Identification of Genetic Factors Underlying the Association between Sodium Intake Habits and Hypertension Risk
Abstract
1. Introduction
2. Materials and Methods
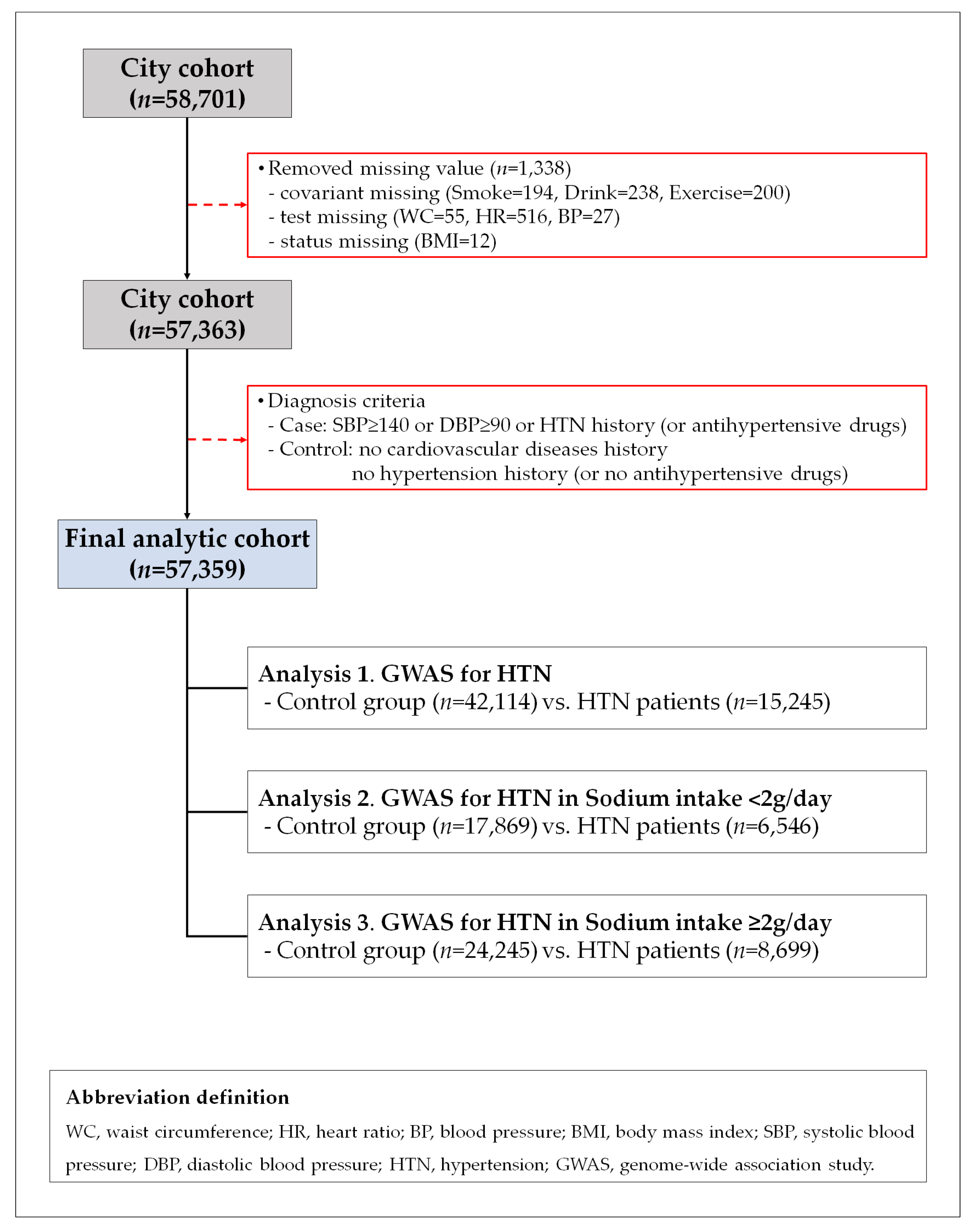
2.1. Study Population
2.2. Assessment of Dietary Sodium and Potassium Intake and Covariates
2.3. Genotyping
2.4. Statistical Analysis
3. Results
3.1. General Characteristics of the Study Population
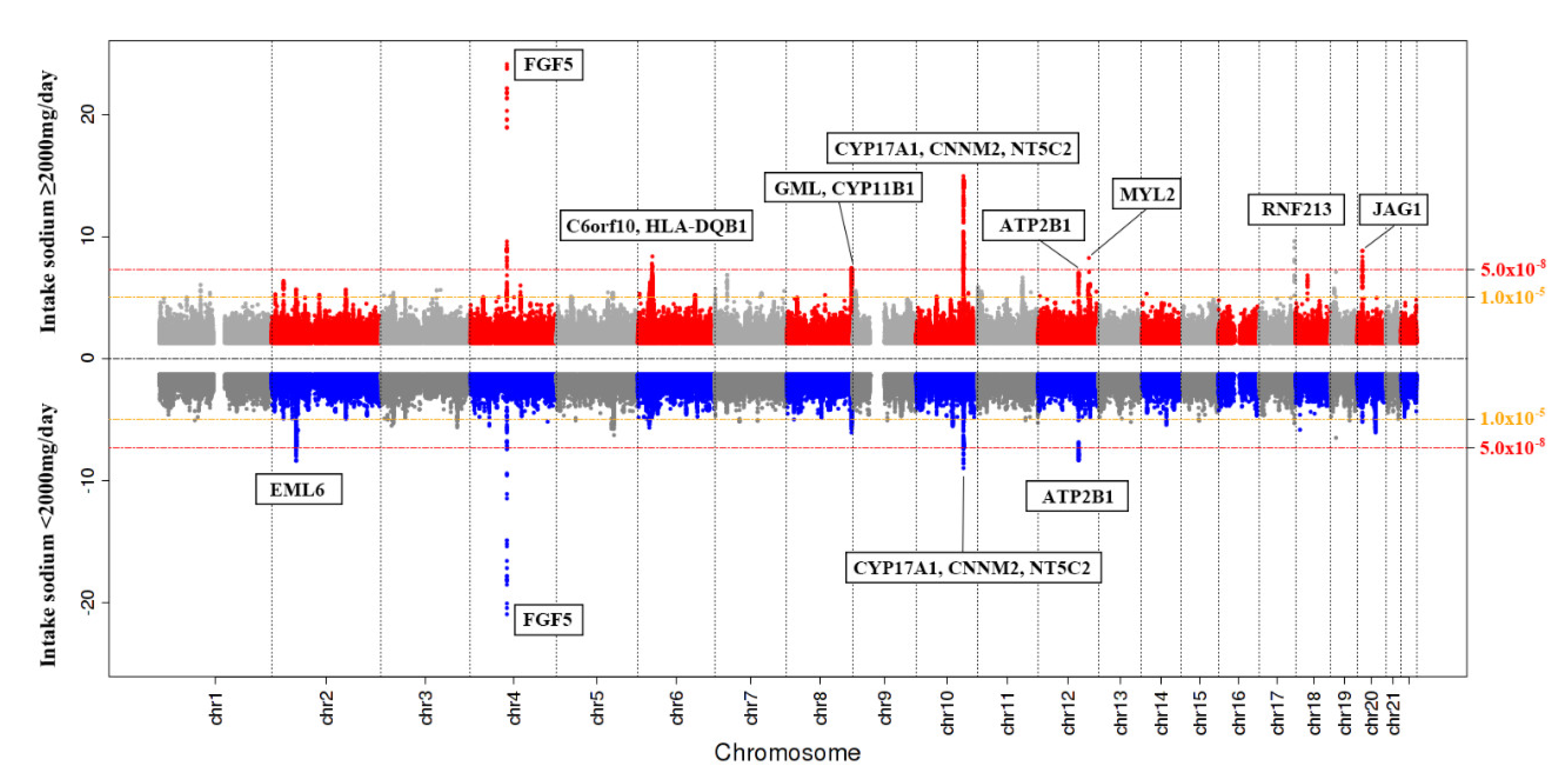
3.2. SNPs Associated with Hypertension Based on Sodium Intake
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shrimanker, I.; Bhattarai, S. Electrolytes. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- GBD 2017 Risk Factor Collaborators. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1923–1994. [Google Scholar]
- Strazzullo, P.; D’Elia, L.; Kandala, N.B.; Cappuccio, F.P. Salt intake, stroke, and cardiovascular disease:meta-analysis of prospective studies. BMJ 2009, 339, b4567. [Google Scholar] [CrossRef]
- He, F.J.; Li, J.; Macgregor, G.A. Effect of longer term modest salt reduction on blood pressure: Cochrane systematic review and meta-analysis of randomised trials. BMJ 2013, 346, f1325. [Google Scholar] [CrossRef] [PubMed]
- Khaw, K.T.; Bingham, S.; Welch, A.; Luben, R.; O’Brien, E.; Wareham, N.; Day, N. Blood pressure and urinary sodium in men and women: The Norfolk Cohort of the European Prospective Investigation into Cancer (EPIC-Norfolk). Am. J. Clin. Nutr. 2004, 80, 1397–1403. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guideline: Sodium Intake for Adults and Children; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Ivy, J.R.; Bailey, M.A. Pressure natriuresis and the renal control of arterial blood pressure. J. Physiol. 2014, 592, 3955–3967. [Google Scholar] [CrossRef]
- Suzuki, H.; Kimmel, P.L. Nutrition and Kidney Disease: A New Era; Karger Medical and Scientific Publishers: Basel, Switzerland, 2007; Volume 155. [Google Scholar]
- Huang, L.; Chu, Y.; Huang, X.; Ma, S.; Lin, K.; Huang, K.; Sun, H.; Yang, Z. Association between gene polymorphisms of voltage-dependent Ca2+ channels and hypertension in the dai people of china: A case-control study. BMC Med. Genet. 2020, 21, 44. [Google Scholar] [CrossRef]
- Mervaala, E.M.; Himberg, J.J.; Laakso, J.; Tuomainen, P.; Karppanen, H. Beneficial effects of a potassium- and magnesium-enriched salt alternative. Hypertension 1992, 19, 535–540. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kathryn, L.M. Pathophysiology: The Biologic Basis for Disease in Adults and Children, 7th ed.; Elsevier, Mosby: St. Louis, MO, USA, 2014. [Google Scholar]
