The High-Fat Diet Based on Extra-Virgin Olive Oil Causes Dysbiosis Linked to Colorectal Cancer Prevention
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Diets
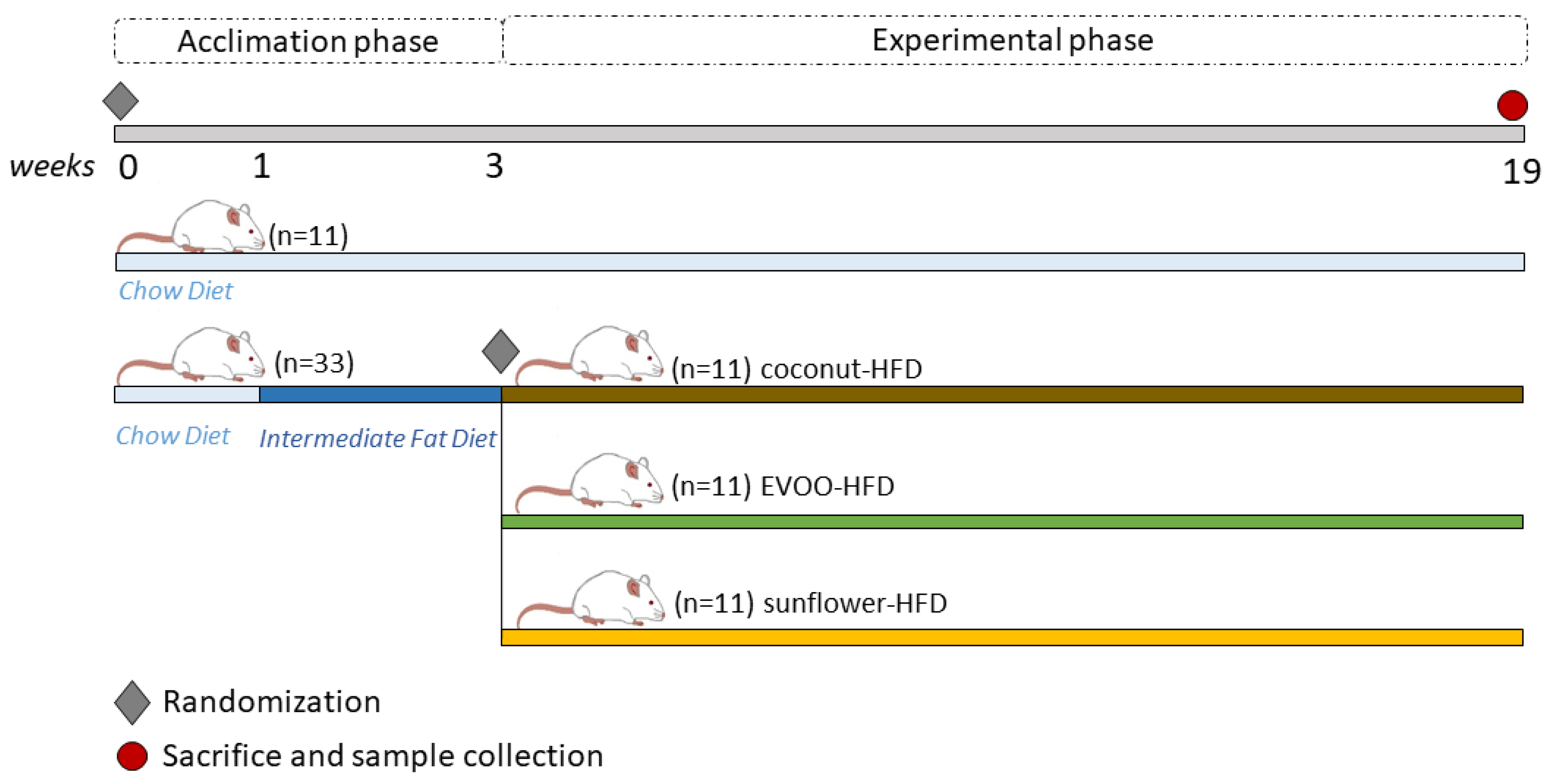
2.3. Experimental Design
- Group 1: Chow diet (chow)
- Group 2: HFD of coconut oil (coconut-HFD)
- Group 3: HFD of sunflower oil (sunflower-HFD)
- Group 4: HFD of extra virgin olive oil (EVOO-HFD)
2.4. Sample Collection and Mucosa-Associated Microbiota Analysis by PCR Amplification and Sequencing of the 16S rRNA Gene
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
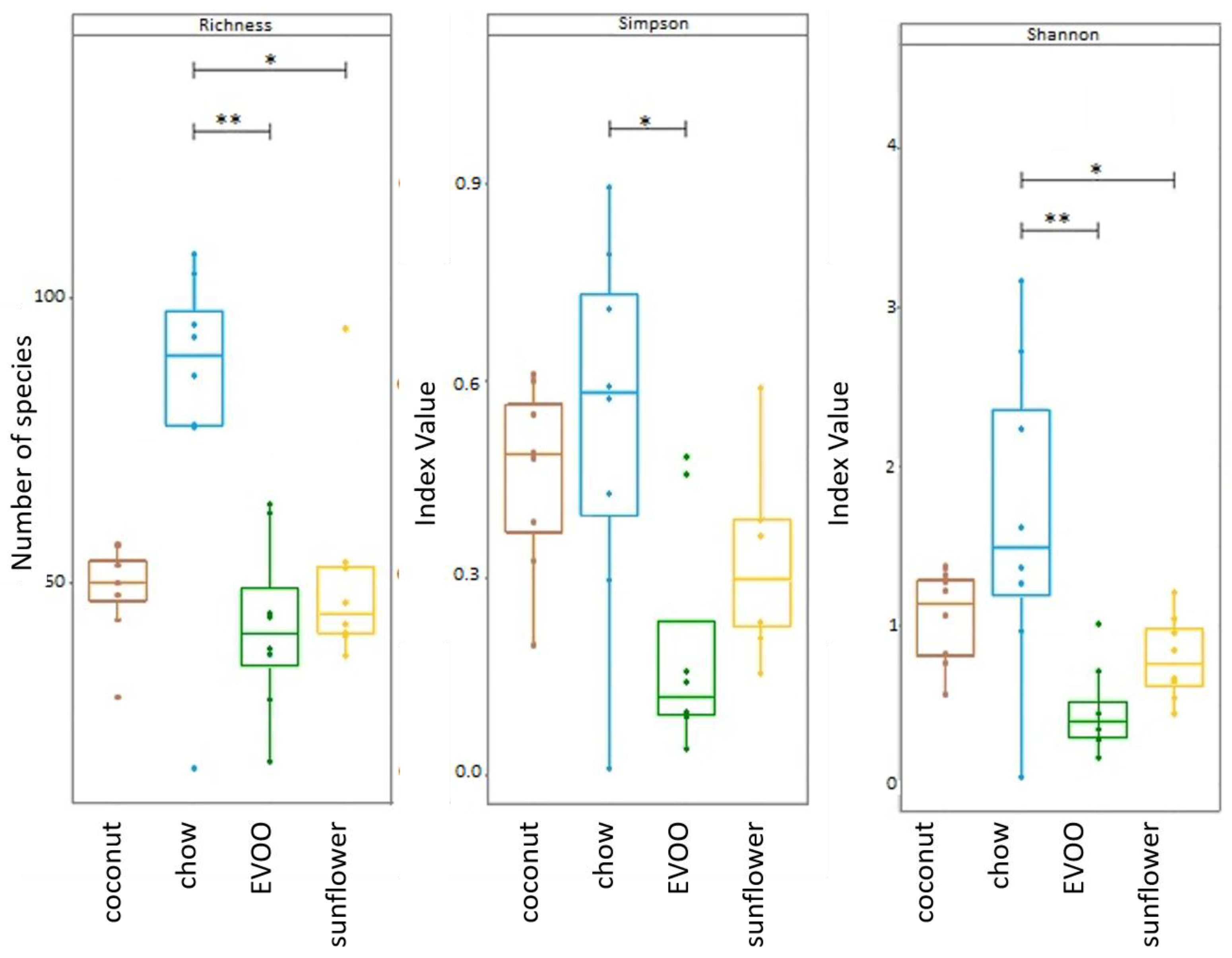
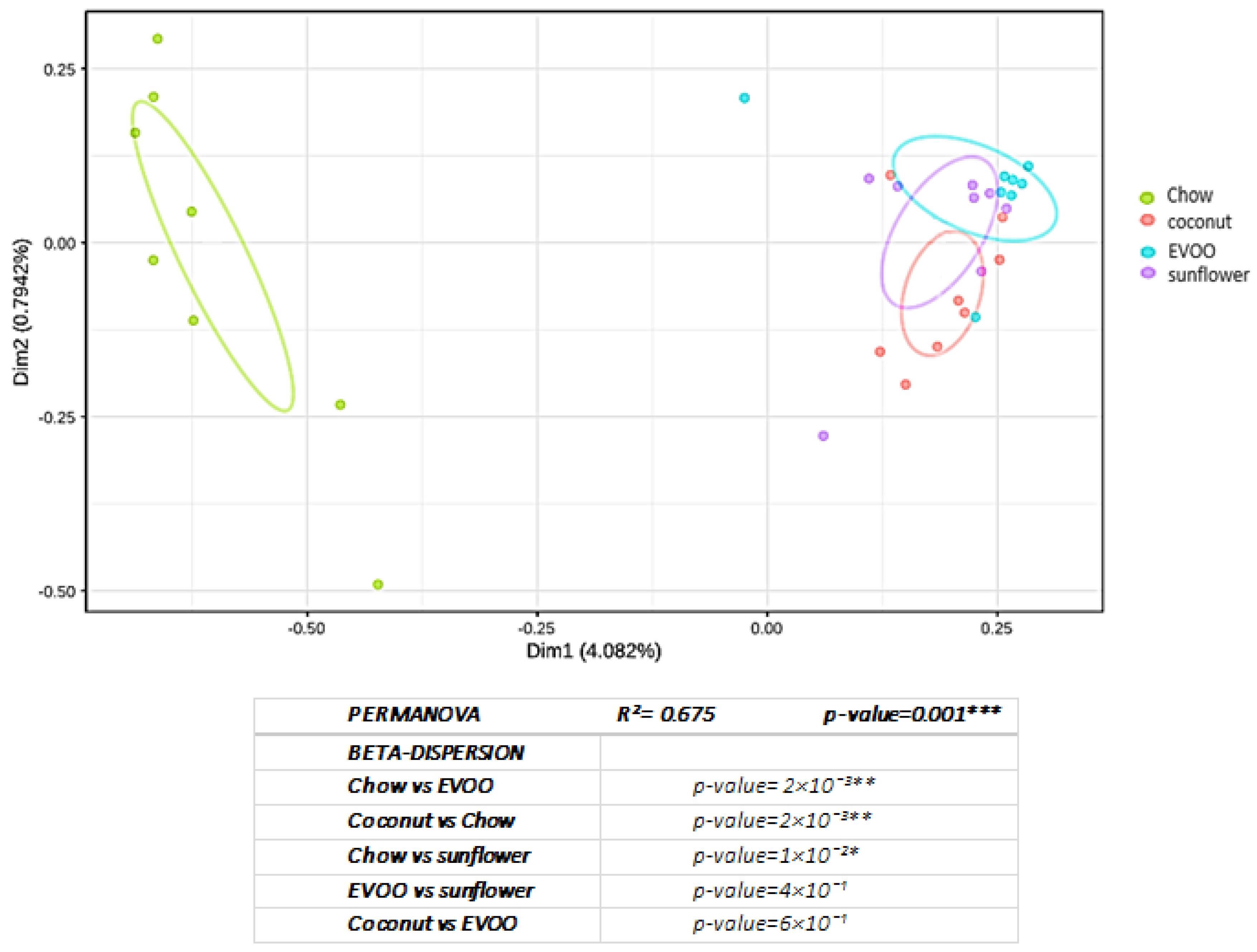
