The Effect of Interactions between Folic Acid Supplementation and One Carbon Metabolism Gene Variants on Small-for-Gestational-Age Births in the Screening for Pregnancy Endpoints (SCOPE) Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Recruitment and Selection
2.2. Ethics
2.3. Sociodemographic, Lifestyle and FAS Data Collection
2.4. Single-Nucleotide Polymorphisms
2.5. Analysis of Single Nucleotide Polymorphisms
2.6. Outcomes and Definitions
2.7. Statistical Analysis
3. Results
3.1. Description of Sample
3.2. Genotype Distribution and Allele Frequency
3.3. Associations between Maternal Polymorphisms and SGA
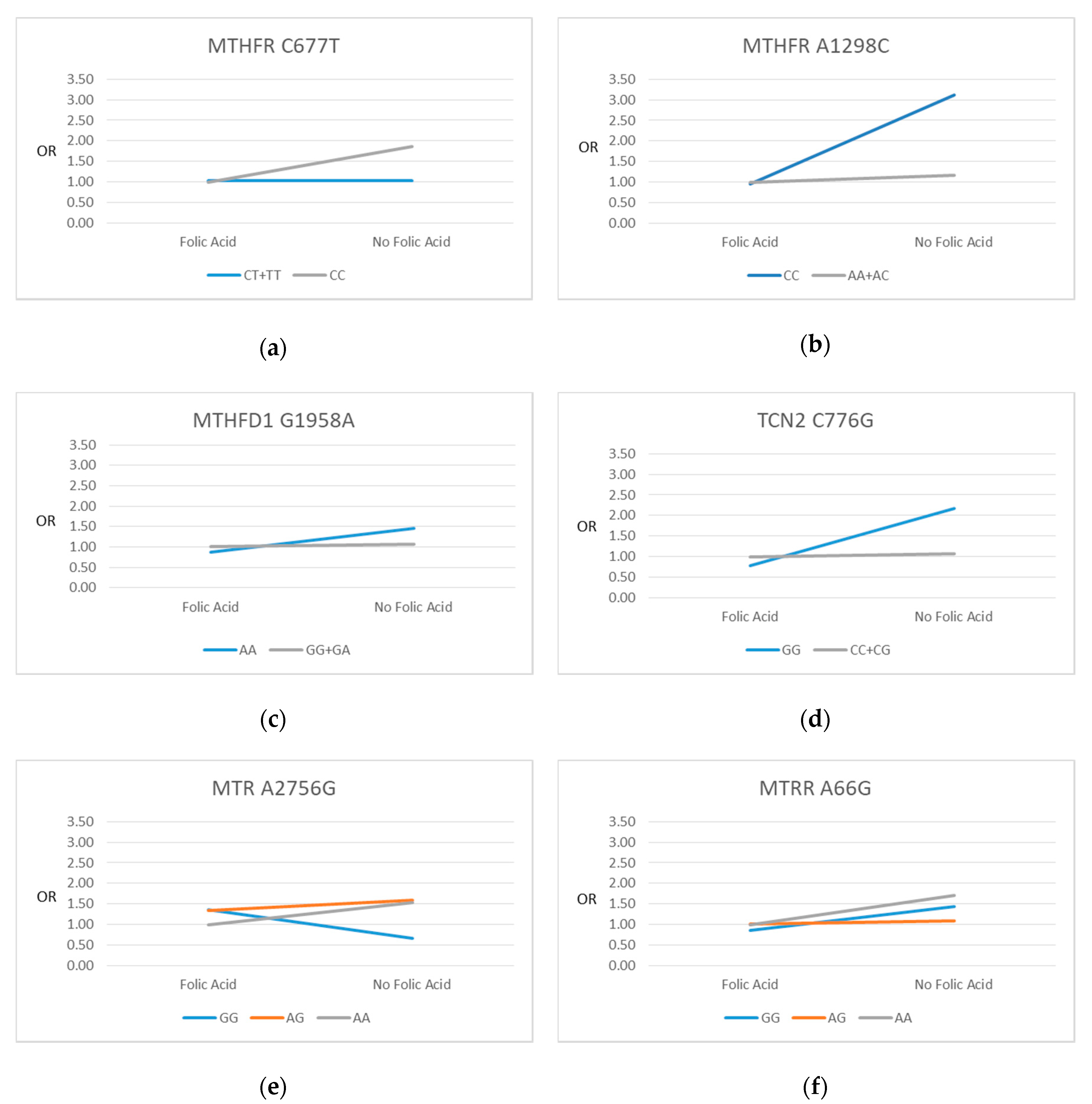
3.4. SNP-FAS Interactions and SGA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Finken, M.J.; van der Steen, M.; Smeets, C.C.; Walenkamp, M.J.; de Bruin, C.; Hokken-Koelega, A.C.; Wit, J.M. Children born small for gestational age: Differential diagnosis, molecular genetic evaluation, and implications. Endocr. Rev. 2018, 39, 851–894. [Google Scholar] [CrossRef] [PubMed]
- McCowan, L.M.; Thompson, J.M.; Taylor, R.S.; Baker, P.N.; North, R.A.; Poston, L.; Roberts, C.T.; Simpson, N.A.; Walker, J.J.; Myers, J. Prediction of small for gestational age infants in healthy nulliparous women using clinical and ultrasound risk factors combined with early pregnancy biomarkers. PLoS ONE 2017, 12, e0169311. [Google Scholar] [CrossRef] [PubMed]
- McCowan, L.; Horgan, R.P. Risk factors for small for gestational age infants. Best Pract. Res. Clin. Obstet. Gynaecol. 2009, 23, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Englund-Ögge, L.; Brantsæter, A.L.; Juodakis, J.; Haugen, M.; Meltzer, H.M.; Jacobsson, B.; Sengpiel, V. Associations between maternal dietary patterns and infant birth weight, small and large for gestational age in the Norwegian Mother and Child Cohort Study. Eur. J. Clin. Nutr. 2018, 73, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Kalhan, S.C. Maternal and Child Nutrition: The First 1,000 Days. In One-Carbon Metabolism, Fetal Growth and Long-Term Consequences; Karger Publishers: Basel, Switzerland, 2013; Volume 74, pp. 127–138. [Google Scholar]
- Nazki, F.H.; Sameer, A.S.; Ganaie, B.A. Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene 2014, 533, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Kim, Y.; Refsum, H. Is folic acid good for everyone? Am. J. Clin. Nutr. 2008, 87, 517–533. [Google Scholar] [CrossRef]
- Talaulikar, V.S.; Arulkumaran, S. Folic acid in obstetric practice: A review. Obstet. Gynecol. Surv. 2011, 66, 240–247. [Google Scholar] [CrossRef]
- Field, M.S.; Kamynina, E.; Agunloye, O.C.; Liebenthal, R.P.; Lamarre, S.G.; Brosnan, M.E.; Brosnan, J.T.; Stover, P.J. Nuclear enrichment of folate cofactors and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) protect de novo thymidylate biosynthesis during folate deficiency. J. Biol. Chem. 2014, 289, 29642–29650. [Google Scholar] [CrossRef]