- Asferg, C.L.; Nielsen, S.J.; Andersen, U.B.; Linneberg, A.; Møller, D.V.; Hedley, P.L.; Christiansen, M.; Goetze, J.P.; Esler, M.; Jeppesen, J.L. Relative atrial natriuretic peptide deficiency and inadequate renin and angiotensin ii suppression in obese hypertensive men. Hypertension 2013, 62, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Stolarz-Skrzypek, K.; Kuznetsova, T.; Thijs, L.; Tikhonoff, V.; Seidlerová, J.; Richart, T.; Jin, Y.; Olszanecka, A.; Malyutina, S.; Casiglia, E.; et al. Fatal and nonfatal outcomes, incidence of hypertension, and blood pressure changes in relation to urinary sodium excretion. JAMA 2011, 305, 1777–1785. [Google Scholar] [CrossRef]
- O’Donnell, M.J.; Yusuf, S.; Mente, A.; Gao, P.; Mann, J.F.; Teo, K.; McQueen, M.; Sleight, P.; Sharma, A.M.; Dans, A.; et al. Urinary sodium and potassium excretion and risk of cardiovascular events. JAMA 2011, 306, 2229–2238. [Google Scholar] [CrossRef]
- O’Donnell, M.; Mente, A.; Rangarajan, S.; McQueen, M.J.; Wang, X.; Liu, L.; Yan, H.; Lee, S.F.; Mony, P.; Devanath, A.; et al. Urinary sodium and potassium excretion, mortality, and cardiovascular events. N. Engl. J. Med. 2014, 371, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Carretero, O.A.; Oparil, S. Essential hypertension. Part I: Definition and etiology. Circulation 2000, 101, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Doris, P.A. The genetics of blood pressure and hypertension: The role of rare variation. Cardiovasc. Ther. 2011, 29, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; DeStefano, A.L.; Larson, M.G.; O’Donnell, C.J.; Lifton, R.P.; Gavras, H.; Cupples, L.A.; Myers, R.H. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension 2000, 36, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.G.; Fan, J.B.; Siao, C.J.; Berno, A.; Young, P.; Sapolsky, R.; Ghandour, G.; Perkins, N.; Winchester, E.; Spencer, J.; et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 1998, 280, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Stover, P.J. Influence of human genetic variation on nutritional requirements. Am. J. Clin. Nutr. 2006, 83, 436S–442S. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, 1350. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, K.; Kim, H.C. Dietary assessment methods in epidemiologic studies. Epidemiol. Health 2014, 36, e2014009. [Google Scholar] [CrossRef]
- Moon, S.; Kim, Y.J.; Han, S.; Hwang, M.Y.; Shin, D.M.; Park, M.Y.; Lu, Y.; Yoon, K.; Jang, H.M.; Kim, Y.K.; et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Sci. Rep. 2019, 9, 1382. [Google Scholar] [CrossRef]
- Guyton, A.C. Blood pressure control—Special role of the kidneys and body fluids. Science 1991, 252, 1813–1816. [Google Scholar] [CrossRef]