3.1. High-Fat Diets Promoted Dysbiosis Independently of Body Weight and the Type of Vegetable Fat Present in the Diet
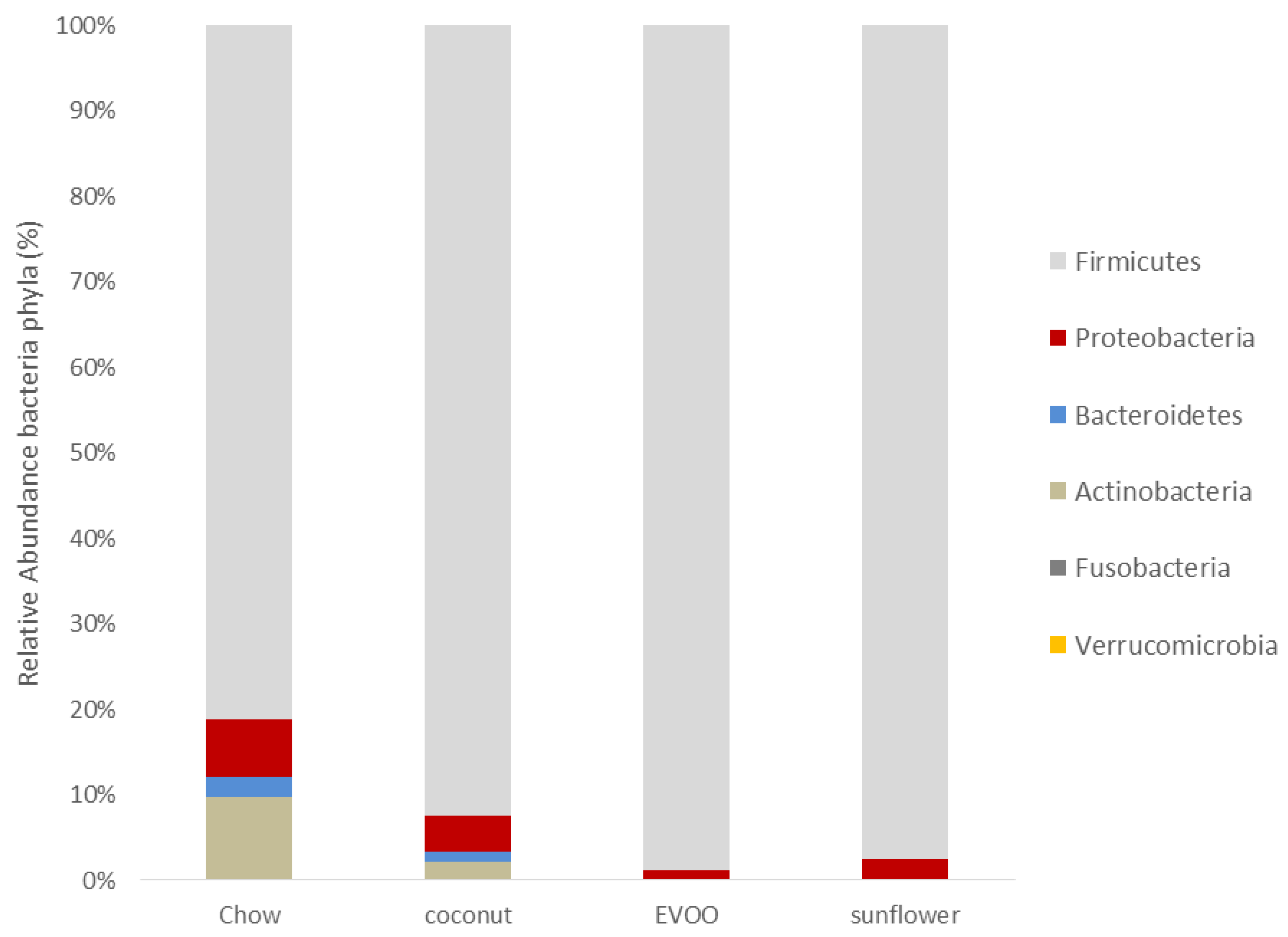
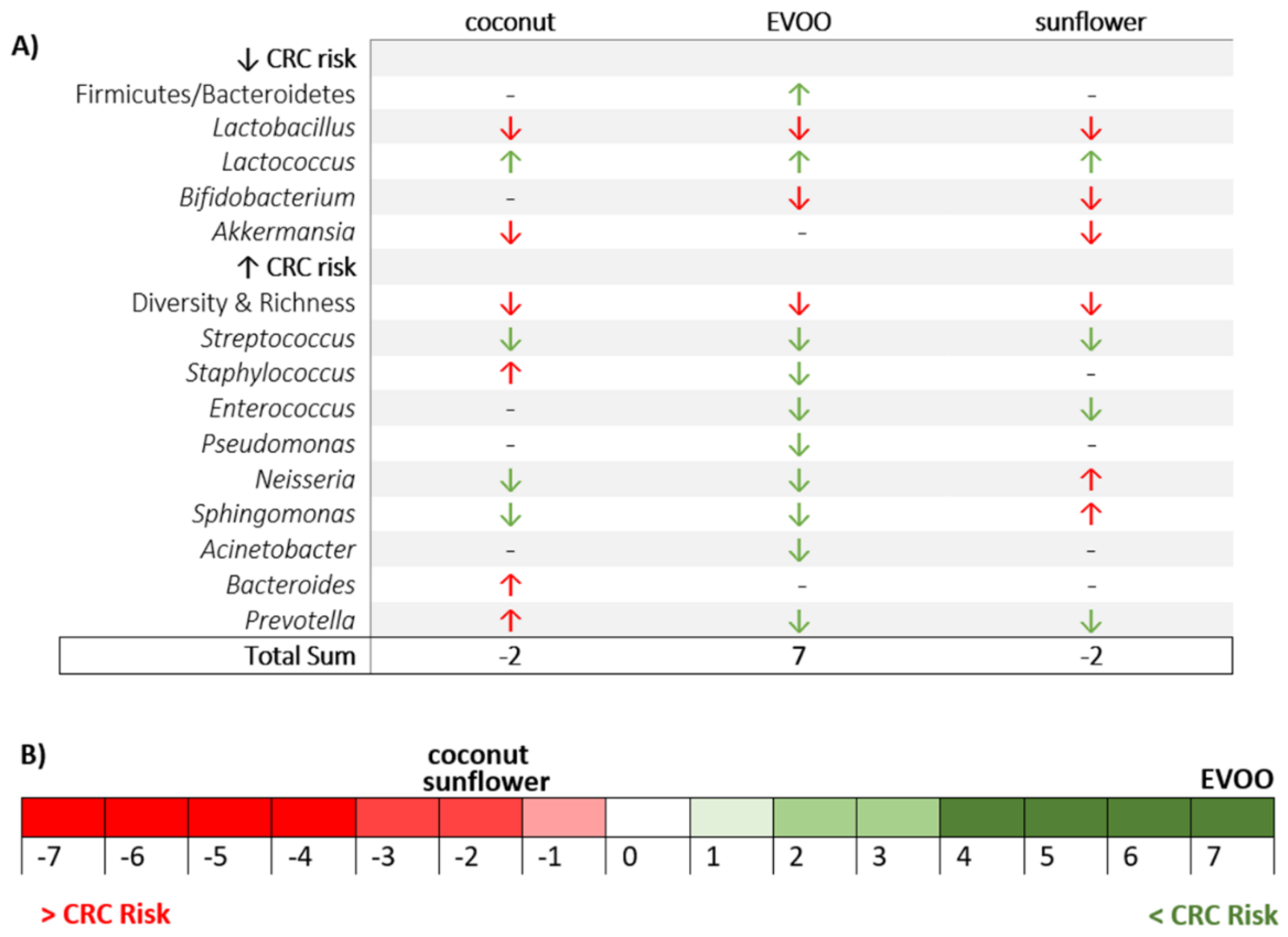
3.2. Main Findings Found at the Phylum Level: EVOO-HFD Specifically Reduced Proteobacteria While Coconut-HFD Decreased Verrucomicrobia
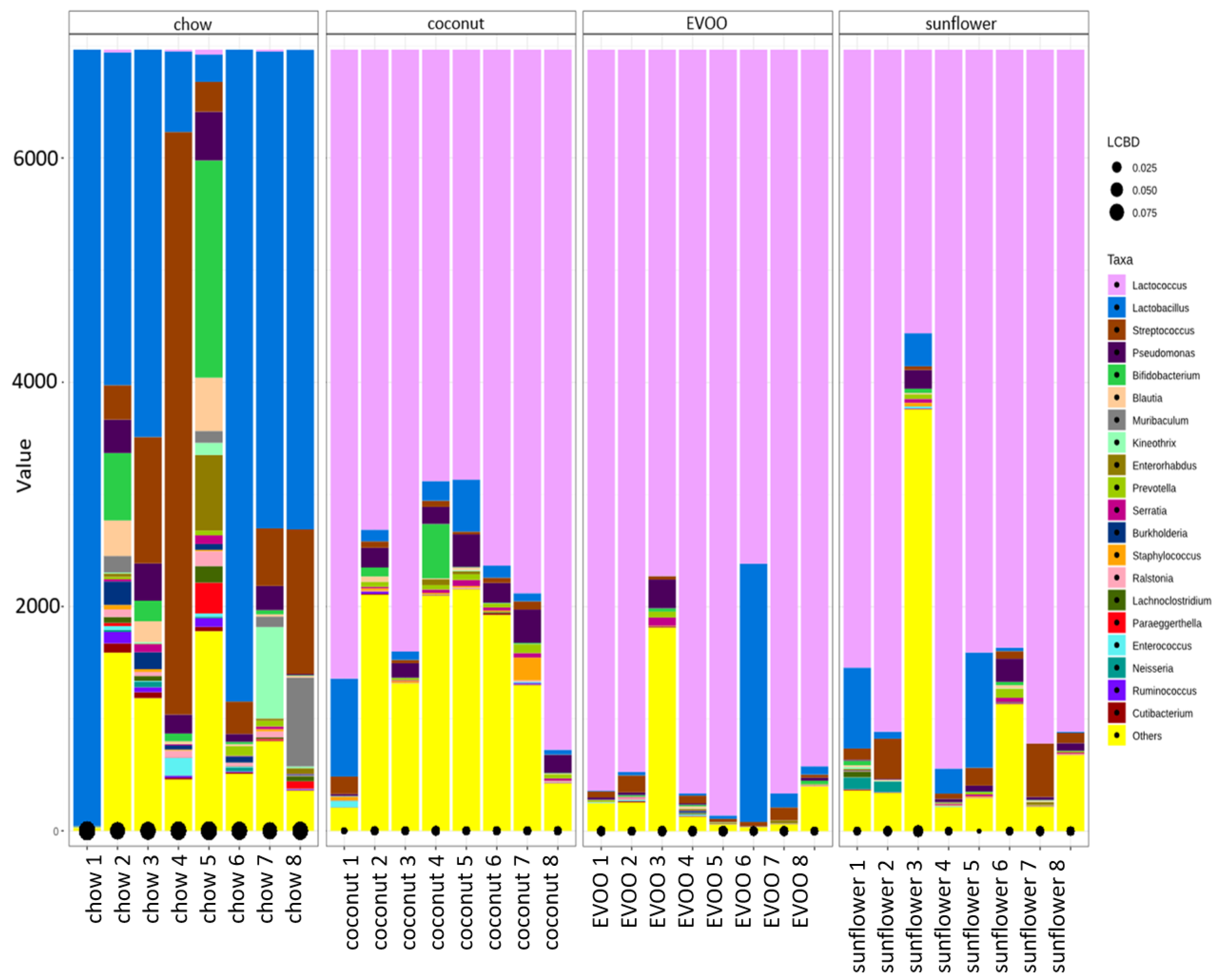
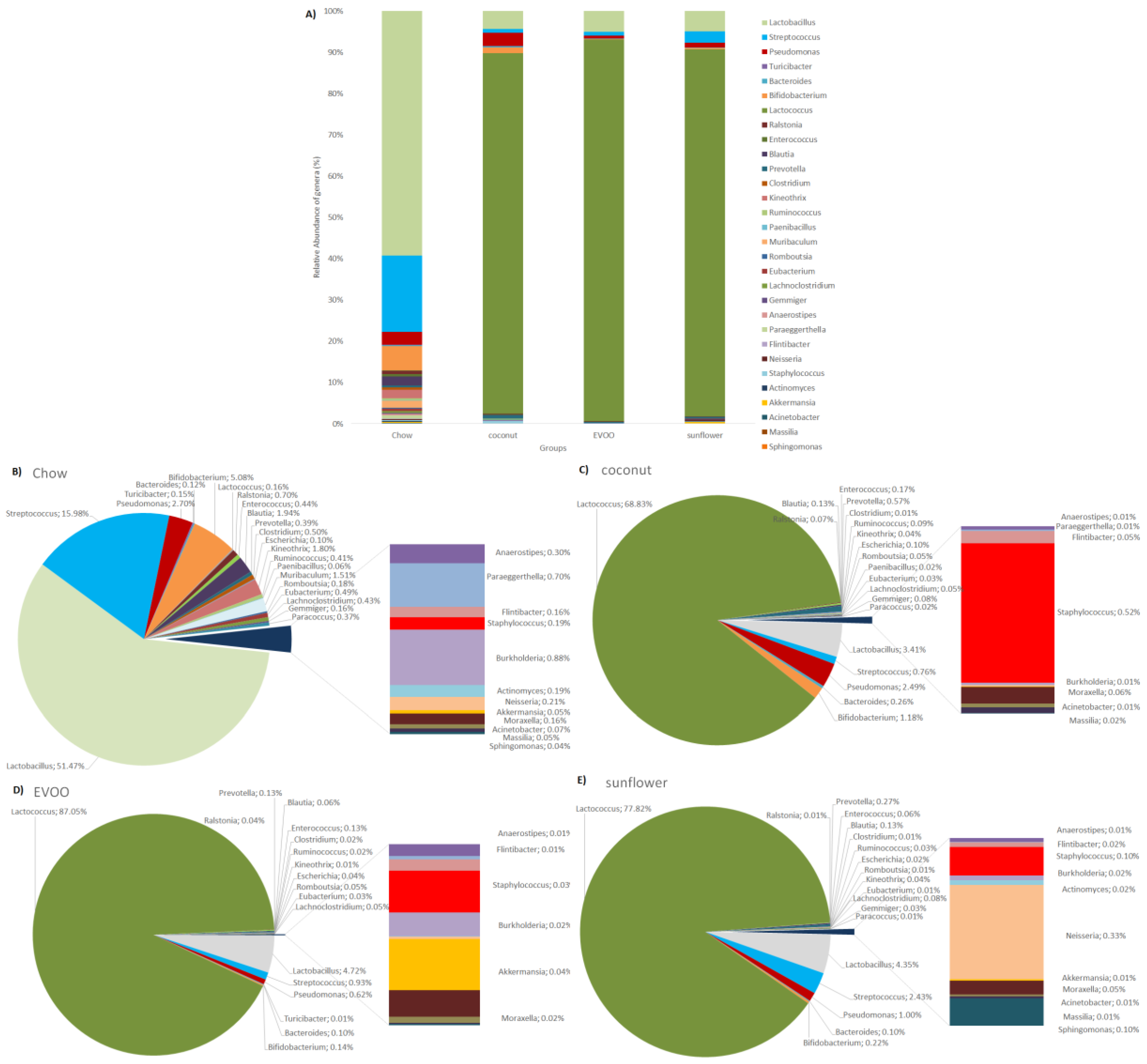
3.3. Main Findings Found at Both Genera and Species Levels: HFD Produced Both Switching Lactobacillus for Lactococcus and Only Coconut-HFD Promoting Staphylococcus Colonisation
3.4. Sunflower-HFD Enhanced Sphingomonas Genus