- Lassi, Z.S.; Salam, R.A.; Haider, B.A.; Bhutta, Z.A. Folic acid supplementation during pregnancy for maternal health and pregnancy outcomes. Cochrane Database Syst. Rev. 2013. [Google Scholar] [CrossRef]
- World Health Organization. Guideline: Daily Iron and Folic Acid Supplementation in Pregnant Women; World Health Organizatio: Geneva, Switzerland, 2012. [Google Scholar]
- Ministry of Health. Food and Nutrition Guidelines for Healthy Pregnant and Breastfeeding Women—A Background Paper; Ministry of Health: Wellington, New Zealand, 2006.
- Williams, P.J.; Bulmer, J.N.; Innes, B.A.; Broughton Pipkin, F. Possible roles for folic acid in the regulation of trophoblast invasion and placental development in normal early human pregnancy. Biol. Reprod. 2011, 84, 1148–1153. [Google Scholar] [CrossRef]
- World Health Organization. Recommendations on Antenatal Care for A Positive Pregnancy Experience; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Hodgetts, V.A.; Morris, R.K.; Francis, A.; Gardosi, J.; Ismail, K.M. Effectiveness of folic acid supplementation in pregnancy on reducing the risk of small-for-gestational age neonates: A population study, systematic review and meta-analysis. BJOG 2015, 122, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, Y.; Xin, X.; Zhang, Y.; Liu, D.; Peng, Z.; He, Y.; Xu, J.; Ma, X. Effect of folic acid supplementation on preterm delivery and small for gestational age births: A systematic review and meta-analysis. Reprod. Toxicol. 2017, 67, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Solanky, N.; Jimenez, A.R.; D’Souza, S.W.; Sibley, C.P.; Glazier, J.D. Expression of folate transporters in human placenta and implications for homocysteine metabolism. Placenta 2010, 31, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Zinck, J.W.; de Groh, M.; MacFarlane, A.J. Genetic modifiers of folate, vitamin B-12, and homocysteine status in a cross-sectional study of the Canadian population. Am. J. Clin. Nutr. 2015, 101, 1295–1304. [Google Scholar] [CrossRef]
- Ashfield-Watt, P.A.; Pullin, C.H.; Whiting, J.M.; Clark, Z.E.; Moat, S.J.; Newcombe, R.G.; Burr, M.L.; Lewis, M.J.; Powers, H.J.; McDowell, I.F. Methylenetetrahydrofolate reductase 677C→ T genotype modulates homocysteine responses to a folate-rich diet or a low-dose folic acid supplement: A randomized controlled trial. Am. J. Clin. Nutr. 2002, 76, 180–186. [Google Scholar] [CrossRef]
- Klai, S.; Fekih-Mrissa, N.; El Housaini, S.; Kaabechi, N.; Nsiri, B.; Rachdi, R.; Gritli, N. Association of MTHFR A1298C polymorphism (but not of MTHFR C677T) with elevated homocysteine levels and placental vasculopathies. Blood Coagul. Fibrinol. 2011, 22, 374–378. [Google Scholar] [CrossRef]