- Gug, C.; Mihaescu, A.; Mozos, I. Two mutations in the thiazide-sensitive nacl co-transporter gene in a romanian gitelman syndrome patient: Case report. Ther. Clin. Risk Manag. 2018, 14, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.; Fraser, S.A.; Galic, S.; Choy, S.W.; Katerelos, M.; Gleich, K.; Kemp, B.E.; Mount, P.F.; Power, D.A. Novel mechanisms of na+ retention in obesity: Phosphorylation of nkcc2 and regulation of spak/osr1 by ampk. Am. J. Physiol. Renal Physiol. 2014, 307, F96–F106. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, M.H.; Miller, J.Z.; Luft, F.C.; Grim, C.E.; Fineberg, N.S. Definitions and characteristics of sodium sensitivity and blood pressure resistance. Hypertension 1986, 8, II127. [Google Scholar] [CrossRef] [PubMed]
- Elijovich, F.; Weinberger, M.H.; Anderson, C.A.; Appel, L.J.; Bursztyn, M.; Cook, N.R.; Dart, R.A.; Newton-Cheh, C.H.; Sacks, F.M.; Laffer, C.L. Salt sensitivity of blood pressure: A scientific statement from the american heart association. Hypertension 2016, 68, e7–e46. [Google Scholar] [CrossRef] [PubMed]
- Flack, J.M.; Ensrud, K.E.; Mascioli, S.; Launer, C.A.; Svendsen, K.; Elmer, P.J.; Grimm, R.H., Jr. Racial and ethnic modifiers of the salt-blood pressure response. Hypertension 1991, 17, I115–I121. [Google Scholar] [CrossRef]
- Weinberger, M.H. Salt sensitivity of blood pressure in humans. Hypertension 1996, 27, 481–490. [Google Scholar] [CrossRef]
- Newton-Cheh, C.; Johnson, T.; Gateva, V.; Tobin, M.D.; Bochud, M.; Coin, L.; Najjar, S.S.; Zhao, J.H.; Heath, S.C.; Eyheramendy, S.; et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009, 41, 666–676. [Google Scholar] [CrossRef]
- Vatner, S.F. FGF induces hypertrophy and angiogenesis in hibernating myocardium. Circ. Res. 2005, 96, 705–707. [Google Scholar] [CrossRef]
- Xi, B.; Shen, Y.; Reilly, K.H.; Wang, X.; Mi, J. Recapitulation of four hypertension susceptibility genes (CSK, CYP17A1, MTHFR, and FGF5) in East Asians. Metabolism 2013, 62, 196–203. [Google Scholar] [CrossRef]
- Rhee, M.Y.; Yang, S.J.; Oh, S.W.; Park, Y.; Kim, C.I.; Park, H.K.; Park, S.W.; Park, C.Y. Novel genetic variations associated with salt sensitivity in the Korean population. Hypertens. Res. 2011, 34, 606–611. [Google Scholar] [CrossRef]
- Cheema, A.N.; Rosenthal, S.L.; Ilyas Kamboh, M. Proficiency of data interpretation: Identification of signaling SNPs/specific loci for coronary artery disease. Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Bragina, E.Y.; Goncharova, I.A.; Garaeva, A.F.; Nemerov, E.V.; Babovskaya, A.A.; Karpov, A.B.; Semenova, Y.V.; Zhalsanova, I.Z.; Gomboeva, D.E.; Saik, O.V. Molecular relationships between bronchial asthma and hypertension as comorbid diseases. J. Integr. Bioinform. 