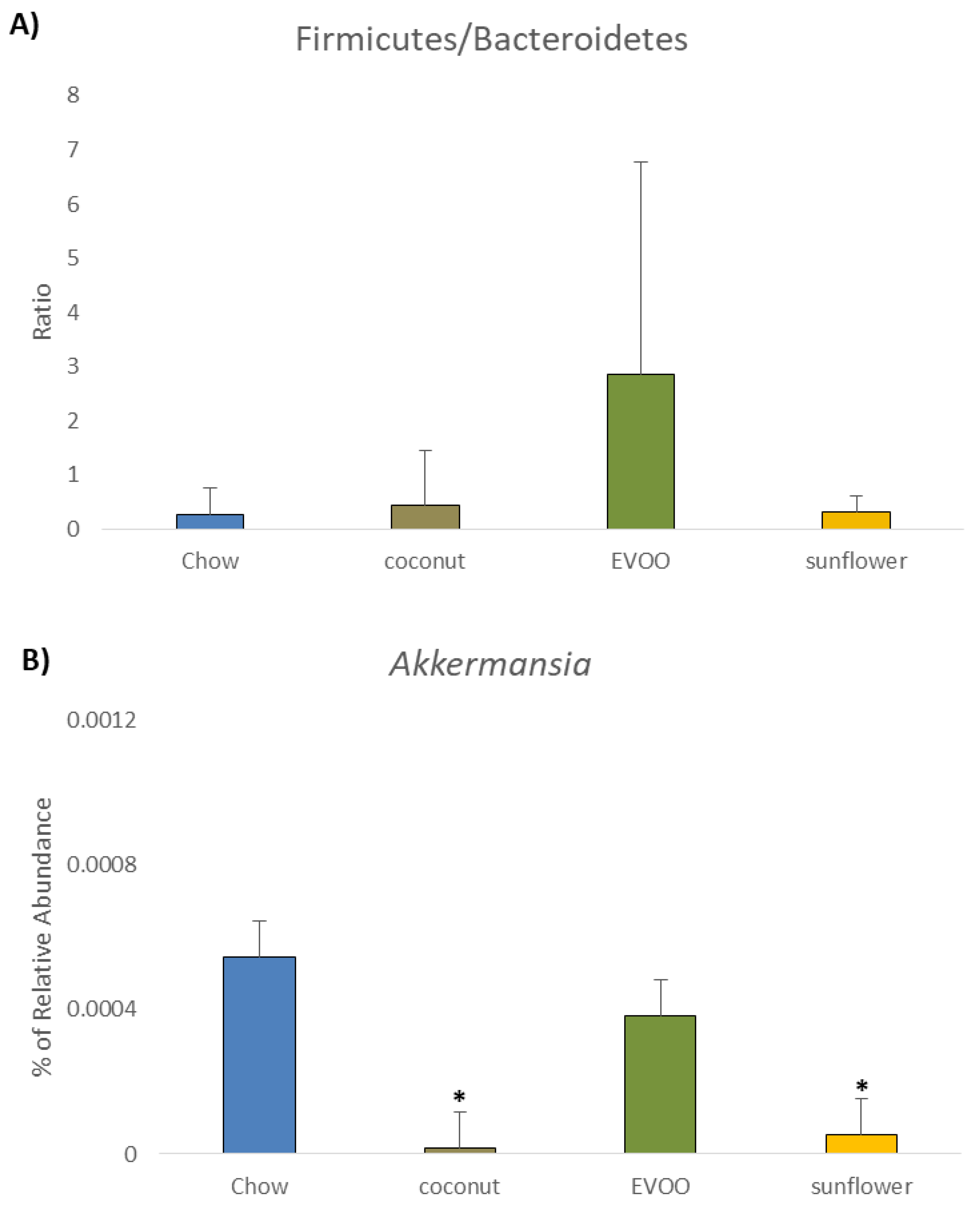
3.5. Coconut and Sunflower-HFD Reduced Akkermansia Muciniphila Abundance
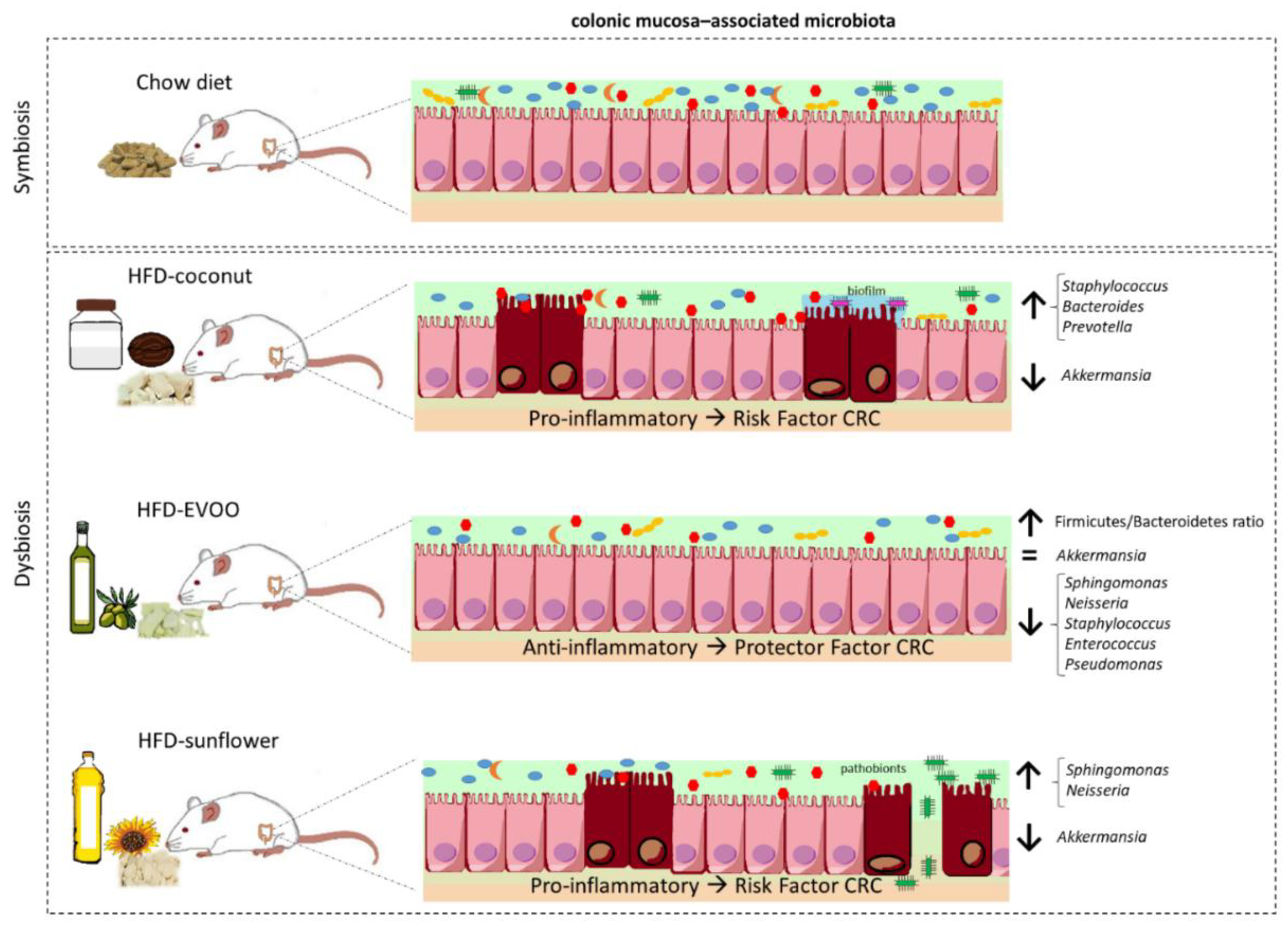
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Guo, Z.; Song, X.; Liu, L.; Dong, W.; Wang, S.; Xu, M.; Yang, C.; Wang, B.-M.; Cao, H. High-fat diet-induced dysbiosis mediates MCP-1/CCR2 axis-dependent M2 macrophage polarization and promotes intestinal adenoma-adenocarcinoma sequence. J. Cell. Mol. Med. 2020, 24, 2648–2662. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.J.D. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Sun, E.W.; Rogers, G.B.; Keating, D. The Influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of gut microbiota in Inflammatory Bowel Disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Chassaing, B.; Gewirtz, A.T. Chapter 35—Gut Microbiome and Metabolism, in Physiology of the Gastrointestinal Tract, 6th ed.; Said, H.M., Ed.; Academic Press, ELSEVIER: New York, NY, USA, 2018; pp. 775–793. [Google Scholar]
- Kriss, M.; Hazleton, K.; Nusbacher, N.M.; Martin, C.G.; Lozupone, C.A. Low diversity gut microbiota dysbiosis: Drivers, functional implications and recovery. Curr. Opin. Microbiol. 2018, 44, 34–40. [Google Scholar] [CrossRef]
- Yu, L.C.-H. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: Exploring a common ground hypothesis. J. Biomed. Sci. 2018, 25, 79. [Google Scholar] [CrossRef]
- Danneskiold-Samsøe, N.B.; Barros, H.D.d.F.Q.; Santos, R.; Bicas, J.L.; Cazarin, C.B.B.; Madsen, L.; Kristiansen, K.; Pastore, G.M.; Brix, S.; Junior, M.R.M. Interplay between food and gut microbiota in health and disease. Food Res. Int. 2019, 115, 23–31. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- ZymoBIOMICS Microbial Community Standard. Available online: https://www.zymoresearch.com/collections/zymobiomics-microbial-community-standards/products/zymobiomics-microbial-community-standard (accessed on 6 May 2020).