- Mtiraoui, N.; Zammiti, W.; Ghazouani, L.; Braham, N.J.; Saidi, S.; Finan, R.R.; Almawi, W.Y.; Mahjoub, T. Methylenetetrahydrofolate reductase C677T and A1298C polymorphism and changes in homocysteine concentrations in women with idiopathic recurrent pregnancy losses. Reproduction 2006, 131, 395–401. [Google Scholar] [CrossRef]
- Furness, D.; Dekker, G.A.; McCormack, C.D.; Nowak, R.C.; Thompson, S.D.; Roberts, C.T. 522. The association of folate pathway enzyme polymorphisms and pregnancy outcome. Reprod. Fertil. Dev. 2009, 21, 121. [Google Scholar] [CrossRef]
- Engel, S.M.; Olshan, A.F.; Siega-Riz, A.M.; Savitz, D.A.; Chanock, S.J. Polymorphisms in folate metabolizing genes and risk for spontaneous preterm and small-for-gestational age birth. Am. J. Obstet. Gynecol. 2006, 195, 1231.e1–1231.e11. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, P.; Geng, X.; Liu, Z.; Cui, L.; Gao, Z.; Jiang, B.; Yang, L. Genetic polymorphism of MTHFR C677T with preterm birth and low birth weight susceptibility: A meta-analysis. Arch. Gynecol. Obstet. 2017, 295, 1105–1118. [Google Scholar] [CrossRef]
- Yila, T.A.; Sasaki, S.; Miyashita, C.; Braimoh, T.S.; Kashino, I.; Kobayashi, S.; Okada, E.; Baba, T.; Yoshioka, E.; Minakami, H. Effects of maternal 5, 10-methylenetetrahydrofolate reductase C677T and A1298C Polymorphisms and tobacco smoking on infant birth weight in a Japanese population. J. Epidemiol. 2012, 22, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, C.A.; Cleves, M.A.; Karim, M.A.; Zhao, W.; MacLeod, S.L. Maternal folate-related gene environment interactions and congenital heart defects. Obstet. Gynecol. 2010, 116, 316. [Google Scholar] [CrossRef] [PubMed]
- Isotalo, P.A.; Wells, G.A.; Donnelly, J.G. Neonatal and fetal methylenetetrahydrofolate reductase genetic polymorphisms: An examination of C677T and A1298C mutations. Am. J. Hum. Genet. 2000, 67, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Cabo, R.; Hernes, S.; Slettan, A.; Haugen, M.; Ye, S.; Blomhoff, R.; Mansoor, M.A. Effect of genetic polymorphisms involved in folate metabolism on the concentration of serum folate and plasma total homocysteine (p-tHcy) in healthy subjects after short-term folic acid supplementation: A randomized, double blind, crossover study. Genes Nutr. 2015, 10, 1–12. [Google Scholar] [CrossRef]
- Torres-Sánchez, L.; López-Carrillo, L.; Blanco-Munoz, J.; Chen, J. Maternal dietary intake of folate, vitamin B 12 and MTHFR 677C> T genotype: Their impact on newborn’s anthropometric parameters. Genes Nutr. 2014, 9, 429. [Google Scholar] [CrossRef][Green Version]
- Australian and New Zealand Clinical Trials Registry. Screening Nulliparous Women to Identify the Combinations of Clinical Risk Factors and/or Biomarkers Required to Predict Preeclampsia, Small for Gestational Age Babies and Spontaneous Preterm Birth. Available online: www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=82254 (accessed on 24 April 2020).