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- Tabara, Y.; Kohara, K.; Kita, Y.; Hirawa, N.; Katsuya, T.; Ohkubo, T.; Hiura, Y.; Tajima, A.; Morisaki, T.; Miyata, T.; et al. Common variants in the ATP2B1 gene are associated with susceptibility to hypertension: The Japanese Millennium Genome Project. Hypertension 2010, 56, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Hirawa, N.; Tabara, Y.; Muraoka, H.; Fujita, M.; Miyazaki, N.; Fujiwara, A.; Ichikawa, Y.; Yamamoto, Y.; Ichihara, N.; et al. Mice lacking hypertension candidate gene ATP2B1 in vascular smooth muscle cells show significant blood pressure elevation. Hypertension 2012, 59, 854–860. [Google Scholar] [CrossRef]
- Giri, A.; Hellwege, J.N.; Keaton, J.M.; Park, J.; Qiu, C.; Warren, H.R.; Torstenson, E.S.; Kovesdy, C.P.; Sun, Y.V.; Wilson, O.D.; et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat. Genet. 2019, 51, 51–62. [Google Scholar] [CrossRef]
- Takeuchi, F.; Akiyama, M.; Matoba, N.; Katsuya, T.; Nakatochi, M.; Tabara, Y.; Narita, A.; Saw, W.Y.; Moon, S.; Spracklen, C.N.; et al. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Zhu, F.; Sun, Y.; Wang, M.; Ma, S.; Chen, X.; Cao, A.; Chen, F.; Qiu, Y.; Liao, Y. Correlation between HLA-DRB1, HLA-DQB1 polymorphism and autoantibodies against angiotensin AT(1) receptors in Chinese patients with essential hypertension. Clin. Cardiol. 2011, 34, 302–308. [Google Scholar] [CrossRef]
- Liu, W.; Morito, D.; Takashima, S.; Mineharu, Y.; Kobayashi, H.; Hitomi, T.; Hashikata, H.; Matsuura, N.; Yamazaki, S.; Toyoda, A.; et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 2011, 6, e22542. [Google Scholar] [CrossRef]
- Park, M.G.; Shin, J.H.; Lee, S.W.; Park, H.R.; Park, K.P. RNF213 rs112735431 polymorphism in intracranial artery steno-occlusive disease and moyamoya disease in Koreans. J. Neurol. Sci. 2017, 375, 331–334. [Google Scholar] [CrossRef]
- Guey, S.; Kraemer, M.; Hervé, D.; Ludwig, T.; Kossorotoff, M.; Bergametti, F.; Schwitalla, J.C.; Choi, S.; Broseus, L.; Callebaut, I.; et al. Rare rnf213 variants in the c-terminal region encompassing the ring-finger domain are associated with moyamoya angiopathy in caucasians. Eur. J. Hum. Genet. 2017, 25, 995–1003. [Google Scholar] [CrossRef]
- Liu, W.; Hitomi, T.; Kobayashi, H.; Harada, K.H.; Koizumi, A. Distribution of moyamoya disease susceptibility polymorphism p.R4810k in rnf213 in east and southeast asian populations. Neurol Med. Chir. 2012, 52, 299–303. [Google Scholar] [CrossRef]