- Mehta, R.S.; Song, M.; Nishihara, R.; Drew, D.; Wu, K.; Qian, Z.R.; Fung, T.T.; Hamada, T.; Masugi, Y.; Da Silva, A.; et al. Dietary patterns and risk of colorectal cancer: Analysis by tumor location and molecular subtypes. Gastroenterology 2017, 152, 1944–1953.e1. [Google Scholar] [CrossRef]
- Moss, A.; Nalankilli, K. The association between diet and colorectal cancer risk: Moving beyond generalizations. Gastroenterology 2017, 152, 1821–1823. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yu, J. The association of diet, gut microbiota and colorectal cancer: What we eat may imply what we get. Protein Cell 2018, 9, 474–487. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.D.; Atay, Ç.; Heringer, J.; Romrig, F.K.; Schwitalla, S.; Aydin, B.; Ziegler, P.K.; Varga, J.; Reindl, W.; Pommerenke, C.; et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 2014, 514, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lang, T.; Shen, J.; Dai, J.; Tian, L.; Wang, X. Core gut bacteria analysis of healthy mice. Front. Microbiol. 2019, 10, 887. [Google Scholar] [CrossRef] [PubMed]
- Castañer, O.; Goday, A.; Park, Y.-M.M.; Lee, S.H.; Magkos, F.; Shiow, S.-A.T.E.; Schröder, H. The gut microbiome profile in obesity: A systematic review. Int. J. Endocrinol. 2018, 2018, 4095789. [Google Scholar] [CrossRef]
- Bishehsari, F.; Engen, P.; Preite, N.Z.; Tuncil, Y.E.; Naqib, A.; Shaikh, M.; Rossi, M.; Wilber, S.; Green, S.J.; Hamaker, B.R.; et al. Dietary fiber treatment corrects the composition of gut microbiota, promotes scfa production, and suppresses colon carcinogenesis. Genes 2018, 9, 102. [Google Scholar] [CrossRef]
- He, B.; Hoang, T.K.; Wang, T.; Ferris, M.; Taylor, C.M.; Tian, X.; Luo, M.; Tran, D.Q.; Zhou, J.; Tatevian, N.; et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency–induced autoimmunity via adenosine A2A receptors. J. Exp. Med. 2016, 214, 107–123. [Google Scholar] [CrossRef]
- Ballal, S.A.; Veiga, P.; Fenn, K.; Michaud, M.; Kim, J.; Gallini, C.A.; Glickman, J.N.; Quéré, G.; Garault, P.; Beal, C.; et al. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc. Natl. Acad. Sci. USA 2015, 112, 7803–7808. [Google Scholar] [CrossRef]
- Souza, B.M.; Preisser, T.M.; Pereira, V.B.; Turk, M.Z.; De Castro, C.P.; Da Cunha, V.P.; De Oliveira, R.P.; Gomes-Santos, A.C.; Faria, A.C.; Cara, D.; et al. Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microb. Cell Factories 2016, 15, 150. [Google Scholar] [CrossRef]
- Besten, G.D.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Bárcena, C.; Valdés-Mas, R.; Mayoral, P.; Garabaya, C.; Durand, S.; Rodríguez, F.; Fernández-García, M.T.; Salazar, N.; Nogacka, A.M.; Garatachea, N.; et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 2019, 25, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Wu, W.; Yang, L.; Lv, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; Wu, J.; Jiang, X.; et al. Administration of akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front. Microbiol. 2019, 10, 2259. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Sun, W.; Shan, X.; Jiang, H.; Cai, C.; Hao, J.; Li, G.; Yu, G. Carrageenan-induced colitis is associated with decreased population of anti-inflammatory bacterium, Akkermansia muciniphila, in the gut microbiota of C57BL/6J mice. Toxicol. Lett. 2017, 279, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Alrafas, H.R.; Busbee, P.B.; Nagarkatti, M.; Nagarkatti, P.S. Resveratrol modulates the gut microbiota to prevent murine colitis development through induction of Tregs and suppression of Th17 cells. J. Leukoc. Biol. 2019, 106, 467–480. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, X.; Cao, S.; Wang, L.; Wang, D.; Yang, H.; Feng, Y.; Wang, S.; Li, L. Caffeic acid ameliorates colitis in association with increased Akkermansia population in the gut microbiota of mice. Oncotarget 2016, 7, 31790–31799. [Google Scholar] [CrossRef]
- Molan, A.L.; Liu, Z.; Plimmer, G. Evaluation of the effect of blackcurrant products on gut microbiota and on markers of risk for colon cancer in humans. Phytother. Res. 2013, 28, 416–422. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Itoh, N.; Sugiyama, T.; Kurai, H. Clinical features of Clostridium bacteremia in cancer patients: A case series review. J. Infect. Chemother. 2020, 26, 92–94. [Google Scholar] [CrossRef]
- Dinakaran, V.; Mandape, S.N.; Shuba, K.; Pratap, S.; Sakhare, S.S.; Tabatabai, M.A.; Smoot, D.T.; Farmer-Dixon, C.M.; Kesavalu, L.N.; Adunyah, S.E.; et al. Identification of specific oral and gut pathogens in full thickness colon of colitis patients: Implications for colon motility. Front. Microbiol. 2019, 9, 3220. [Google Scholar] [CrossRef]
- Sánchez, E.; Donat, E.; Ribes-Koninckx, C.; Fernández-Murga, L.; Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013, 79, 5472–5479. [Google Scholar] [CrossRef]
- Vieira, S.M.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.M.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef]
- De La Visitación, N.; Robles-Vera, I.; Toral, M.; Duarte, J. Protective effects of probiotic consumption in cardiovascular disease in systemic lupus erythematosus. Nutrients 2019, 11, 2676. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.A.; Koren, O.; Goodrich, J.K.; Johansson, M.E.; Nalbantoglu, I.; Aitken, J.D.; Su, Y.; Chassaing, B.; Walters, W.A.; González, A.; et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 2012, 12, 139–152. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD—What role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Awdisho, A.; Zulfikar, R. A case report of Neisseria mucosa peritonitis in a chronic ambulatory peritoneal dialysis patient. Infect. Dis. Rep. 2017, 8, 6950. [Google Scholar] [CrossRef]