- North, R.A.; McCowan, L.M.; Dekker, G.A.; Poston, L.; Chan, E.H.; Stewart, A.W.; Black, M.A.; Taylor, R.S.; Walker, J.J.; Baker, P.N. Clinical risk prediction for pre-eclampsia in nulliparous women: Development of model in international prospective cohort. BMJ 2011, 342, d1875. [Google Scholar] [CrossRef]
- Bulloch, R.E.; McCowan, L.M.; Thompson, J.M.; Houghton, L.A.; Wall, C.R. Plasma folate and its association with folic acid supplementation, socio-demographic and lifestyle factors among New Zealand pregnant women. Br. J. Nutr. 2019, 122, 910–918. [Google Scholar] [CrossRef]
- Friso, S.; Choi, S. Gene-nutrient interactions in one-carbon metabolism. Curr. Drug Metab. 2005, 6, 37–46. [Google Scholar] [CrossRef]
- Boyles, A.L.; Billups, A.V.; Deak, K.L.; Siegel, D.G.; Mehltretter, L.; Slifer, S.H.; Bassuk, A.G.; Kessler, J.A.; Reed, M.C.; Nijhout, H.F. Neural tube defects and folate pathway genes: Family-based association tests of gene–gene and gene–environment interactions. Environ. Health Perspect. 2006, 114, 1547–1552. [Google Scholar] [CrossRef]
- Barbosa, P.R.; Stabler, S.P.; Machado, A.; Braga, R.C.; Hirata, R.; Hirata, M.H.; Sampaio-Neto, L.F.; Allen, R.H.; Guerra-Shinohara, E.M. Association between decreased vitamin levels and MTHFR, MTR and MTRR gene polymorphisms as determinants for elevated total homocysteine concentrations in pregnant women. Eur. J. Clin. Nutr. 2008, 62, 1010–1021. [Google Scholar] [CrossRef]
- Chedraui, P.; Salazar-Pousada, D.; Villao, A.; Escobar, G.S.; Ramirez, C.; Hidalgo, L.; Pérez-López, F.R.; Genazzani, A.; Simoncini, T. Polymorphisms of the methylenetetrahydrofolate reductase gene (C677T and A1298C) in nulliparous women complicated with preeclampsia. Gynecol. Endocrinol. 2014, 30, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Bufalino, A.; Ribeiro Paranaíba, L.M.; Nascimento de Aquino, S.; Martelli-Júnior, H.; Oliveira Swerts, M.S.; Coletta, R.D. Maternal polymorphisms in folic acid metabolic genes are associated with nonsyndromic cleft lip and/or palate in the Brazilian population. Birth Defects Res. Part A Clin. Mol. Teratol. 2010, 88, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Christensen, K.E.; Rohlicek, C.V.; Andelfinger, G.U.; Michaud, J.; Bigras, J.; Richter, A.; MacKenzie, R.E.; Rozen, R. The MTHFD1 p. Arg653Gln variant alters enzyme function and increases risk for congenital heart defects. Hum. Mutat. 2009, 30, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Oussalah, A.; Levy, J.; Filhine-Trésarrieu, P.; Namour, F.; Guéant, J. Association of TCN2 rs1801198 c. 776G> C polymorphism with markers of one-carbon metabolism and related diseases: A systematic review and meta-analysis of genetic association studies. Am. J. Clin. Nutr. 2017, 106, 1142–1156. [Google Scholar] [CrossRef]
