.; Adelmant, G.; Stafford, J.; Kahn, C.R.; Granner, D.K.; et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001, 413, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Patti, M.E.; Butte, A.J.; Crunkhorn, S.; Cusi, K.; Berria, R.; Kashyap, S.; Miyazaki, Y.; Kohane, I.; Costello, M.; Saccone, R.; et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 2003, 100, 8466–8471. [Google Scholar] [CrossRef]
- Chen, M.; Macpherson, A.; Owens, J.; Wittert, G.; Heilbronn, L.K. Obesity alone or with type 2 diabetes is associated with tissue specific alterations in DNA methylation and gene expression of PPARGC1A and IGF2. J. Diabetes Res. Clin. Metab. 2012, 1, 16. [Google Scholar] [CrossRef][Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Michael, L.F.; Wu, Z.; Cheatham, R.B.; Puigserver, P.; Adelmant, G.; Lehman, J.J.; Kelly, D.P.; Spiegelman, B.M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 2001, 98, 3820–3825. [Google Scholar] [CrossRef]
- Lin, J.; Wu, H.; Tarr, P.T.; Zhang, C.-Y.; Wu, Z.; Boss, O.; Michael, L.F.; Puigserver, P.; Isotani, E.; Olson, E.N.; et al. Transcriptional Co-Activator PGC-1 Alpha Drives the Formation of Slow-Twitch Muscle Fibres. Nature 2002, 418, 797–801. [Google Scholar] [CrossRef]
- Oropeza, D.; Jouvet, N.; Bouyakdan, K.; Perron, G.; Ringuette, L.-J.; Philipson, L.H.; Kiss, R.S.; Poitout, V.; Alquier, T.; Estall, J.L. PGC-1 coactivators in β-cells regulate lipid metabolism and are essential for insulin secretion coupled to fatty acids. Mol. Metab. 2015, 4, 811–822. [Google Scholar] [CrossRef]
- Yoon, J.C.; Xu, G.; Deeney, J.T.; Yang, S.-N.; Rhee, J.; Puigserver, P.; Levens, A.R.; Yang, R.; Zhang, C.-Y.; Lowell, B.B. Suppression of β cell energy metabolism and insulin release by PGC-1α. Dev. Cell 2003, 5, 73–83. [Google Scholar] [CrossRef]
- Cataldo, L.R.; Mizgier, M.L.; Bravo-Sagua, R.; Jaña, F.; Cárdenas, C.; Llanos, P.; Busso, D.; Olmos, P.; Galgani, J.E.; Santos, J.L. Prolonged activation of the Htr2b serotonin receptor impairs glucose stimulated insulin secretion and mitochondrial function in MIN6 cells. PLoS ONE 2017, 12, e0170213. [Google Scholar] [CrossRef]