- Fujimura, M.; Sonobe, S.; Nishijima, Y.; Niizuma, K.; Sakata, H.; Kure, S.; Tominaga, T. Genetics and Biomarkers of Moyamoya Disease: Significance of RNF213 as a Susceptibility Gene. J. Stroke 2014, 16, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, K.; Sakai, Y.; Inoue, H.; Akamine, S.; Ishizaki, Y.; Matsushita, Y.; Sanefuji, M.; Torisu, H.; Ihara, K.; Sardiello, M.; et al. Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells. Sci. Rep. 2015, 5, 13191. [Google Scholar] [CrossRef] [PubMed]
- Toda, N.; Arakawa, K. Salt-induced hemodynamic regulation mediated by nitric oxide. J. Hypertens. 2011, 29, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, A.W.; Lewis, S.A.; McKeever, T.M.; Britton, J.R. Is higher sodium intake associated with elevated systemic inflammation? A population-based study. Am. J. Clin. Nutr. 2009, 89, 1901–1904. [Google Scholar] [CrossRef]
- Koizumi, A.; Kobayashi, H.; Liu, W.; Fujii, Y.; Senevirathna, S.T.; Nanayakkara, S.; Okuda, H.; Hitomi, T.; Harada, K.H.; Takenaka, K.; et al. P.R4810K, a polymorphism of RNF213, the susceptibility gene for moyamoya disease, is associated with blood pressure. Environ. Health Prev. Med. 2013, 18, 121–129. [Google Scholar] [CrossRef]
- Kim, Y.J.; Go, M.J.; Hu, C.; Hong, C.B.; Kim, Y.K.; Lee, J.Y.; Hwang, J.Y.; Oh, J.H.; Kim, D.J.; Kim, N.H.; et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 2011, 43, 990–995. [Google Scholar] [CrossRef]
- Heo, S.G.; Hwang, J.Y.; Uhmn, S.; Go, M.J.; Oh, B.; Lee, J.Y.; Park, J.W. Male-specific genetic effect on hypertension and metabolic disorders. Hum. Genet. 2014, 133, 311–319. [Google Scholar] [CrossRef]
- Claes, G.R.; van Tienen, F.H.; Lindsey, P.; Krapels, I.P.; Helderman-van den Enden, A.T.; Hoos, M.B.; Barrois, Y.E.; Janssen, J.W.; Paulussen, A.D.; Sels, J.W.; et al. Hypertrophic remodelling in cardiac regulatory myosin light chain (MYL2) founder mutation carriers. Eur. Heart J. 2016, 37, 1815–1822. [Google Scholar] [CrossRef]
- Lu, X.; Huang, J.; Wang, L.; Chen, S.; Yang, X.; Li, J.; Cao, J.; Chen, J.; Li, Y.; Zhao, L.; et al. Genetic predisposition to higher blood pressure increases risk of incident hypertension and cardiovascular diseases in Chinese. Hypertension 2015, 66, 786–792. [Google Scholar] [CrossRef][Green Version]
- Lu, X.; Wang, L.; Lin, X.; Huang, J.; Charles Gu, C.; He, M.; Shen, H.; He, J.; Zhu, J.; Li, H.; et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum. Mol. Genet. 2015, 24, 865–874. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Sodium Intake | p1,† | p2,† | p3,† | Sodium Intake <2 g/Day | Sodium Intake ≥2 g/Day | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| <2 g/Day (n = 24,415) | ≥2 g/Day (n = 32,944) | Without HTN (n = 17,869) | With HTN (n = 6546) | p† | Without HTN (n = 24,245) | With HTN (n = 8699) | p† | ||||