- McCowan, L.; Stewart, A.W.; Francis, A.; Gardosi, J. A customised birthweight centile calculator developed for a New Zealand population. Aust. N. Z. J. Obstet. Gynaecol. 2004, 44, 428–431. [Google Scholar] [CrossRef]
- Shaw, G.M.; Iovannisci, D.M.; Yang, W.; Finnell, R.H.; Carmichael, S.L.; Cheng, S.; Lammer, E.J. Risks of human conotruncal heart defects associated with 32 single nucleotide polymorphisms of selected cardiovascular disease-related genes. Am. J. Med. Genet. Part A 2005, 138, 21–26. [Google Scholar] [CrossRef]
- Garcia-Closas, M.; Ye, Y.; Rothman, N.; Figueroa, J.D.; Malats, N.; Dinney, C.P.; Chatterjee, N.; Prokunina-Olsson, L.; Wang, Z.; Lin, J. A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Hum. Mol. Genet. 2011, 20, 4282–4289. [Google Scholar] [CrossRef]
- Fekete, K.; Berti, C.; Cetin, I.; Hermoso, M.; Koletzko, B.V.; Decsi, T. Perinatal folate supply: Relevance in health outcome parameters. Matern. Child Nutr. 2010, 6, 23–38. [Google Scholar] [CrossRef]
- Finnell, R.H.; Shaw, G.M.; Lammer, E.J.; Rosenquist, T.H. Gene–nutrient interactions: Importance of folic acid and vitamin B12 during early embryogenesis. Food Nutr. Bull. 2008, 29, S86–S98. [Google Scholar] [CrossRef]
- Hiraoka, M.; Kagawa, Y. Genetic polymorphisms and folate status. Congenit. Anom. 2017, 57, 142–149. [Google Scholar] [CrossRef]
- Amigou, A.; Rudant, J.; Orsi, L.; Goujon-Bellec, S.; Leverger, G.; Baruchel, A.; Bertrand, Y.; Nelken, B.; Plat, G.; Michel, G. Folic acid supplementation, MTHFR and MTRR polymorphisms, and the risk of childhood leukemia: The ESCALE study (SFCE). Cancer Causes Control 2012, 23, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Hogeveen, M.; Blom, H.J.; Den Heijer, M. Maternal homocysteine and small-for-gestational-age offspring: Systematic review and meta-analysis. Am. J. Clin. Nutr. 2012, 95, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Zhu, J.; Hao, L.; Yang, Q.; Yang, T.P.; Gindler, J.; Maneval, D.R.; Quinlivan, E.P.; Li, Z.; Bailey, L.B. MTHFR 677C→ T genotype is associated with folate and homocysteine concentrations in a large, population-based, double-blind trial of folic acid supplementation. Am. J. Clin. Nutr. 2011, 93, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Beresford, S.A.; McLerran, D.; Lampe, J.W.; Deeb, S.; Feng, Z.; Motulsky, A.G. Response of serum and red blood cell folate concentrations to folic acid supplementation depends on methylenetetrahydrofolate reductase C 677 T genotype: Results from a crossover trial. Mol. Nutr. Food Res. 2013, 57, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Miyaki, K.; Murata, M.; Kikuchi, H.; Takei, I.; Nakayama, T.; Watanabe, K.; Omae, K. Assessment of tailor-made prevention of atherosclerosis with folic acid supplementation: Randomized, double-blind, placebo-controlled trials in each MTHFR C677T genotype. J. Hum. Genet. 2005, 50, 241–248. [Google Scholar] [CrossRef]