- Ling, C.; Del Guerra, S.; Lupi, R.; Rönn, T.; Granhall, C.; Luthman, H.; Masiello, P.; Marchetti, P.; Groop, L.; Del Prato, S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 2008, 51, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Vandenbeek, R.; Khan, N.P.; Estall, J.L. Linking metabolic disease with the PGC-1α Gly482Ser polymorphism. Endocrinology 2018, 159, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Pihlajamäki, J.; Kinnunen, M.; Ruotsalainen, E.; Salmenniemi, U.; Vauhkonen, I.; Kuulasmaa, T.; Kainulainen, S.; Laakso, M. Haplotypes of PPARGC1A are associated with glucose tolerance, body mass index and insulin sensitivity in offspring of patients with type 2 diabetes. Diabetologia 2005, 48, 1331–1334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ek, J.; Andersen, G.; Urhammer, S.A.; Gaede, P.H.; Drivsholm, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type ii diabetes mellitus. Diabetologia 2001, 44, 2220–2226. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Tobe, K.; Okada, T.; Kadowaki, H.; Akanuma, Y.; Ito, C.; Kimura, S.; Kadowaki, T. A genetic variation in the PGC-1 gene could confer insulin resistance and susceptibility to type II diabetes. Diabetologia 2002, 45, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Andrulionytè, L.; Zacharova, J.; Chiasson, J.-L.; Laakso, M. STOP-NIDDM Study Group. Common polymorphisms of the PPAR-γ2 (Pro12Ala) and PGC-1α (Gly482Ser) genes are associated with the conversion from impaired glucose tolerance to type 2 diabetes in the STOP-NIDDM trial. Diabetologia 2004, 47, 2176–2184. [Google Scholar] [CrossRef]
- Clarke-Harris, R.; Wilkin, T.J.; Hosking, J.; Pinkney, J.; Jeffery, A.N.; Metcalf, B.S.; Godfrey, K.M.; Voss, L.D.; Lillycrop, K.A.; Burdge, G.C. PGC1α promoter methylation in blood at 5–7 years predicts adiposity from 9 to 14 Years (EarlyBird 50). Diabetes 2014, 63, 2528–2537. [Google Scholar] [CrossRef]
- Gillberg, L.; Jacobsen, S.C.; Ribel-Madsen, R.; Gjesing, A.P.; Boesgaard, T.W.; Ling, C.; Pedersen, O.; Hansen, T.; Vaag, A. Does DNA methylation of PPARGC1A influence insulin action in first degree relatives of patients with type 2 diabetes? PLoS ONE 2013, 8, e58384. [Google Scholar] [CrossRef]