| Age (years,) | 54.05 ± 7.90 | 53.58 ± 8.07 | <0.0001 | <0.0001 | 0.002 | 52.70 ± 7.70 | 57.73 ± 7.25 | <0.0001 | 52.23 ± 7.85 | 57.36 ± 7.44 | <0.0001 |
| Sex (male, %) | 7453 (30.5) | 12,348 (37.5) | <0.0001 ‡ | <0.0001 ‡ | <0.0001 ‡ | 4929 (27.6) | 2524 (38.6) | <0.0001 ‡ | 8264 (34.1) | 4084 (46.9) | <0.0001 ‡ |
| BMI (kg/m2) | 24.02 ± 46.63 | 24.20 ± 34.68 | 0.606 | 0.743 | <0.0001 | 23.29 ± 2.70 | 24.91 ± 2.92 | <0.0001 | 23.59 ± 2.74 | 25.17 ± 2.95 | <0.0001 |
| WC (cm,) | 80.08 ± 8.54 | 81.26 ± 8.65 | <0.0001 | <0.0001 | <0.0001 | 78.72 ± 8.23 | 83.78 ± 8.29 | <0.0001 | 79.93 ± 8.35 | 84.97 ± 8.40 | <0.0001 |
| SBP (mmHg) | 122.14 ± 14.80 | 122.57 ± 14.73 | 0.001 | <0.0001 | 0.042 | 117.57 ± 11.73 | 134.62 ± 15.12 | <0.0001 | 118.07 ± 11.65 | 135.12 ± 15.14 | <0.0001 |
| DBP (mmHg) | 75.44 ± 9.71 | 75.94 ± 9.72 | <0.0001 | <0.0001 | 0.001 | 72.89 ± 8.18 | 82.43 ± 10.11 | <0.0001 | 73.42 ± 8.21 | 82.96 ± 10.15 | <0.0001 |
| HR (bpm) | 69.30 ± 9.28 | 69.06 ± 9.00 | 0.002 | 0.012 | 0.09 | 68.94 ± 8.93 | 70.27 ± 10.11 | <0.0001 | 68.73 ± 8.70 | 70.00 ± 9.74 | <0.0001 |
| TC (mg/dL) | 197.19 ± 36.00 | 197.50 ± 35.45 | 0.307 | 0.424 | 0.558 | 197.88 ± 35.46 | 195.33 ± 37.35 | <0.0001 | 198.16 ± 35.11 | 195.68 ± 36.30 | <0.0001 |
| TG (mg/dL) | 122.06 ± 83.51 | 127.35 ± 87.03 | <0.0001 | <0.0001 | 0.0001 | 115.17 ± 77.32 | 140.86 ± 95.93 | <0.0001 | 120.52 ± 82.17 | 146.35 ± 96.79 | <0.0001 |
| HDL-C (mg/dL) | 54.23 ± 13.30 | 53.43 ± 13.04 | <0.0001 | <0.0001 | <0.0001 | 55.05 ± 13.34 | 52.00 ± 12.91 | <0.0001 | 54.23 ± 13.14 | 51.19 ± 12.49 | <0.0001 |
| LDL-C (mg/dL) | 119.14 ± 32.36 | 119.40 ± 31.98 | 0.344 | 0.531 | 0.52 | 120.28 ± 31.72 | 116.00 ± 33.89 | <0.0001 | 120.48 ± 31.46 | 116.36 ± 33.21 | <0.0001 |
| CRP (mg/dL) | 0.14 ± 0.45 | 0.14 ± 0.34 | 0.333 | 0.432 | 0.639 | 0.13 ± 0.46 | 0.17 ± 0.41 | <0.0001 | 0.13 ± 0.34 | 0.16 ± 0.34 | <0.0001 |
| Exercise status | |||||||||||
| Yes (%) | 13,103 (53.7) | 18,250 (55.4) | <0.0001 ‡ | <0.0001 ‡ | 0.441 | 9422 (52.7) | 3681 (56.2) | <0.0001 ‡ | 13,304 (54.9) | 4946 (56.9) | <0.0001 ‡ |
| No (%) | 11,312 (46.3) | 14,694 (44.6) | 8447 (47.3) | 2865 (43.8) | 10,941 (45.1) | 3753 (43.1) | |||||
| Smoking status | |||||||||||
| Non-smokers (%) | 18,638 (76.3) | 23,386 (71.0) | <0.0001 ‡ | <0.0001 ‡ | <0.0001 ‡ | 13,957 (78.1) | 4681 (71.5) | <0.0001 ‡ | 17,779 (73.3) | 5607 (64.5) | <0.0001 ‡ |
| Ex-smokers (%) | 3484 (14.3) | 5586 (17.0) | 2235 (12.5) | 1249 (19.1) | 3562 (14.7) | 2024 (23.3) | |||||
| Current smokers (%) | 2293 (9.4) | 3972 (12.1) | 1677 (9.4) | 616 (9.4) | 2904 (12.0) | 1068 (12.3) | |||||
| Drinking status | |||||||||||
| Non-drinker (%) | 13,411 (54.9) | 16,369 (49.7) | <0.0001 ‡ | <0.0001 ‡ | <0.0001 ‡ | 9880 (55.3) | 3531 (53.9) | <0.0001 ‡ | 12,333 (50.9) | 4036 (46.4) | <0.0001 ‡ |