- El Aidy, S.; Derrien, M.; Aardema, R.; Hooiveld, G.J.; Richards, S.; Dane, A.; Dekker, J.; Vreeken, R.J.; Levenez, F.; Doré, J.; et al. Transient inflammatory-like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ-free mice. Benef. Microbes 2014, 5, 67–77. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; Da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018, 9, 131–142. [Google Scholar] [CrossRef]
- Sánchez-Fidalgo, S.; Cárdeno, A.; Sánchez-Hidalgo, M.; Soto, M.A.; Villegas, I.; Rosillo, M.A.; De La Lastra, A. Dietary unsaponifiable fraction from extra virgin olive oil supplementation attenuates acute ulcerative colitis in mice. Eur. J. Pharm. Sci. 2013, 48, 572–581. [Google Scholar] [CrossRef]
- Morvaridi, M.; Jafarirad, S.; Seyedian, S.S.; Alavinejad, P.; Cheraghian, B. The effects of extra virgin olive oil and canola oil on inflammatory markers and gastrointestinal symptoms in patients with ulcerative colitis. Eur. J. Clin. Nutr. 2020. [Google Scholar] [CrossRef]
- Sánchez-Fidalgo, S.; Villegas, I.; Cárdeno, A.; Talero, E.; Sánchez-Hidalgo, M.; Motilva, V.; De La Lastra, A.; Sánchez, V.M. Extra-virgin olive oil-enriched diet modulates DSS-colitis-associated colon carcinogenesis in mice. Clin. Nutr. 2010, 29, 663–673. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, B.; Ross, R.P.; Jin, Y.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Orally administered CLA ameliorates DSS-induced colitis in mice via intestinal barrier improvement, oxidative stress reduction, and inflammatory cytokine and gut microbiota modulation. J. Agric. Food Chem. 2019, 67, 13282–13298. [Google Scholar] [CrossRef] [PubMed]
- Praengam, K.; Sahasakul, Y.; Kupradinun, P.; Sakarin, S.; Sanitchua, W.; Rungsipipat, A.; Rattanapinyopituk, K.; Angkasekwinai, P.; Changsri, K.; Mhuantong, W.; et al. Brown rice and retrograded brown rice alleviate inflammatory response in dextran sulfate sodium (DSS)-induced colitis mice. Food Funct. 2017, 8, 4630–4643. [Google Scholar] [CrossRef] [PubMed]
- Hofer, U. Pro-inflammatory Prevotella? Nat. Rev. Genet. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Cheng, N.; Zhou, W.; Chen, S.; Wang, Q.; Gao, H.; Xue, X.; Wu, L.; Cao, W. Honey polyphenols ameliorate DSS-induced ulcerative colitis via modulating gut microbiota in rats. Mol. Nutr. Food Res. 2019, 63, e1900638. [Google Scholar] [CrossRef]
- Li, A.; Ni, W.; Zhang, Q.; Li, Y.; Zhang, X.; Wu, H.; Du, P.; Hou, J.; Zhang, Y. Effect of cinnamon essential oil on gut microbiota in the mouse model of dextran sodium sulfate-induced colitis. Microbiol. Immunol. 2019, 64, 23–32. [Google Scholar] [CrossRef]
- Bisanz, J.E.; Upadhyay, V.; Turnbaugh, J.A.; Ly, K.; Turnbaugh, P. Diet induces reproducible alterations in the mouse and human gut microbiome. bioRxiv 2019, 541797. [Google Scholar] [CrossRef]
| Weight | Chow | Coconut | EVOO | Sunflower |
|---|---|---|---|---|
| Initial (g) | 29.49 ± 0.37 | 28.65 ± 0.47 | 29.42 ± 0.76 | 31.04 ± 0.77 |
| Final (g) | 45.48 ± 1.85 | 44.88 ± 1.47 | 55.89 ± 2.65 ** | 55.3 ± 3.33 ** |
| Phylum | Chow | Coconut | EVOO | Sunflower |
|---|---|---|---|---|
| Firmicutes | 78.695 ± 20.27 | 75.014 ± 13.08 | 93.739 ± 10.56 | 86.081 ± 18.37 |
| Proteobacteria | 6.578 ± 5.34 | 3.447 ± 1.86 | 0.958 ± 1.61 *** | 2.17 ± 1.35 |
| Bacteroidetes | 2.331 ± 3.04 | 0.948 ± 0.49 * | 0.283 ± 0.48 *** | 0.532 ± 0.46 *** |
| Actinobacteria | 9.186 ± 15.32 | 1.712 ± 2.78 ** | 0.437 ± 0.22 *** | 0.654 ± 0.45 *** |
| Fusobacteria | 0.083 ± 0.14 | 0.011 ± 0.02 | 0.007 ± 0.01 | 0.046 ± 0.09 |
| Verrucomicrobia | 0.055 ± 0.12 | 0.002 ± 0.003 ** | 0.038 ± 0.06 | 0.011 ± 0.02 |
| Genera | Chow | Coconut | EVOO | Sunflower |
|---|---|---|---|---|
| FIRMICUTES | ||||
| Lactobacillus | 51.471 ± 32.92 | 3.412 ± 3.99 1 | 4.717 ± 11.76 1 | 4.351 ± 5.58 1 |
| Streptococcus | 15.982 ± 24.53 | 0.76 ± 0.57 1 | 0.928 ± 0.66 1 | 2.428 ± 2.35 1 |
| Turicibacter | 0.152 ± 0.23 | 0.001 ± 0.003 1 | 0.006 ± 0.006 1 | 0.004 ± 0.004 1 |
| Lactococcus | 0.156 ± 0.19 | 68.83 ± 12.28 1 | 87.055 ± 13.74 1 | 77.819 ± 17.58 1 |
| Enterococcus | 0.444 ± 0.84 | 0.173 ± 0.4 | 0.125 ± 0.24 1 | 0.06 ± 0.06 1 |
| Blautia | 1.942 ± 2.71 | 0.129 ± 0.18 1 | 0.058 ± 0.06 1 | 0.129 ± 0.12 1 |
| Clostridium | 0.505 ± 0.64 | 0.01 ± 0.02 1 | 0.024 ± 0.02 1 | 0.01 ± 0.02 1 |
| Kineothrix | 1.796 ± 3.8 | 0.036 ± 0.07 1 | 0.014 ± 0.02 1 | 0.037 ± 0.05 1 |
| Ruminococcus | 0.411 ± 0.48 | 0.094 ± 0.11 1 | 0.023 ± 0.03 1 | 0.034 ± 0.03 1 |
| Paenibacillus | 0.061 ± 0.11 | 0.022 ± 0.05 | 0.003 ± 0.004 1 | 0.0003 ± 0.0007 1 |
| Romboutsia | 0.178 ± 0.24 | 0.046 ± 0.06 1 | 0.048 ± 0.06 | 0.014 ± 0.02 1 |
| Eubacterium | 0.495 ± 0.59 | 0.034 ± 0.05 1 | 0.03 ± 0.06 1 | 0.013 ± 0.01 1 |
| Lachnoclostridium | 0.426 ± 0.47 | 0.053 ± 0.07 1 | 0.046 ± 0.05 1 | 0.079 ± 0.2 1 |
| Gemmiger | 0.157 ± 0.23 | 0.081 ± 0.19 | 0.004 ± 0.004 1 | 0.033 ± 0.05 |
| Anaerostipes | 0.296 ± 0.45 | 0.012 ± 0.02 1 | 0.009 ± 0.01 1 | 0.014 ± 0.02 1 |
| Flintibacter | 0.162 ± 0.23 | 0.047 ± 0.06 | 0.009 ± 0.01 1 | 0.017 ± 0.02 1 |
| Staphylococcus | 0.194 ± 0.16 | 0.522 ± 0.82 | 0.031 ± 0.04 1,2 | 0.101 ± 0.1 |
| PROTEOBACTERIA | ||||
| Pseudomonas | 2.702 ± 2.23 | 2.489 ± 1.33 | 0.618 ± 1.19 1 | 0.999 ± 1.09 |
| Ralstonia | 0.696 ± 0.57 | 0.071 ± 0.74 1 | 0.038 ± 0.06 1 | 0.014 ± 0.01 1 |
| Escherichia | 0.095 ± 0.12 | 0.102 ± 0.04 | 0.038 ± 0.06 | 0.023 ± 0.02 |
| Paracoccus | 0.375 ± 0.98 | 0.024 ± 0.02 | 0.003 ± 0.004 | 0.006 ± 0.01 |
| Burkholderia | 0.882 ± 1.07 | 0.01 ± 0.02 1 | 0.018 ± 0.05 1 | 0.018 ± 0.05 1 |
| Moraxella | 0.159 ± 0.35 | 0.063 ± 0.14 | 0.02 ± 0.04 | 0.049 ± 0.11 |
| Neisseria | 0.214 ± 0.24 | 0.004 ± 0.008 1 | 0.002 ± 0.003 1 | 0.335 ± 0.61 2,3 |
| Acinetobacter | 0.073 ± 0.14 | 0.013 ± 0.01 | 0.005 ± 0.009 1 | 0.007 ± 0.01 |