- Kelstrup, L.; Hjort, L.; Houshmand-Oeregaard, A.; Clausen, T.D.; Hansen, N.S.; Broholm, C.; Borch-Johnsen, L.; Mathiesen, E.R.; Vaag, A.A.; Damm, P. Gene expression and DNA methylation of PPARGC1A in muscle and adipose tissue from adult offspring of women with diabetes in pregnancy. Diabetes 2016, 65, 2900–2910. [Google Scholar] [CrossRef]
- Ribel-Madsen, R.; Fraga, M.F.; Jacobsen, S.; Bork-Jensen, J.; Lara, E.; Calvanese, V.; Fernandez, A.F.; Friedrichsen, M.; Vind, B.F.; Højlund, K. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS ONE 2012, 7, e51302. [Google Scholar] [CrossRef]
- Arner, P.; Sahlqvist, A.-S.; Sinha, I.; Xu, H.; Yao, X.; Waterworth, D.; Rajpal, D.; Loomis, A.K.; Freudenberg, J.M.; Johnson, T. The Epigenetic signature of systemic insulin resistance in obese women. Diabetologia 2016, 59, 2393–2405. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.L.; Yévenes, I.; Cataldo, L.R.; Morales, M.; Galgani, J.; Arancibia, C.; Vega, J.; Olmos, P.; Flores, M.; Valderas, J.P.; et al. Development and assessment of the disposition index based on the oral glucose tolerance test in subjects with different glycaemic status. J. Physiol. Biochem. 2016, 72, 121–131. [Google Scholar] [CrossRef]
- Matsuda, M.; DeFronzo, R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: Comparison with the euglycemic insulin clamp. Diabetes Care 1999, 22, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Kang, S.J.; Lee, M.K.; Park, S.E.; Rhee, E.J.; Park, C.Y.; Oh, K.W.; Park, S.W.; Lee, W.Y. C-peptide-based index is more related to incident type 2 diabetes in non-diabetic subjects than insulin-based index. Endocrinol. Metab. (Seoul) 2016, 31, 320–327. [Google Scholar] [CrossRef]
- Tura, A.; Sbrignadello, S.; Succurro, E.; Groop, L.; Sesti, G.; Pacini, G. An empirical index of insulin sensitivity from short IVGTT: Validation against the minimal model and glucose clamp indices in patients with different clinical characteristics. Diabetologia 2010, 53, 144–152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marcelli-Tourvieille, S.; Hubert, T.; Pattou, F.; Vantyghem, M.C. Acute insulin response (AIR): Review of protocols and clinical interest in islet transplantation. Diabetes Metab. 2006, 32, 295–303. [Google Scholar] [CrossRef]