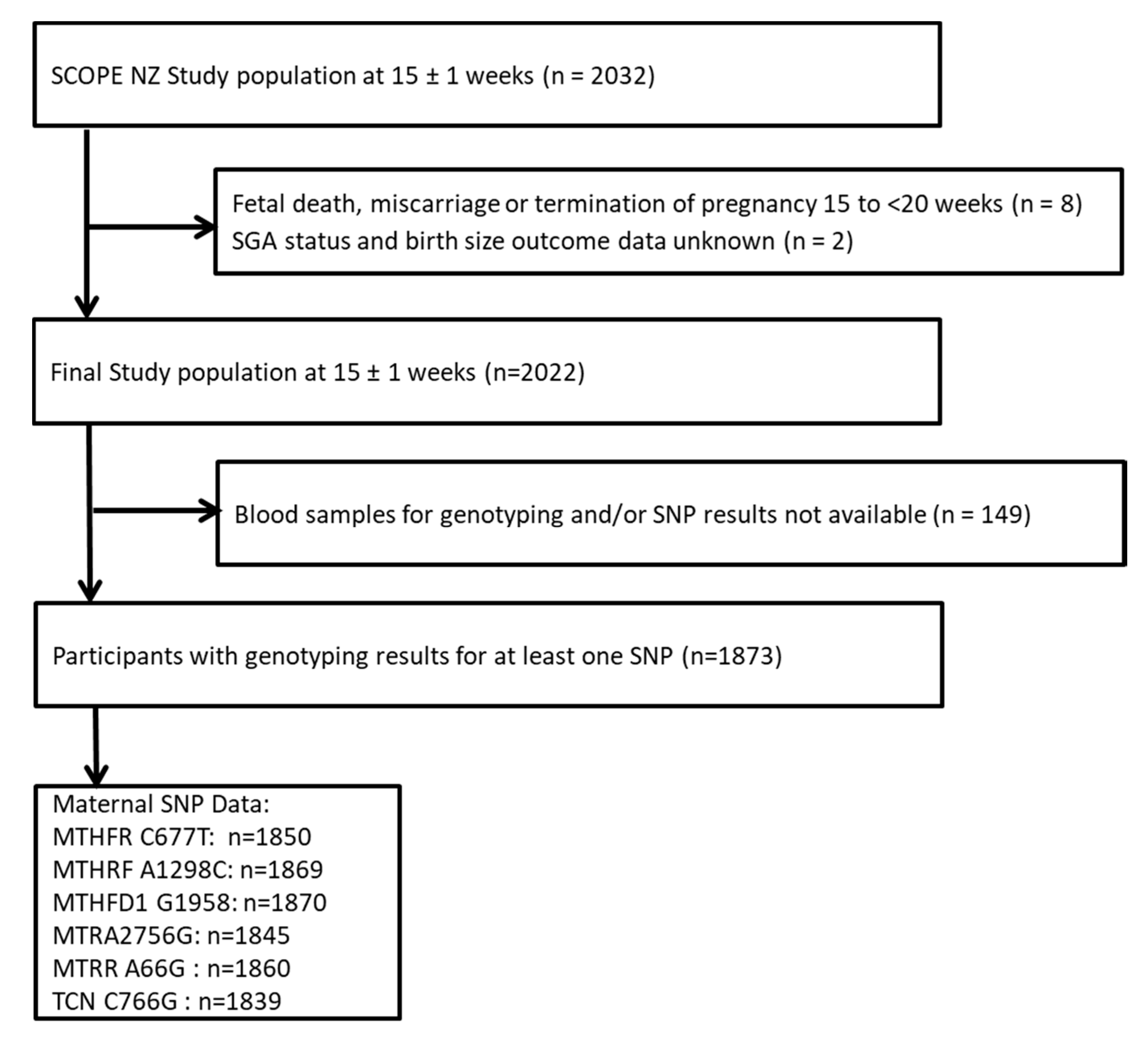
| Characteristic | Mean ± SD or n (%) |
|---|---|
| Total n (%) | 1873 (100) |
| Maternal age (years) | 30.4 ± 4.7 |
| Ethnicity: | |
| NZ/Other European | 1577 (84.2) |
| Māori | 61 (3.3) |
| Pacific | 35 (1.9) |
| Asian | 97 (5.2) |
| Indian | 74 (4.0) |
| Other Non-European | 29 (1.6) |
| Socioeconomic Index * | 48 ± 15 |
| Education > 12 years | 1160 (64.8) |
| Marital partner (yes) | 1811 (96.7) |
| BMI at 15 weeks’ research visit (kg/m2) | 24.8 (±4.2) |
| Smoking (at 15 weeks’) | 73 (3.9) |
| Folic Acid Supplement Users $ | 1367 (73.0) |
| Folic Acid Supplement Dose (µg/day) | 564 (14) ¥ |
| Plasma Folate (nmol/L) § | 48.6 (1.6) ¥ |
| Fetal Sex: | |
| Male | 973 (52.0) |
| Female | 900 (48.0) |
| Final Delivery Gestation (weeks) | 39.6 (±2.1) |
| Birthweight (grams) | 3414 (±570) |
| Customised Birthweight Centile | 49 (±29) |
| SGA | 189 (10.1) |
| Spontaneous Preterm Birth | 77 (4.1) |
| SNP: | Total n (%) | SGA n (%) | OR (95% CI) | p-Value (p > z) | p-Value for Overall Genotype Effect |
|---|---|---|---|---|---|
| MTHFR C677T | 1850 (100) | ||||
| Genotype: | 0.457 | ||||
| MTHFR 677 CC (Ref) | 887 (48.0) | 96 (10.8) | 1.00 (Ref) | ||
| MTHFR 677 CT | 786 (42.5) | 15 (9.3) | 0.84 (0.61–1.16) | 0.299 | |
| MTHFR 677 TT | 177 (9.6) | 73 (8.5) | 0.76 (0.43–1.35) | 0.352 | |
| Dominant Model | 0.227 | ||||
| MTHFR 677 CC (Ref) | 887 (48.0) | 96 (10.8) | 1.00 (Ref) | ||
| MTHFR 677 CT + TT | 963 (52.1) | 88 (9.1) | 0.83 (0.61–1.12) | 0.227 | |
| MTHFR A1298C | 1869 (100) | ||||
| Genotype: | 0.141 | ||||
| MTHFR 1298 AA (Ref) | 932 (49.9) | 93 (10.0) | 1.00 (Ref) | ||
| MTHFR 1298 AC | 778 (41.6) | 72 (9.3) | 0.92 (0.67–1.27) | 0.614 | |
| MTHFR 1298 CC | 159 (8.5) | 23 (14.5) | 1.53 (0.93–2.49) | 0.092 | |
| Recessive Model | 0.055 | ||||
| MTHFR 1298 AA + AC (Ref) | 1710 (91.5) | 165 (9.6) | 1.00 (Ref) | ||
| MTHFR 1298 CC | 159 (8.5) | 23 (14.5) | 1.58 (0.99–2.53) | 0.055 | |
| MTHFD1 G1958A | 1870 (100) | ||||
| Genotype: | 0.766 | ||||
| MTHFD1 1958 GG (Ref) | 621 (33.2) | 67 (10.8) | 1.00 (Ref) | ||
| MTHFD1 1958 GA | 920 (49.2) | 91 (9.9) | 0.91 (0.65–1.27) | 0.569 | |
| MTHFD1 1958 AA | 329 (17.6) | 31 (9.4) | 0.86 (0.55–1.35) | 0.510 | |
| Recessive Model | 0.650 | ||||
| MTHFD1 1958 GG + GA (Ref) | 1541 (82.4) | 158 (10.3) | 1.00 (Ref) | ||
| MTHFD1 1958 AA | 329 (17.6) | 31 (9.4) | 0.91 (0.61–1.37) | 0.650 | |
| MTR A2756G | 1845 (100) | ||||