| Ex-drinker (%) | 921 (3.8) | 1212 (3.7) | 605 (3.4) | 316 (4.8) | 767 (3.2) | 445 (5.1) | |||||
| Current drinker (%) | 10083 (41.3) | 15363 (46.6) | 7384 (41.3) | 2699 (41.2) | 11145 (46.0) | 4218 (48.5) | |||||
| Total intake energy (kcal/day) | 1498.32 ± 394.32 | 1923.57 ± 577.56 | <0.0001 | <0.0001 | <0.0001 | 1502.56 ± 397.52 | 1486.77 ± 385.27 | 0.0056 | 1938.06 ± 588.37 | 1883.05 ± 544.22 | <0.0001 |
| Sugar (g/day) | 275.09 ± 74.34 | 335.71 ± 92.28 | <0.0001 | <0.0001 | <0.0001 | 274.83 ± 74.99 | 275.80 ± 72.54 | 0.3667 | 337.21 ± 93.79 | 331.51 ± 87.81 | <0.0001 |
| Fat (g/day) | 21.03 ± 10.72 | 32.85 ± 19.94 | <0.0001 | <0.0001 | <0.0001 | 21.54 ± 10.94 | 19.62 ± 9.96 | <0.0001 | 33.57 ± 20.32 | 30.81 ± 18.71 | <0.0001 |
| Protein (g/day) | 46.51 ± 14.39 | 68.25 ± 27.58 | <0.0001 | <0.0001 | <0.0001 | 46.83 ± 14.50 | 45.64 ± 14.03 | <0.0001 | 68.90 ± 28.03 | 66.45 ± 26.22 | <0.0001 |
| Sugar ratio | 73.48 ± 6.45 | 70.41 ± 7.05 | <0.0001 | <0.0001 | <0.0001 | 12.53 ± 2.23 | 12.32 ± 2.12 | <0.0001 | 14.12 ± 2.55 | 14.02 ± 2.69 | <0.0001 |
| Fat ratio | 5.58 ± 2.28 | 6.60 ± 2.40 | <0.0001 | <0.0001 | <0.0001 | 5.71 ± 2.32 | 5.24 ± 2.12 | <0.0001 | 6.70 ± 2.39 | 6.32 ± 2.40 | <0.0001 |
| Protein ratio | 12.47 ± 2.20 | 14.09 ± 2.59 | <0.0001 | <0.0001 | <0.0001 | 73.19 ± 6.57 | 74.28 ± 6.05 | <0.0001 | 70.20 ± 7.01 | 71.01 ± 7.12 | <0.0001 |
| Na (mg/day) | 1323.39 ± 435.98 | 3254.94 ± 1251.19 | <0.0001 | <0.0001 | <0.0001 | 1328.30 ± 433.08 | 1310.05 ± 443.58 | 0.0038 | 3258.11 ± 1258.90 | 3245.85 ± 1229.30 | 0.4329 |
| K (mg/day) | 1591.80 ± 561.36 | 2694.00 ± 1046.87 | <0.0001 | <0.0001 | <0.0001 | 1607.78 ± 564.81 | 1548.20 ± 549.57 | <0.0001 | 2719.81 ± 1069.77 | 2621.77 ± 976.41 | <0.0001 |
| Na/K ratio | 0.87 ± 0.28 | 1.26 ± 0.34 | <0.0001 | <0.0001 | <0.0001 | 0.86 ± 0.28 | 0.88 ± 0.29 | <0.0001 | 1.25 ± 0.34 | 1.29 ± 0.35 | <0.0001 |
| SNP | Chr:BP | A1 | MAF | Gene | Feature | Cluster SNP * | OR (95% CI) | p | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Present Study | EAS | EUR | AMR | ||||||||
| Participants with sodium intake <2 g/day | |||||||||||
| rs67617923 | 2:54968517 | A | 0.063 | 0.074 | 0.160 | 0.098 | EML6 | intron variant | rs72806698; rs67246257; rs67514855 | 1.294 (1.187–1.410) | 4.29 × 10−9 |
| rs16998073 | 4:81184341 | T | 0.347 | 0.360 | 0.268 | 0.267 | FGF5 | upstream gene variant | rs12509595; rs10857147 | 1.245 (1.190–1.302) | 1.14 × 10−21 |
| rs11191582 | 10:104913653 | A | 0.227 | 0.265 | 0.089 | 0.193 | NT5C2 | intron variant | rs11191479; rs11191484; rs72050190; rs145010450; rs10883815 | 0.849 (0.806–0.895) | 1.08 × 10−9 |