- Arpón, A.; Milagro, F.I.; Ramos-Lopez, O.; Mansego, M.L.; Santos, J.L.; Riezu-Boj, J.-I.; Martínez, J.A. Epigenome-wide association study in peripheral white blood cells involving insulin resistance. Sci. Rep. 2019, 9, 2445. [Google Scholar] [CrossRef]
- Arpón, A.; Santos, J.L.; Milagro, F.I.; Cataldo, L.R.; Bravo, C.; Riezu-Boj, J.-I.; Martínez, J.A. Insulin sensitivity is associated with lipoprotein lipase (LPL) and Catenin Delta 2 (CTNND2) DNA methylation in peripheral white blood cells in non-diabetic young women. Int. J. Mol. Sci. 2019, 20, 2928. [Google Scholar] [CrossRef]
- Martínez-Redondo, V.; Pettersson, A.T.; Ruas, J.L. The Hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia 2015, 58, 1969–1977. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Goni, L.; Cuervo, M.; Martinez, J.A. Association of the Gly482Ser PPARGC1A gene variant with different cholesterol outcomes in response to two energy-restricted diets in subjects with excessive weight. Nutrition 2018, 47, 83–89. [Google Scholar] [CrossRef]
- Koch, A.; Jeschke, J.; Van Criekinge, W.; van Engeland, M.; De Meyer, T. MEXPRESS update 2019. Nucleic Acids Res. 2019, 47, W561–W565. [Google Scholar] [CrossRef] [PubMed]
- Cuzmar, V.; Alberti, G.; Uauy, R.; Pereira, A.; García, C.; De Barbieri, F.; Corvalán, C.; Santos, J.L.; Mericq, V.; Villarroel, L.; et al. Early obesity: Risk factor for fatty liver disease. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, B.T.; Gudnason, H.; Aspelund, T.; Harris, T.B.; Launer, L.J.; Eiriksdottir, G.; Smith, A.V.; Gudnason, V. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS ONE 2012, 7, e46705. [Google Scholar] [CrossRef] [PubMed]
- Muller, Y.L.; Bogardus, C.; Pedersen, O.; Baier, L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor γ coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes 2003, 52, 895–898. [Google Scholar] [CrossRef]
- Lacquemant, C.; Chikri, M.; Boutin, P.; Samson, C.; Froguel, P. No association between the G482S polymorphism of the proliferator-activated receptor-γ coactivator-1 (PGC-1) gene and type ii diabetes in French caucasians. Diabetologia 2002, 45, 602–603. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, W.; Li, X.; Tang, Y.; Xie, P.; Ji, Y.; Fan, L.; Chen, Q. Association between PPARGC1A gene polymorphisms and coronary artery disease in a Chinese population. Clin. Exp. Pharmacol. Physiol. 2008, 35, 1172–1177. [Google Scholar] [CrossRef]