| Genotype: | 0.438 | ||||
| MTR 2756 AA (Ref) | 1206 (65.4) | 116 (9.6) | 1.00 (Ref) | ||
| MTR 2756 AG | 569 (30.8) | 66 (11.6) | 1.23 (0.90–1.70) | 0.200 | |
| MTR 2756 GG | 70 (3.8) | 7 (10.0) | 1.04 (0.47–2.33) | 0.916 | |
| Recessive Model | 0.945 | ||||
| MTR 2756 AA + AG (Ref) | 1775 (96.2) | 182 (10.3) | 1.00 (Ref) | ||
| MTR 2756 GG | 70 (3.8) | 7 (10.0) | 0.97 (0.44–2.16) | 0.945 | |
| MTRR A66G | 1860 (100) | ||||
| Genotype: | 0.714 | ||||
| MTRR 66 AA (Ref) | 450 (24.2) | 50 (11.1) | 1.00 (Ref) | ||
| MTRR 66 AG | 892 (48.0) | 88 (9.9) | 0.88 (0.61–1.26) | 0.478 | |
| MTRR 66 GG | 518 (27.9) | 50 (9.7) | 0.85 (0.57–1.29) | 0.457 | |
| Dominant Model | 0.418 | ||||
| MTRR 66 AA (Ref) | 450 (24.2) | 50 (11.1) | 1.00 (Ref) | ||
| MTRR 66 AG + GG | 1410 (75.8) | 138 (9.8) | 0.87 (0.62–1.22) | 0.418 | |
| TCN2 C766G | 1839 (100) | ||||
| Genotype: | 0.420 | ||||
| TCN2 766 CC (Ref) | 540 (29.4) | 51 (9.4) | 1.00 (Ref) | 0.597 | |
| TCN2 766 CG | 919 (50.0) | 96 (10.8) | 1.16 (1.16–0.26) | 0.435 | |
| TCN2 766 GG | 380 (20.7) | 41 (10.8) | 1.12 (0.75–1.790) | ||
| Recessive Model | 0.682 | ||||
| TCN2 766 CC + CG (Ref) | 1459 (79.3) | 147 (10.1) | 1.00 (Ref) | ||
| TCN2 766 GG | 380 (20.7) | 41 (10.8) | 1.08 (0.75–1.56) | 0.682 |
| SNP | All n (%) | FAS Yes n (%) $ | FAS No n (%) $ | FAS Yes aOR (95% CI) * | FAS No aOR (95% CI) * | p-Value * |
|---|---|---|---|---|---|---|
| MTHFR C677T | 1850 (100) | |||||
| CC (Ref) | 887 (48.0) | 628 (33.9) | 259 (14.0) | 1.00 (Ref) | 1.87 (1.21–2.88) | 0.072 |
| CT | 786 (42.5) | 587 (31.7) | 199 (10.8) | 1.06 (0.71–1.56) | 1.02 (0.58–1.77) | |
| TT | 177 (9.6) | 135 (7.3) | 42 (2.3) | 0.91 (0.46–1.78) | 1.08 (0.37–3.12) | |
| CC (Ref) | 887 (48.0) | 628 (33.9) | 259 (14.0) | 1.00 (Ref) | 1.87 (1.21–2.88) | 0.019 |
| CT + TT | 963 (52.1) | 722 (39.0) | 241 (13.0) | 1.03 (0.71–1.49) | 1.03 (0.61–1.72) | |
| MTHFR A1298C | 1869 (100) | |||||
| AA (Ref) | 932 (49.9) | 702 (37.6) | 230 (12.3) | 1.00 (Ref) | 1.07 (0.65–1.75) | 0.020 |
| AC | 778 (41.6) | 560 (30.0) | 218 (11.7) | 0.86 (0.58–1.27) | 1.13 (0.69–1.86) | |
| CC | 159 (8.5) | 101 (5.4) | 58 (3.1) | 0.90 (0.43–1.86) | 2.92 (1.52–5.60) | |
| AA + AC (Ref) | 1710 (91.5) | 1262 (67.5) | 448 (24.0) | 1.00 (Ref) | 1.17 (0.82–1.67) | 0.005 |
| CC | 159 (8.5) | 101 (5.4) | 58 (3.1) | 0.96 (0.47–1.95 | 3.11 (1.66–5.85) | |
| MTHFD1 G1958A | 1870 (100) | |||||
| GG (Ref) | 621 (33.2) | 439 (23.5) | 182 (9.7) | 1.00 (Ref) | 1.40 (0.82–2.38) | 0.133 |
| GA | 920 (49.2) | 680 (36.4) | 240 (12.8) | 1.01 (0.67–1.51) | 1.02 (0.60–1.73) | |
| AA | 329 (17.6) | 245 (13.1) | 84 (4.5) | 0.69 (0.38–1.23) | 1.84 (0.96–3.54) | |
| GG + GA (Ref) | 1541 (82.4) | 1119 (59.8) | 422 (22.6) | 1.00 (Ref) | 1.18 (0.82–1.69) | 0.062 |