| rs11105378 | 12:90090741 | T | 0.372 | 0.310 | 0.141 | 0.112 | ATP2B1–LINC00936 | intergenic region | rs2681485; rs7136259; rs11105377; rs1401982; rs1689040 | 0.874 (0.836–0.915) | 4.67 × 10−9 |
| Participants with sodium intake ≥2 g/day | |||||||||||
| rs12509595 | 4:81182554 | C | 0.347 | 0.361 | 0.267 | 0.267 | PRDM8–FGF5 | intergenic region | rs16998073; rs10857147 | 1.228 (1.181–1.277) | 7.46 × 10−25 |
| rs6913309 | 6:32339840 | A | 0.212 | 0.139 | 0.310 | 0.244 | C6orf10–HLA-DQB1 | upstream gene variant | N/A | 1.145 (1.094–1.197) | 4.23 × 10−9 |
| rs112735431 | 17:78358945 | A | 0.011 | 0.002 | 0.000 | 0.000 | RNF213 | missense variant | rs138309870 | 1.706 (1.446–2.012) | 2.38 × 10−10 |
| rs3819496 | 8:143923891 | G | 0.312 | 0.321 | 0.421 | BP0.442 | GML–CYP11B1 | intron variant | rs3753123; rs143247792; rs4527848; rs4606038; rs28524031 | 0.892 (0.857.0.929) | 3.73 × 10−8 |
| rs140473396 | 10:104795885 | GAC | 0.247 | 0.285 | 0.097 | 0.197 | CNNM2–NT5C2 | intron variant | rs11191479; rs11191484; rs72050190; rs145010450; rs10883815 | 0.836 (0.800–0.873) | 1.11 × 10−15 |
| rs12229654 | 12:111414461 | G | 0.141 | 0.159 | 0.000 | 0.000 | MYL2–CUX2 | intergenic region | rs149607519; rs148177611; rs2188380; rs12227162 | 0.834 (0.787–0.883) | 5.25 × 10−10 |
| rs1887320 | 20:10965998 | G | 0.478 | 0.540 | 0.461 | 0.264 | JAG1 | intergenic region | rs6108787; rs1327235; rs6108789; rs913220 | 0.892 (0.859–0.925) | 1.45x10−9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, Y.-J.; Kim, J.O.; Park, J.-M.; Choi, J.-E.; Park, D.-H.; Song, Y.; Kim, S.-J.; Lee, J.-W.; Hong, K.-W. Identification of Genetic Factors Underlying the Association between Sodium Intake Habits and Hypertension Risk. Nutrients 2020, 12, 2580. https://doi.org/10.3390/nu12092580
Kwon Y-J, Kim JO, Park J-M, Choi J-E, Park D-H, Song Y, Kim S-J, Lee J-W, Hong K-W. Identification of Genetic Factors Underlying the Association between Sodium Intake Habits and Hypertension Risk. Nutrients. 2020; 12(9):2580. https://doi.org/10.3390/nu12092580
Chicago/Turabian StyleKwon, Yu-Jin, Jung Oh Kim, Jae-Min Park, Ja-Eun Choi, Da-Hyun Park, Youhyun Song, Seong-Jin Kim, Ji-Won Lee, and Kyung-Won Hong. 2020. "Identification of Genetic Factors Underlying the Association between Sodium Intake Habits and Hypertension Risk" Nutrients 12, no. 9: 2580. https://doi.org/10.3390/nu12092580
APA StyleKwon, Y.-J., Kim, J. O., Park, J.-M., Choi, J.-E., Park, D.-H., Song, Y., Kim, S.-J., Lee, J.-W., & Hong, K.-W. (2020). Identification of Genetic Factors Underlying the Association between Sodium Intake Habits and Hypertension Risk. Nutrients, 12(9), 2580. https://doi.org/10.3390/nu12092580