- Kunej, T.; Globocnik Petrovic, M.; Dovc, P.; Peterlin, B.; Petrovic, D. A Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1) gene is associated with type 2 diabetes in caucasians. Folia Biol. (Praha) 2004, 50, 157–158. [Google Scholar]
- Jemaa, Z.; Kallel, A.; Sleimi, C.; Mahjoubi, I.; Feki, M.; Ftouhi, B.; Slimane, H.; Jemaa, R.; Kaabachi, N. The Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) is associated with type 2 diabetes in Tunisian population. Diabetes Metab. Syndr. 2015, 9, 316–319. [Google Scholar] [CrossRef]
- Mirzaei, K.; Hossein-nezhad, A.; Emamgholipour, S.; Ansar, H.; Khosrofar, M.; Tootee, A.; Alatab, S. An exonic peroxisome proliferator-activated receptor-γ coactivator-1α variation may mediate the resting energy expenditure through a potential regulatory role on important gene expression in this pathway. J. Nutrigenet. Nutrigenom. 2012, 5, 59–71. [Google Scholar] [CrossRef]
- Franks, P.W.; Christophi, C.A.; Jablonski, K.A.; Billings, L.K.; Delahanty, L.M.; Horton, E.S.; Knowler, W.C.; Florez, J.C.; Diabetes Prevention Program Research Group. Common variation at ppargc1a/b and change in body composition and metabolic traits following preventive interventions: The diabetes prevention program. Diabetologia 2014, 57, 485–490. [Google Scholar] [CrossRef]
| Mean ± Standard Deviation | |
|---|---|
| Age (years) | 27.4 ± 6.9 |
| Weight (kg) | 60.1 ± 9.3 |
| Height (m) | 1.59 ± 0.07 |
| BMI (kg/m2) | 23.8 ± 3.5 |
| HOMA-S index | 77.9 ± 44.0 |
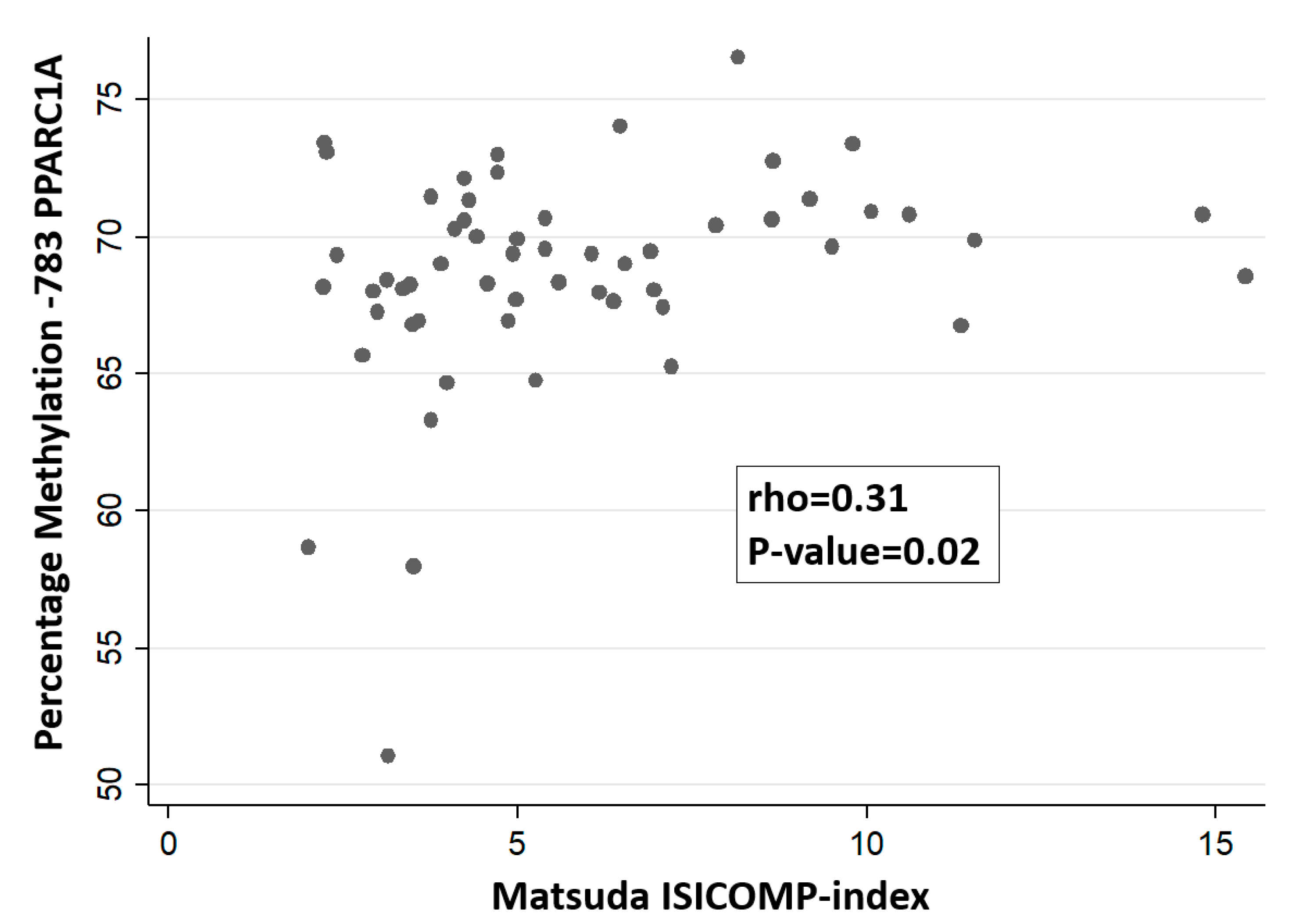
| OGTT-based Matsuda-ISICOMP index | 5.7 ± 3.0 |
| OGTT-based insulinogenic index | 0.43 ± 0.22 |
| OGTT-based C-peptidogenic index | 0.31 ± 1.34 |
| OGTT-based Disposition Index (ODI using C-peptide) | 0.20 ± 0.12 |
| OGTT-based Disposition Index (ODI using insulin) | 3.06 ± 1.14 |
| IVGTT-based CSi Index | 6.46 ± 4.01 |
| IVGTT-based AIR Index | 59.7 ± 33.62 |
| IVGTT-based Disposition Index (DI) | 337.3 ± 202.4 |
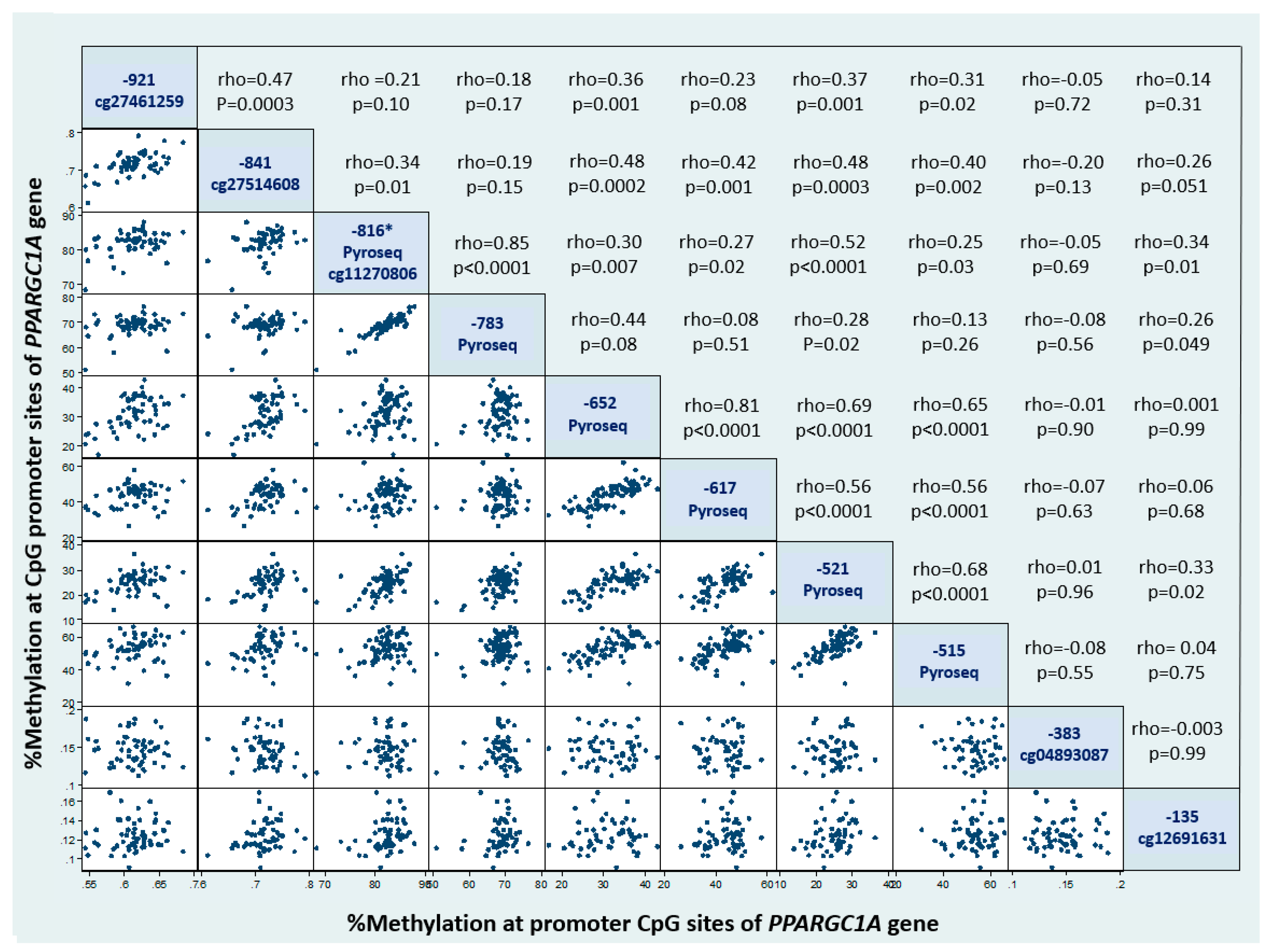