| AA | 329 (17.6) | 245 (13.1) | 84 (4.5) | 0.68 (0.40–1.16) | 1.83 (1.00–3.36) | |
| MTR A2756G | 1845 (100) | |||||
| AA (Ref) | 1206 (65.4) | 879 (47.6) | 327 (17.7) | 1.00 (Ref) | 1.54 (1.03–2.30) | 0.253 |
| AG | 569 (30.8) | 415 (22.5) | 154 (8.3) | 1.34 (0.91–1.97) | 1.60 (0.95–2.71) | |
| GG | 70 (3.8) | 53 (2.9) | 17 (0.9) | 1.37 (0.57–3.31) | 0.67 (0.09–5.12) | |
| MTRR A66G | 1860 (100) | |||||
| AA (Ref) | 450 (24.2) | 333 (17.9) | 117 (6.2) | 1.00 (Ref) | 1.71 (0.92–3.18) | 0.245 |
| AG | 892 (48.0) | 650 (34.9) | 242 (13.0) | 1.01 (0.65–1.58) | 1.08 (0.62–1.88) | |
| GG | 518 (27.9) | 375 (20.2) | 143 (7.7) | 0.85 (0.51–1.42) | 1.44 (0.79–2.64) | |
| TCN2 C776G | 1839 (100) | |||||
| CC (Ref) | 540 (29.4) | 401 (21.8) | 139 (7.6) | 1.00 (Ref) | 1.23 (0.65–2.32) | 0.057 |
| CG | 919 (50.0) | 660 (35.9) | 259 (14.1) | 1.18 (0.77–1.81) | 1.18 (0.70–2.00) | |
| GG | 380 (20.7) | 281 (15.3) | 99 (5.4) | 0.86(0.50 –1.50) | 2.41 (1.31–4.41) | |
| CC + CG (Ref) | 1459 (79.3) | 1061 (57.7) | 398 (21.6) | 1.00 (Ref) | 1.07 (0.74–1.57) | 0.017 |
| GG | 380 (20.7) | 281 (15.3) | 99 (5.4) | 0.77 (0.48–1.25) | 2.16 (1.26–3.71) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulloch, R.E.; Wall, C.R.; McCowan, L.M.E.; Taylor, R.S.; Roberts, C.T.; Thompson, J.M.D. The Effect of Interactions between Folic Acid Supplementation and One Carbon Metabolism Gene Variants on Small-for-Gestational-Age Births in the Screening for Pregnancy Endpoints (SCOPE) Cohort Study. Nutrients 2020, 12, 1677. https://doi.org/10.3390/nu12061677
Bulloch RE, Wall CR, McCowan LME, Taylor RS, Roberts CT, Thompson JMD. The Effect of Interactions between Folic Acid Supplementation and One Carbon Metabolism Gene Variants on Small-for-Gestational-Age Births in the Screening for Pregnancy Endpoints (SCOPE) Cohort Study. Nutrients. 2020; 12(6):1677. https://doi.org/10.3390/nu12061677
Chicago/Turabian StyleBulloch, Rhodi E., Clare R. Wall, Lesley M. E. McCowan, Rennae S. Taylor, Claire T. Roberts, and John M. D. Thompson. 2020. "The Effect of Interactions between Folic Acid Supplementation and One Carbon Metabolism Gene Variants on Small-for-Gestational-Age Births in the Screening for Pregnancy Endpoints (SCOPE) Cohort Study" Nutrients 12, no. 6: 1677. https://doi.org/10.3390/nu12061677
APA StyleBulloch, R. E., Wall, C. R., McCowan, L. M. E., Taylor, R. S., Roberts, C. T., & Thompson, J. M. D. (2020). The Effect of Interactions between Folic Acid Supplementation and One Carbon Metabolism Gene Variants on Small-for-Gestational-Age Births in the Screening for Pregnancy Endpoints (SCOPE) Cohort Study. Nutrients, 12(6), 1677. https://doi.org/10.3390/nu12061677