| Massilia | 0.049 ± 0.08 | 0.022 ± 0.06 | 0.0009 ± 0.002 1 | 0.007 ± 0.01 |
| Sphingomonas | 0.036 ± 0.06 | 0.002 ± 0.005 | 0.001 ± 0.001 | 0.097± 0.17 2,3 |
| BACTEROIDETES | ||||
| Bacteroides | 0.125 ± 0.13 | 0.264 ± 0.21 | 0.103 ± 0.2 | 0.099 ± 0.12 |
| Prevotella | 0.389 ± 0.48 | 0.57 ± 0.29 | 0.134 ± 0.24 | 0.273 ± 0.33 |
| Parabacteroides | 0.004 ± 0.003 | 0.017 ± 0.02 | 0.008 ± 0.01 | 0.008 ± 0.01 |
| Muribaculum | 1.509 ± 3.26 | 0.003 ± 0.007 1 | 0.0003 ± 0.0007 1 | ND 1 |
| ACTINOBACTERIA | ||||
| Bifidobacterium | 5.075 ± 9.59 | 1.175 ± 2.52 | 0.138 ± 0.13 1 | 0.224 ± 0.24 1 |
| Paraeggerthella | 0.701 ± 1.38 | 0.005 ± 0.01 1 | 0.003 ± 0.005 1 | 0.002 ± 0.003 1 |
| Actinomyces | 0.192 ± 0.47 | 0.0001 ± 0.0003 1 | 0.0006 ± 0.001 1 | 0.016 ± 0.03 1 |
| VERRUCOMICROBIA | ||||
| Akkermansia | 0.055 ± 0.12 | 0.001 ± 0.003 1 | 0.038 ± 0.06 | 0.005 ± 0.008 1 |
| Species | Chow | Coconut | EVOO | Sunflower |
|---|---|---|---|---|
| Lactobacillus reuteri | 42.45 ± 29.95 | 0.335 ± 0.57 1 | 1.106 ± 2.84 1 | 0.658 ± 0.77 1 |
| Lactobacillus taiwanensis | 5.979 ± 7.31 | 0.002 ± 0.003 1 | 0.006 ± 0.01 | 0.002 ± 0.004 1 |
| Lactobacillus animalis | 1.71 ± 2.29 | 1.449 ± 2.1 | 0.415 ± 0.45 | 1.533 ± 2.76 |
| Lactobacillus caviae | 0.501 ± 0.34 | 0.004 ± 0.01 1 | 0.019 ± 0.05 1 | 0.009 ± 0.01 1 |
| Lactobacillus gasseri | 0.129 ± 0.16 | 0.017 ± 0.02 | 2.85 ± 8.05 | 0.019 ± 0.05 |
| Lactobacillus secaliphilus | 0.078 ± 0.14 | 0.0002 ± 0.0007 1 | 0.023 ± 0.06 | 0.001 ± 0.002 1 |
| Lactobacillus faecis | 0.026 ± 0.04 | 0.011 ± 0.02 | 0.01 ± 0.02 | 0.019 ± 0.03 |
| Lactobacillus johnsonii | 0.004 ± 0.004 | 1.351 ± 2.06 | 0.195 ± 0.38 | 2.012 ± 4.70 |
| Lactobacillus satsumensis | 0.002 ± 0.003 | 0.008 ± 0.01 | 0.016 ± 0.03 | 0.062 ± 0.12 |
| Streptococcus danieliae | 15.229 ± 24.65 | 0.711 ± 0.56 1 | 0.76 ± 0.6 1 | 1.774 ± 2.05 1 |
| Streptococcus oralis | 0.255 ± 0.30 | 0.022 ± 0.03 1 | 0.021 ± 0.04 1 | 0.474 ± 1.11 2,3 |
| Streptococcus sanguinis | 0.192 ± 0.43 | 0.0001 ± 0.0003 1 | 0.006 ± 0.01 1 | 0.055 ± 0.14 |
| Lactococcus lactis | 0.153 ± 0.19 | 68.407 ± 12.20 1 | 86.28 ± 13.62 1 | 77.241 ± 17.41 1 |
| Lactococcus taiwanensis | 0.003 ± 0.002 | 0.419 ± 0.11 1 | 0.761 ± 0.25 1 | 0.557 ± 0.21 1 |
| Enterococcus gallinarum | 0.361 ± 0.85 | 0.022 ± 0.06 | 0.003 ± 0.004 1 | 0.028 ± 0.05 1 |
| Blautia wexlerae | 0.613 ± 0.88 | 0.039 ± 0.06 1 | 0.012 ± 0.01 1 | 0.047 ± 0.07 1 |
| Blautia luti | 0.559 ± 0.69 | 0.042 ± 0.09 1 | 0.014 ± 0.02 1 | 0.015 ± 0.02 1 |
| Blautia obeum | 0.416 ± 0.90 | 0.012 ± 0.02 1 | 0.002 ± 0.003 1 | 0.004 ± 0.003 1 |
| Blautia faecis | 0.045 ± 0.06 | 0.009 ± 0.01 | 0.001 ± 0.001 1 | 0.005 ± 0.01 1 |
| Clostridium scindens | 0.088 ± 0.09 | 0.014 ± 0.02 | 0.027 ± 0.04 | 0.029 ± 0.07 |
| Clostridium spiroforme | 0.101 ± 0.17 | 0.012 ± 0.03 1 | 0.004 ± 0.01 1 | 0.0004 ± 0.0005 1 |
| Clostridioides difficile | ND | 0.001 ± 0.003 | 0.016 ± 0.03 | 0.003 ± 0.006 |
| Ruminococcus gnavus | 0.306 ± 0.8 | 0.021 ± 0.04 1 | 0.024 ± 0.04 1 | 0.051 ± 0.09 1 |
| Eubacterium hallii | 0.38 ± 0.47 | 0.012 ± 0.02 1 | 0.005 ± 0.01 1 | 0.008 ± 0.01 1 |
| Anaerostipes hadrus | 0.291 ± 0.45 | 0.012 ± 0.02 1 | 0.009 ± 0.01 1 | 0.014 ± 0.02 1 |
| Staphylococcus epidermidis | 0.065 ± 0.09 | 0.328 ± 0.87 | 0.003 1,2 | 0.031 ± 0.04 2 |
| Pseudomonas migulae | 1.538 ± 1.56 | 1.199 ± 0.93 | 0.428 ± 0.89 1 | 0.493 ± 0.58 |
| Pseudomonas trivialis | 0.277 ± 0.36 | 0.238 ± 0.20 | 0.075 ± 0.16 | 0.169 ± 0.25 |
| Pseudomonas helmanticensis | 0.521 ± 1.17 | 0.888 ± 1.64 1 | 0.04 ± 0.1 1 | 0.277 ± 0.76 1 |
| Pseudomonas aeruginosa | 0.04 ± 0.07 | 0.001 ± 0.001 1 | 0.001 ± 0.001 1 | ND 1 |
| Neisseria mucosa | 0.055 ± 0.15 | 0.003 ± 0.007 1 | 0.0003 ± 0.0007 1 | 0.176 ± 0.32 2,3 |
| Ralstonia insidiosa | 0.685 ± 0.57 | 0.071 ± 0.07 1 | 0.038 ± 0.06 1 | 0.013 ± 0.01 1 |
| Prevotella oralis | 0.003 ± 0.01 | 0.004 ± 0.01 | 0.005 ± 0.01 | 0.0002 ± 0.0007 |
| Prevotella copri | 0.217 ± 0.32 | 0.543 ± 0.27 | 0.113 ± 0.22 | 0.244 ± 0.32 |
| Muribaculum intestinale | 1.509 ± 3.26 | 0.003 ± 0.01 1 | 0.0003 ± 0.0007 1 | ND 1 |
| Bifidobacterium longum | 0.683 ± 0.88 | 0.131 ± 0.22 1 | 0.033 ± 0.05 1 | 0.057 ± 0.08 1 |
| Bifidobacterium animalis | 4.022 ± 8.49 | 0.124 ± 0.14 1 | 0.094 ± 0.13 1 | 0.12 ± 0.12 1 |
| Bifidobacterium thermophilum | 0.053 ± 0.11 | 0.006 ± 0.02 1 | 0.001 ± 0.001 1 | 0.0001 ± 0.0003 1 |
| Bifidobacterium adolescentis | 0.189 ± 0.21 | 0.096 ± 0.23 | 0.005 ± 0.005 1 | 0.022 ± 0.04 1 |
| Bifidobacterium bifidum | 0.052 ± 0.09 | 0.014 ± 0.03 1 | 0.002 ± 0.005 1 | 0.004 ± 0.01 1 |
| Akkermansia muciniphila | 0.055 ± 0.12 | 0.001 ± 0.02 1 | 0.038 ± 0.06 | 0.005 ± 0.01 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-García, C.; Sánchez-Quesada, C.; Algarra, I.; Gaforio, J.J. The High-Fat Diet Based on Extra-Virgin Olive Oil Causes Dysbiosis Linked to Colorectal Cancer Prevention. Nutrients 2020, 12, 1705. https://doi.org/10.3390/nu12061705
Rodríguez-García C, Sánchez-Quesada C, Algarra I, Gaforio JJ. The High-Fat Diet Based on Extra-Virgin Olive Oil Causes Dysbiosis Linked to Colorectal Cancer Prevention. Nutrients. 2020; 12(6):1705. https://doi.org/10.3390/nu12061705
Chicago/Turabian StyleRodríguez-García, Carmen, Cristina Sánchez-Quesada, Ignacio Algarra, and José J. Gaforio. 2020. "The High-Fat Diet Based on Extra-Virgin Olive Oil Causes Dysbiosis Linked to Colorectal Cancer Prevention" Nutrients 12, no. 6: 1705. https://doi.org/10.3390/nu12061705
APA StyleRodríguez-García, C., Sánchez-Quesada, C., Algarra, I., & Gaforio, J. J. (2020). The High-Fat Diet Based on Extra-Virgin Olive Oil Causes Dysbiosis Linked to Colorectal Cancer Prevention. Nutrients, 12(6), 1705. https://doi.org/10.3390/nu12061705