| % methylation (position -922) (Illumina 450K array) | 61.5 ± 3 |
| % methylation (position -841) (Illumina 450K array) | 71.7 ± 3 |
| % methylation (position -816) (Pyrosequencing) | 82 ± 3.1 |
| % methylation (position -783) (Pyrosequencing) | 68.7 ± 3.6 |
| % methylation (position -652) (Pyrosequencing) | 31.5 ± 5.6 |
| % methylation (position -617) (Pyrosequencing) | 43.3 ± 6.6 |
| % methylation (position -521) (Pyrosequencing) | 24.8 ± 4.3 |
| % methylation (position -515) (Pyrosequencing) | 53.1 ± 7.0 |
| % methylation (position -383) (Illumina 450K array) | 14.7 ± 0.02 |
| % methylation (position -136) (Illumina 450K array) | 12.4 ± 0.02 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, J.L.; Krause, B.J.; Cataldo, L.R.; Vega, J.; Salas-Pérez, F.; Mennickent, P.; Gallegos, R.; Milagro, F.I.; Prieto-Hontoria, P.; Riezu-Boj, J.I.; et al. PPARGC1A Gene Promoter Methylation as a Biomarker of Insulin Secretion and Sensitivity in Response to Glucose Challenges. Nutrients 2020, 12, 2790. https://doi.org/10.3390/nu12092790
Santos JL, Krause BJ, Cataldo LR, Vega J, Salas-Pérez F, Mennickent P, Gallegos R, Milagro FI, Prieto-Hontoria P, Riezu-Boj JI, et al. PPARGC1A Gene Promoter Methylation as a Biomarker of Insulin Secretion and Sensitivity in Response to Glucose Challenges. Nutrients. 2020; 12(9):2790. https://doi.org/10.3390/nu12092790
Chicago/Turabian StyleSantos, José L., Bernardo J. Krause, Luis Rodrigo Cataldo, Javier Vega, Francisca Salas-Pérez, Paula Mennickent, Raúl Gallegos, Fermín I. Milagro, Pedro Prieto-Hontoria, J. Ignacio Riezu-Boj, and et al. 2020. "PPARGC1A Gene Promoter Methylation as a Biomarker of Insulin Secretion and Sensitivity in Response to Glucose Challenges" Nutrients 12, no. 9: 2790. https://doi.org/10.3390/nu12092790
APA StyleSantos, J. L., Krause, B. J., Cataldo, L. R., Vega, J., Salas-Pérez, F., Mennickent, P., Gallegos, R., Milagro, F. I., Prieto-Hontoria, P., Riezu-Boj, J. I., Bravo, C., Salas-Huetos, A., Arpón, A., Galgani, J. E., & Martínez, J. A. (2020). PPARGC1A Gene Promoter Methylation as a Biomarker of Insulin Secretion and Sensitivity in Response to Glucose Challenges. Nutrients, 12(9), 2790. https://doi.org/10.3390/nu12092790







